To The Editor:
Hemophilia A is the most common X-linked coagulation disorder, with an incidence of about one in 5,000 males. About half the families with severe disease have a large genomic inversion of the factor VIII gene that separates the first 22 exons from the final 4 exons. This inversion results from a hotspot of recombination between a 9.5-kb region in intron 22 (int22h1) and either of two extragenic, distal homologs, int22h2 and int22h3; int22h2 andint22h3 are more than 99% identical to one another. Recombination produces an inversion because the extragenic homologs are in the opposite orientation relative toint22h1.1,2
The inversions are detected by Southern blotting, which is slow and labor-intensive. A rapid and inexpensive test is of particular clinical utility, because carrier testing is often paid out-of-pocket due to insurance issues and confidentiality; a low-cost test may facilitate more optimal use of genetic services. This difficult 9.5-kb region previously has been refractory to polymerase chain reaction (PCR) amplification, presumably due to the presence of a 3.5-kb GC island, which includes a 1-kb segment with a GC content of 79%. In addition, an optimal PCR-based assay requires more genomic sequence flanking the homologs.
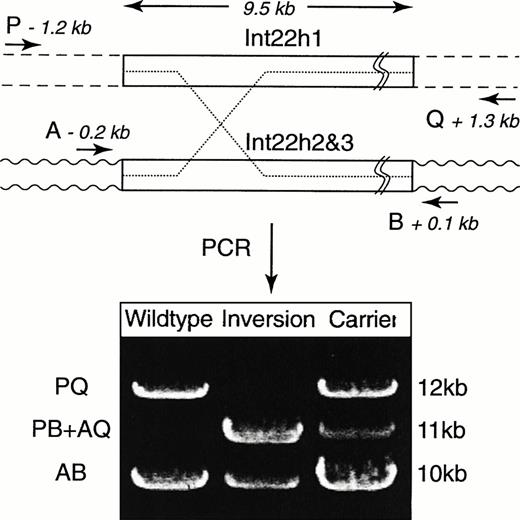
We have developed a single-tube PCR assay that combines overlapping PCR3 with long-distance PCR4 to achieve the genetic diagnosis of inversions causing hemophilia A (Fig1). The inversion was detected by performing PCR directly from genomic DNA with four primers that differentiate the wild-type, inversion, and carrier. Two primers, P and Q, are located within the factor VIII gene at positions −1212 bp and +1334 bp flanking int22h1. Two primers, A and B, are located at −167 bp and +118 bp flanking int22h2 and int22h3.Segments PQ (12 kb) and AB (10 kb) are produced in hemizygous wild-type males. Males with hemophilia due to the inversion produce PB (11 kb) and AQ segments (11 kb) along with the 10-kb AB segment from the nonrecombined extragenic homolog. Female carriers produce PQ, PB + AQ, and AB segments. In all cases, an AB segment serves as an internal control because at least one copy of int22h2 or int22h3remains intact. The three segment sizes are readily separated on a 0.6% agarose gel. High yield and reproducible amplification depended on three unusual modifications to standard long-distance PCR protocols: (1) high concentrations of dimethylsulfoxide (DMSO) additive; (2) substantially increased amounts of Taq andPwo DNA polymerases; and (3) 50% deaza-dGTP.
Schematic of the PCR assay. The four primers (P, Q, A, and B) are represented by arrows and their positions are indicated. The upper box represents int22h1, and the dashed lines indicate flanking sequences. The lower box represents int22h2 andint22h3, and the wavy lines indicate the flanking sequences. Deleterious inversions can occur by recombination betweenint22h1 and either int22h2 or int22h3 (dotted lines). Amplified products in male patients with the wild-type and the inverted factor VIII genes and a carrier female. PCR was performed in 25 μL with 250 ng of genomic DNA, a mixture containing 50 mmol/L Tris.HCl, pH 9.2, 2.25 mmol/L MgCl2, 7.5% DMSO, 16 mmol/L (NH4)2SO4, 250 mmol/L each of dGTP and deaza-dGTP, 500 mmol/L of the other dNTPs, and 3.3 U of Expand Long Template DNA polymerases (Boehringer Mannheim, Mannheim, Germany). After 2 minutes of denaturation at 94°C, 30 cycles were performed at 94°C for 12 seconds, 65°C for 30 seconds, and 68°C for 12 minutes for 10 cycles, (the remaining 20 cycles were performed for 12 minutes with 20 more seconds for each additional cycle). The primers used are as follows: P = GCC CTG CCTG TCC ATT ACA CTG AT GAC ATT ATG CTG AC; Q = GGC CCT ACA ACC ATT CTG CCT TTC ACT TTC AGT GCA ATA; A = CAC AAG GGG GAA GAG TGT GAG GGT GTG GGA TAA GAA; B = CCC CAA ACT ATA ACC AGC ACC TTG AAC TTA CCC TCT. Primer concentrations were 0.4, 0.4, 0.12, and 0.12 μmol/L, respectively.
Schematic of the PCR assay. The four primers (P, Q, A, and B) are represented by arrows and their positions are indicated. The upper box represents int22h1, and the dashed lines indicate flanking sequences. The lower box represents int22h2 andint22h3, and the wavy lines indicate the flanking sequences. Deleterious inversions can occur by recombination betweenint22h1 and either int22h2 or int22h3 (dotted lines). Amplified products in male patients with the wild-type and the inverted factor VIII genes and a carrier female. PCR was performed in 25 μL with 250 ng of genomic DNA, a mixture containing 50 mmol/L Tris.HCl, pH 9.2, 2.25 mmol/L MgCl2, 7.5% DMSO, 16 mmol/L (NH4)2SO4, 250 mmol/L each of dGTP and deaza-dGTP, 500 mmol/L of the other dNTPs, and 3.3 U of Expand Long Template DNA polymerases (Boehringer Mannheim, Mannheim, Germany). After 2 minutes of denaturation at 94°C, 30 cycles were performed at 94°C for 12 seconds, 65°C for 30 seconds, and 68°C for 12 minutes for 10 cycles, (the remaining 20 cycles were performed for 12 minutes with 20 more seconds for each additional cycle). The primers used are as follows: P = GCC CTG CCTG TCC ATT ACA CTG AT GAC ATT ATG CTG AC; Q = GGC CCT ACA ACC ATT CTG CCT TTC ACT TTC AGT GCA ATA; A = CAC AAG GGG GAA GAG TGT GAG GGT GTG GGA TAA GAA; B = CCC CAA ACT ATA ACC AGC ACC TTG AAC TTA CCC TCT. Primer concentrations were 0.4, 0.4, 0.12, and 0.12 μmol/L, respectively.
To design PCR primers, additional sequences flanking the int22hhomologs were required. Fifty basepairs of available sequence flanking the 5′ end of int22h1 was extended by sequencing a 6-kb PCR segment that spans the distance between exon 22 and the 5′ end ofint22h1. Additional sequences flanking int22h2 andint22h3 were obtained by a method of inverse PCR.5
A blinded analysis was performed with 40 blood samples provided by hemophilia centers in the United States. Genomic DNA was isolated from whole blood by phenol extraction.6 Thirteen samples were hemizygous for the inversion, 6 carried the inversion, and 21 were wild-type, as determined by standard Southern blot analysis. The PCR analysis was performed successfully in less than 1 day, with complete concordance with the Southern blot results. The PCR was successful with as little as 25 ng genomic DNA per 25 μL reaction, which is 400-fold less than that required for the Southern blot analysis. Currently, the PCR assay is being used in our Molecular Diagnosis Laboratory for screening severe cases of hemophilia A.
In conclusion, a PCR assay for factor VIII gene inversions has been developed for patient screening, carrier testing, and prenatal diagnosis of severe hemophilia A. The method is simple, rapid, reproducible, inexpensive, nonisotopic, and amenable to automation. This approach may be helpful in analyzing other inversions, deletions, and translocations in the genome.
ACKNOWLEDGMENT
We thank Stylianos Antonarakis for helpful discussions.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal