To The Editor:
An atypical myeloproliferative disorder has been described that is associated with T-cell leukemia/lymphoma and peripheral blood eosinophilia.1 All these cases are associated with a translocation between 8p11 and 13q11-12 or two rare variant translocations involving 8p11 and 9q322-4 or 8p11 and 6q27.5 Recently, we localized the breakpoint on chromosome 8p11 to a region close to the FGFR1 gene.6 Further analysis has shown that the 8p11 translocation breakpoint involved in the t(8;13)(p11;q11-12) and the variant translocations t(8;9)(p11;32) and t(6;8)(q27;p11) disrupts the FGFR1 gene.7 Subsequently, the rearranged gene, named ZNF1988 or RAMP9 on chromosome 13, was cloned by 5′ RACE from primers located within the FGFR1 cDNA. In the 5 cases analyzed by these investigators, the translocation breakpoints cluster within intron 8 of the FGFR1 gene and result in the fusion of the FGFR1 tyrosine kinase domain to the zinc finger domains of the gene located in 13q11.8 9 This fusion is thought to result in constitutive activation of the tyrosine kinase domain by oligomerization of the fusion protein.
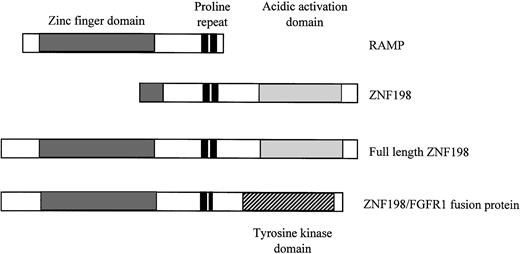
Examination of the cloned ZNF198 and RAMP cDNAs shows a number of discrepancies between the two sequences (Fig1). The ZNF198 cDNA product is 3,478 bp in length.8 Conceptual translation of the 2,271-bp open reading frame (ORF) predicts a protein of 757 amino acids consisting of a domain containing two zinc fingers of the atypical type Cys-X2-Cys-X19-22-Cys-X3-Cys- X13-17-Cys-X2-Cys-X19-25-Cys- X3-Cys, two proline repeats, and a carboxy terminal acidic domain. The RAMP cDNA product derived by Smedley et al9 is 2,553 bp long, with an ORF of 699 amino acids. This predicted protein contains three extra N-terminal zinc fingers, but lacks the acidic activation domain and terminates 48 amino acids after the second proline repeat. Our reconstruction of the ZNF198/RAMP gene has confirmed the presence of six sequencing errors in the RAMP sequence that lead to the introduction of a stop codon, resulting in a truncation of the ORF. The 5′ end of the sequence of the ZNF198 cDNA, described as the 5′ untranslated region, appears to represent an unspliced intron from the ZNF198 gene. These errors have probably occurred during the cloning of the products of the 3′ and 5′ RACE reactions, respectively. A recent report by Popovici et al10 shows that the complete ZNF198/RAMP cDNA sequence, which they have named FIM, is, therefore, 5,008 bp long, which is in agreement with the predicted mRNA transcript size detected by Northern blot analysis.8 10 Thus, the gene encodes a protein of 1379 amino acids consisting of an N-terminal domain containing five zinc fingers, two proline repeats, and a carboxy terminal acidic activation domain (Fig 1). The translocation, therefore, fuses the N-terminal zinc finger domain to the FGFR1 tyrosine kinase domain, resulting in a protein of 1309 amino acids with a predicted molecular mass of 146 kD.
Comparison of partial RAMP and ZNF198 ORFs with the revised, completed ZNF198 protein and ZNF198/FGFR1 fusion product. This completed sequence contains the five atypical zinc fingers reported by Smedley et al9 and the proline repeats and potential acidic activation domain first reported by Xiao et al.8 The translocation therefore fuses the five zinc fingers and one complete proline repeat to the tyrosine kinase domain of FGFR1.
Comparison of partial RAMP and ZNF198 ORFs with the revised, completed ZNF198 protein and ZNF198/FGFR1 fusion product. This completed sequence contains the five atypical zinc fingers reported by Smedley et al9 and the proline repeats and potential acidic activation domain first reported by Xiao et al.8 The translocation therefore fuses the five zinc fingers and one complete proline repeat to the tyrosine kinase domain of FGFR1.
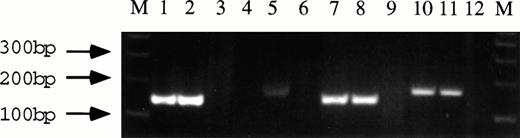
Fluorescence in situ hybridization (FISH) analysis of metaphase spreads from patients carrying the t(8;13) translocation with YACs from different regions of chromosome 13 suggests that multiple, distinct breakpoints are involved in the t(8;13) translocation.11,12 We have recently shown that YAC 943E4, which is used for FISH analysis of the 13q11 breakpoint in three cases of this leukemia, contains DNA from both the telomeric region of the 13q11-q12 YAC contig13 and the pericentromeric region close to D13S183E (Still et al, manuscript in preparation). This results in the detection of signal on both derivative chromosomes, when this YAC is used. Interestingly the patient described by Smedley et al12 was determined to have a breakpoint in 13q12, defined by markers D13S267 and D13S220. The breakpoint in this patient was localized by these investigators within the ZNF198/RAMP/FIM gene,9 which is located on the 967B1 and 911H8 YACs located in 13q1110 (Still et al, manuscript in preparation), suggesting that this patient may carry a complex translocation. To determine whether the breakpoints in two recently identified patients, RB and SR,6,14 are similar to those previously described, we have performed reverse transcription-polymerase chain reaction (RT-PCR) using RNA isolated from somatic cell hybrid RBF16 and a bone marrow aspirate sample from patient SR.13 As shown in Fig2, identical PCR products are obtained when primers are used to detect the ZNF198/FGFR1 fusion cDNA in RBF1 (lane 1) and SR (lane 2). This demonstrates that the breakpoints occur in the same introns of the FGFR1 gene and of the ZNF198 gene, as already described by Xiao et al,8 and therefore the fusion cDNA generated in patient RB and SR are identical to the 7 other patients already characterized.8-10 Interestingly, we were also able to detect the reciprocal FGFR1/ZNF198 fusion product in patient SR, albeit at a lower level than the ZNF198/FGFR1 product (Fig 2, lane 5). This demonstrates that all 9 cases molecularly characterized to date generate the same ZNF198/FGFR1 fusion protein and that there is only a single gene in 13q11 disrupted by the t(8;13) translocation.
Detection of the ZNF198/FGFR1 fusion product in patient SR13 and somatic cell hybrid RBF1.6First-strand cDNA synthesis was generated from 1 μg of total RNA, using random hexamers as primers according to standard protocols. Oligonucleotides were designed to sites in the ZNF198 and FGFR1 cDNA flanking the translocation breakpoints: ZNF198F (5′-ccctgtgcctgtgtatatcccag-3′), ZNF198R (5′-tgcaggaatcttctcactgc-3′), FGFRX7F (5′-gatcatcgtctacaagatg-3′), and FGFR1979R (5′-gtgatggccgaaccagaagaac-3′). Lanes 1 and 2, ZNF198/FGFR1 fusion products detected in RBF1 and SR, respectively, by primers ZNF198F/FGF1979R. Lanes 4 and 5, potential FGFR1/ZNF198 fusion products detected by primers FGFX7F/ZNF198R in RBF1 and SR. Lanes 7 and 8, expression of the normal ZNF198 mRNA in RBF1 and SR, detected using primers ZNF198F/ZNF198R. Lanes 10 and 11, FGFR1 expression in RBF1 and SR, respectively, detected with primers FGFRX7F/FGF1979R. Lanes 3, 6, 9, and 12, negative controls.
Detection of the ZNF198/FGFR1 fusion product in patient SR13 and somatic cell hybrid RBF1.6First-strand cDNA synthesis was generated from 1 μg of total RNA, using random hexamers as primers according to standard protocols. Oligonucleotides were designed to sites in the ZNF198 and FGFR1 cDNA flanking the translocation breakpoints: ZNF198F (5′-ccctgtgcctgtgtatatcccag-3′), ZNF198R (5′-tgcaggaatcttctcactgc-3′), FGFRX7F (5′-gatcatcgtctacaagatg-3′), and FGFR1979R (5′-gtgatggccgaaccagaagaac-3′). Lanes 1 and 2, ZNF198/FGFR1 fusion products detected in RBF1 and SR, respectively, by primers ZNF198F/FGF1979R. Lanes 4 and 5, potential FGFR1/ZNF198 fusion products detected by primers FGFX7F/ZNF198R in RBF1 and SR. Lanes 7 and 8, expression of the normal ZNF198 mRNA in RBF1 and SR, detected using primers ZNF198F/ZNF198R. Lanes 10 and 11, FGFR1 expression in RBF1 and SR, respectively, detected with primers FGFRX7F/FGF1979R. Lanes 3, 6, 9, and 12, negative controls.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal