Abstract
The negative regulation of transcription of the human von Willebrand factor (vWF) gene was investigated in human umbilical vein endothelial cells (HUVECs) and HeLa cells. A fragment spanning −89 to +244 nucleotides (nt), containing the first exon, is active in HUVECs only but not in HeLa cells. The activity of this promoter is sharply reduced by mutagenesis of the GATA binding site at +221. Extension of the upstream sequences from nt −89 to −142 and to −496 results in progressive reduction of the activity of the −89 to +244 promoter identifying a negative regulatory element between nt −142 and −89. A factor present in nuclear extracts from endothelial and nonendothelial cells binds to an AT-rich sequence located between nt −133 and −125. Mutagenesis of the AT-rich sequence interferes with nuclear protein binding and restores the activity of the −142 to +244 fragment to the level of the −89 to +244 promoter. Binding of the nuclear protein to the vWF AT-rich sequence in mobility shift assays is inhibited by competition with a consensus Oct-1 binding site and with a silencer octamer-like sequence from the vascular cell adhesion molecule-1 (VCAM-1) promoter. Subsequent supershift experiments identified Oct-1 as the transcription factor that binds to vWF and VCAM-1 silencer elements. These results indicate that Oct-1 acts as a transcriptional repressor of promoters of genes expressed in endothelial cells.
© 1998 by The American Society of Hematology.
VON WILLEBRAND FACTOR (vWF) is a multimeric adhesive glycoprotein that binds platelets to the subendothelium at sites of vascular damage at high shear rates.1,2 vWF is also the carrier protein for the coagulation factor VIII, stabilizing its activity in plasma. The cellular distribution of vWF in vivo is restricted to endothelial cells, megakaryocytes, and platelets.3-5 However, its expression varies among different subpopulations of endothelial cells. In adult mammals, endothelial cells exhibit differential expression of vWF based on the location of the cell in the vascular system, whereas in mouse embryos, the vWF gene is expressed early in vascular development in a limited subset of endothelial cells.6 mRNA studies have shown that the heterogeneity of vWF expression is regulated at the transcriptional level.7 8
The human vWF gene sequence upstream of the transcription initiation site has been cloned9-12 and several studies have addressed the transcriptional regulation of the human promoter in bovine endothelial cells.13-15 We have shown previously that an ubiquitous core promoter, spanning nucleotides (nt) −89 to +19, was repressed in all cell types by an upstream negative regulatory region.13 A region spanning 487 nt of the 5′-flanking region and 247 nt of the first nontranslated exon of the human vWF gene was identified by another group as a cell-type specific promoter in bovine aorta endothelial cells (BAECs).14,15 This region includes the core promoter, a negative regulatory region spanning nt −312 to −487, and a cell-specific positive regulatory region in the first exon. The positive regulatory region contains a GATA transcription factor binding site that is required for expression. The inhibitory effect of the negative regulatory region was subsequently attributed to an NF1-like cis-acting element spanning nt −440 to −470.15 In transgenic mouse embryos, the −487 to +246 promoter fragment was active in the yolk sac vasculature, but in adult animals only brain endothelial cells expressed the transgene.16
The aim of this study was to analyze the transcriptional activity of human vWF gene promoter fragments in human umbilical vein endothelial cells (HUVECs). We report that a promoter fragment spanning nt −496 to +244 is not active in HUVECs and identify a novel negative regulatory element (NRE) that represses basal promoter activity in human endothelial and HeLa cells. The core sequence of this NRE is located directly 5′ of a cell-specific promoter fragment spanning nt −89 to +244 and contains an AT-rich octamer-like binding sequence. The vWF promoter AT-rich NRE is characterized here as an octamer-like binding site related to the previously described vascular cell adhesion molecule-1 (VCAM-1) promoter silencer sequences.17 The transcription factor binding in vitro to the vWF and VCAM-1 NREs is identified as the ubiquitously expressed POU family protein Oct-1.
MATERIALS AND METHODS
Plasmid constructions.
The pGvWCAT plasmids, containing 147, 420, and 1286 nt upstream of the transcription initiation site and 19 nt of the first exon of the vWF gene, have been described previously.13 The plasmids containing inserts −89 to +19 and −500 to +19 have been constructed following the same methodology. To construct the plasmid pGvW(−496/+244), a −496/+244 fragment was synthesized by polymerase chain reaction (PCR) from the parental plasmid pBvWE610 with the forward primer (Xba I site underlined): 5′-GAT CTA GAA TTC AAG ACC TTC ATC TTT AGC CGA T-3′ and the reverse primer (Sac I site underlined): 5′-TAG AGC TCG AGC TGC AAA TGA GGG CTG CGG CT-3′. The PCR fragment was cut by Xba I and Sac I, gel-purified, and inserted between the Xba I and Sac I sites of the multiple cloning site of the vector pGCAT-A.18To create the plasmid pGvW(−89/+244), the construct pGvW(−496/+244) was cut with HindIII at −89 inside the vWF gene 5′-flanking sequence and the restriction site was treated with Klenow DNA polymerase. After cutting with Sac I, the resulting vWF promoter fragment −89/+244 was cloned between the Sma I and Sac I sites of the multiple cloning site of pGCAT-A. To construct pGvW(−142/+19) and pGvW(−142/+244), two fragments −142/+19 and −142/+244 were generated by PCR from the parental plasmids pGvW(−500/+19) and pGvW(−496/+244), respectively. The forward primer 5′-CAA GGC AGT TAA TTA AGG CAG C-3′ was complementary to the vWF promoter, whereas the reverse primer 5′-GAG CAA ATG ACT GAA ATG CCT-3′ was complementary to the CAT gene. The amplified hybrid vWF-CAT fragment was cloned into pCRScript(Amp) (Stratagene Inc, La Jolla, CA). The vWF gene fragment was isolated by BamHI andKpn I digestion and inserted into the multiple cloning site of pGCAT-A. Plasmids pGvW(−142m/+19) and pGvW(−142m/+244) were constructed in the same manner with the upstream primer 5′-CAA GGC AGT TAC CTA AGG CAG C-3′ containing point mutations at nt −130 and −129 (mutations of the wild-type sequence are underlined). To generate plasmid pGvW(−89/+244)GATAm, a mutation of the GATA cis-element at +221 in the first exon of the vWF promoter was obtained with the transformer mutagenesis kit (Clontech, Palo Alto, CA) using the reverse mutagenesis primer 5′-GGG CTG CGG CTG TTT CCA AGG TCC C-3′ (mutations of the wild-type sequence are underlined). The control plasmid pSVGCAT-A has been described.18
Fragments of the 5′-flanking sequence of the vWF gene were inserted in front of the Herpes simplex virus thymidine kinase (TK) promoter of plasmid pBLCAT2.19 The fragments spanning nt −147 to −89 and nt −420 to −89 were isolated from the pGvW(−147/+19) and pGvW(−420/+19) constructs by digesting 5′ in the multiple cloning site of pGCAT-A withBamHI and 3′ with HindIII at nt −89 inside the vWF gene 5′-flanking sequence. The cohesive ends were filled in with Klenow DNA polymerase and ligated into the blunt-endedBamHI site of pBLCAT2.
The orientation and boundaries of all constructs and complete sequences of fragments obtained by PCR were confirmed by sequencing with the T7 polymerase sequencing kit from Pharmacia LKB Biotechnology Inc (Uppsala, Sweden).
Cell cultures and transfections.
HeLa, COS-7, and megakaryoblastic Dami cells were from the American Type Culture Collection (ATCC; Rockville, MD). The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (FCS), penicillin (100 U/mL), and streptomycin (100 μg/mL). Calf pulmonary artery endothelial (CPAE) cells (ATCC) and BAECs were cultured in OptiMEM (Life Technologies, Cergy-Pontoise, France) supplemented with 10% FCS, penicillin (100 U/mL), and streptomycin (100 μg/mL). HUVECs were isolated from human umbilical cord veins as described.5 The cells were cultured in growth medium consisting of MCDB 107, containing 20 U/mL penicillin, 20 μg/mL streptomycin, 30 μg/mL endothelial cell growth supplement (Collaborative Biomedical Products, Becton Dickinson, Bedford, MA), 90 μg/mL heparin (Sigma, St Quentin, France), and 10% FCS (Hyclone Europe, Aalst, Belgium). MCDB 10720 is prepared by adding 15.1 mg/L glycine and 149 mg/L KCl to medium MCDB 105 (Sigma).
Transfections of BAECs, CPAE cells, and HeLa cells were performed by the calcium phosphate-DNA coprecipitation method.21 The precipitate, obtained with 15 μg of the various CAT reporter plasmids and 0.5 μg of the control plasmid pCMVβ (Clontech), coding for β-galactosidase, was incubated with the cells for 24 hours. The medium was changed and cells were cultured for another 24 hours.
HUVECs were transfected by electroporation as described previously.22 Briefly, cells at passage 2 were synchronized in the G2/M phase, harvested, and resuspended in electroporation buffer (MCDB 107 without any addition but 5% FCS) at 4 × 106 cells/mL. The cell suspension (0.75 mL) and the plasmid solution were added to 4-mm electrode gap cuvettes (Bio-Rad, Ivry, France; or Eurogentec, Liège, Belgium) containing 20 or 30 μg of reporter plasmid, 0.5 μg of pCMVβ as internal control plasmid, and 30 μg of plasmid pBS− (Strategene, La Jolla, CA) as carrier DNA. Electroporation was performed at room temperature at a voltage of 360 V and a capacitance of 1,500 μF with a Cellject electroporator (Eurogentec, Liège, Belgium). After electroporation, the cells were immediately transferred into 4 mL of complete growth medium in 60-mm diameter culture dishes coated with gelatin (1% solution in phosphate-buffered saline [PBS]).
After 48 hours of incubation, transfected cells were lysed by four freeze-thaw cycles in TE buffer (100 mmol/L Tris-HCl, pH 7.8, 5 mmol/L EDTA). β-Galactosidase activity was assayed as described.22 CAT activity was measured by a dual-phase diffusion assay23 in the presence of 1.4 μmol/L of [3H] acetyl-coenzyme A (200 mCi/mmol/L; Amersham, Little Chalfont, Bucks, UK), and 1 mmol/L chloramphenicol. The [3H] acetylchloramphenicol partitioned into the organic phase and was measured over a period of several hours in a TopCount counter (Canberra-Packard, Aalst, Belgium) and the rate of CAT enzyme activity during the linear phase was expressed as counts per minute (cpm). CAT activity was normalized by the β-galactosidase activity and expressed as relative CAT activity.
Mobility shift assays and DNase I footprints.
To perform mobility shift assays and footprints, nuclear extracts from HUVECs, CPAE cells, Dami cells, and HeLa cells were prepared by the method of Dignam et al.24 Mobility shift assays were performed with the indicated wild-type or mutated vWF promoter fragment spanning nt −142 to −89, which were labeled with Klenow DNA polymerase and [α-32P] dCTP (3,000 Ci/mmol/L; Amersham) or double-stranded oligonucleotides end-labeled with [γ-32P]ATP or [γ-33P]ATP (3,000 Ci/mmol/L; Amersham) using polynucleotide kinase (Boehringer, Mannheim, Germany), annealed, and purified by polyacrylamide gel electrophoresis. Two micrograms of HeLa or 4 μg of HUVEC nuclear extracts was preincubated for 5 minutes at 4°C in the absence or in the presence of competitors in binding buffer consisting of 2 μg of poly(dI-dC), 1 mmol/L dithiothreitol (DTT), 10 mmol/L HEPES, pH 7.8, 50 mmol/L KCl, 1 mmol/L MgCl2, 0.1 mmol/L EDTA, 0.1% NP-40, 1 mmol/L spermidine, and 10% glycerol. The probes were added and the reaction mixture was then incubated for another 15 minutes at 24°C. The samples were loaded on a nondenaturing 5% polyacrylamide gel run in TGE (25 mmol/L Tris, pH 8.3, 250 mmol/L glycine, 2 mmol/L EDTA).25 Double-stranded oligonucleotides used as competitors were as follows: ROR: 5′-TCG ACT CGT TAT AAC TAG GTC AAG CGC TG-3′, which contains an AT-rich binding site for the orphan nuclear receptor RORα26,27; MLP: 5′-CTA CAC CTA TAA ACC AAT CAC CTG T-3′, containing the CCAAT sequence of the Adenovirus major late promoter28; Oct: 5′-TGT CGA ATG CAA ATC ACT AGA A-3′ (Promega, Madison, WI), the binding site for Oct-1; and VCAM: 5′-TAG TGA ATT TAC ATG ATG ATG A-3′, the octamer-like silencer sequence of the VCAM-1 promoter.17
In supershift assays, the antibodies antihuman Oct-1/2 and anti–Oct-1 (Upstate Biotechnologies, Lake Placid, NY) or Pbx 1/2/3 (Santa Cruz Biotechnology, Santa Cruz, CA) were incubated (1/20 dilution) with the nuclear extract for 20 minutes at room temperature before the addition of the probes.
For DNase I footprinting, a fragment spanning nt −178 to +48 was synthesized by PCR and cloned into the Sma I site of plasmid pBS−. The plasmid was cut with EcoRI and labeled with the Klenow DNA polymerase and [α-32P] dCTP, and the DNA probe was liberated by restriction enzyme digestion with Xba I. Nuclear extract (90 μg) was preincubated for 15 minutes at 24°C in a binding buffer consisting of 2.5 μg of poly(dI-dC), 25 mmol/L HEPES, pH 7.9, 50 mmol/L KCl, 0,1 mmol/L EDTA, 1 mmol/L DTT, 10% glycerol. The labeled probe was added and the reaction mixture was incubated for 30 minutes. Samples were digested for 1 minute with 1 to 4 U of RQ1 DNase I (Promega) in the presence of 5 mmol/L MgCl2 and 0.5 mmol/L CaCl2. DNase I was inactivated by the addition of 12.5 mmol/L EDTA and repeated extraction with phenol-chloroform. The samples were loaded on a 6% sequencing gel to visualize the DNase I digestion pattern. The gel was dried and exposed to x-ray film.
RESULTS
Identification of positive and negative regulatory regions in the vWF gene promoter.
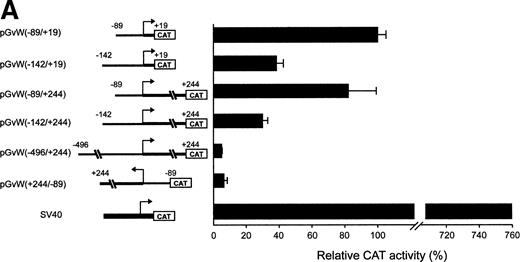
To identify important regions for the basal transcription of the human vWF gene in HUVECs, pGvWCAT constructs containing deletions of the 5′-flanking sequence were transfected into HUVECs, together with a plasmid expressing the lacZ gene as an internal control for transfection efficiency. To normalize between experiments, the CAT activity of the constructs is calculated as a percentage of the mean CAT expression obtained with plasmid pGvW(−89/+19). Figure 1A shows that the fragment spanning nt −89 to +19 represents a functional promoter in HUVECs. The activity of this promoter is 7.6-fold lower in HUVECs than the SV40 promoter construct pSVGCAT-A. Extending the promoter region to nt −142 in the construct pGvW(−142/+19) resulted in a 60% reduction of transcriptional activity compared with plasmid pGvW(−89/+19). This result suggests the presence of an NRE located between nt −89 and −142.
Expression of CAT reporter gene constructs containing deletions of the 5′-flanking region of the vWF gene and 19 nt or 244 nt of the first exon. (A) Depicted on the left side of the figures are the vWF promoter-CAT deletion constructs containing 19 nt or 244 nt of the first exon (pGvW vectors) and pSVGCAT-A (containing the SV40 early promoter). Thirty micrograms of the CAT plasmids was transfected into HUVECs in the presence of 0.5 μg of pCMVβ plasmid and 30 μg of carrier plasmid pBS−. Forty-eight hours later, extracts were prepared and the level of CAT activity was measured, normalized for β-galactosidase activity, and expressed relative to the CAT activity of plasmid pGvW(−89/+19). Plasmid pGvW(+244/−89) was transfected into HUVECs as a reference for background CAT activity. The results are the mean ± SD of 4 to 6 experiments. (B) HUVECs and HeLa cells were transfected with the pGvW vectors indicated on the left side as described in the Materials and Methods. The level of CAT activity was measured, normalized for β-galactosidase activity, and expressed relative to the CAT activity of plasmid pGvW(−89/+19). (▪) HUVECs; () HeLa cells.
Expression of CAT reporter gene constructs containing deletions of the 5′-flanking region of the vWF gene and 19 nt or 244 nt of the first exon. (A) Depicted on the left side of the figures are the vWF promoter-CAT deletion constructs containing 19 nt or 244 nt of the first exon (pGvW vectors) and pSVGCAT-A (containing the SV40 early promoter). Thirty micrograms of the CAT plasmids was transfected into HUVECs in the presence of 0.5 μg of pCMVβ plasmid and 30 μg of carrier plasmid pBS−. Forty-eight hours later, extracts were prepared and the level of CAT activity was measured, normalized for β-galactosidase activity, and expressed relative to the CAT activity of plasmid pGvW(−89/+19). Plasmid pGvW(+244/−89) was transfected into HUVECs as a reference for background CAT activity. The results are the mean ± SD of 4 to 6 experiments. (B) HUVECs and HeLa cells were transfected with the pGvW vectors indicated on the left side as described in the Materials and Methods. The level of CAT activity was measured, normalized for β-galactosidase activity, and expressed relative to the CAT activity of plasmid pGvW(−89/+19). (▪) HUVECs; () HeLa cells.
To investigate whether the first exon could alleviate the repression mediated by the NRE situated between nt −142 and −89, we transfected HUVECs with plasmids containing the insert −89 to +244, −142 to +244, or −496 to +244 (Fig 1A). The level of CAT activity of construct pGvW(−89/+244) in HUVECs is comparable to the CAT activity of plasmid pGvW(−89/+19), which contains the core promoter alone. The promoter activity of plasmid pGvW(−142/+244) is markedly reduced in HUVECs, whereas the activity of pGvW(−496/+244) is close to the background observed with a control plasmid pGvW(+244/−89). Therefore, the NRE, identified between nt −89 and −142, represses the −89 to +244 promoter to the same extent as the −89 to +19 core promoter in HUVECs. Sequences in the first exon are not sufficient to relieve the inhibition mediated by this NRE. The results also suggest that other NREs are located upstream of nt −142.
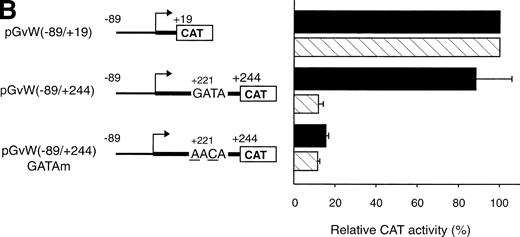
The −89 to +244 vWF gene promoter fragment that contains a GATA sequence at position +221 exhibits transcriptional activity in HUVECs (Fig 1A). The study of the human endothelin promoter showed the importance of GATA binding sites for the control of endothelial cell-specific expression.29,30 To examine further the role of the GATA sequence in the first exon of the vWF gene, this element was mutated to AACA to construct the plasmid pGvW(−89/+244)GATAm. As shown in Fig 1B, CAT activity of the mutant construct in HUVECs is reduced to 20% compared with the activity of plasmids pGvW(−89/+19) and pGvW(−89/+244). This result demonstrates that the GATA element at +221 is essential for the transcriptional activity of the −89 to +244 promoter in HUVECs. The construct pGvW(−89/+19) is also active in HeLa cells13 14 (Fig 1B). However, the vectors pGvW(−89/+244) and pGvW(−89/+244)GATAm are not active in HeLa cells. These data suggest that the GATA binding site is not functional in HeLa cells and that silencing sequences, which are located in the first exon and which are distinct from the GATA binding site, contribute to prevent expression of the vWF promoter.
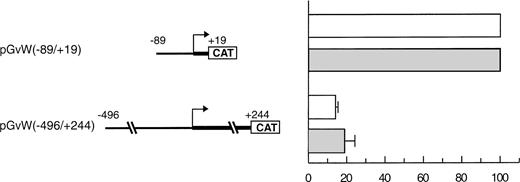
In a study performed by another group in BAECs, the activity of a −90 to +20 core promoter linked to the growth hormone gene was also repressed due to an upstream NRE located between nt −312 and −487.15 However, the activities of a −487 to +247 promoter and of the core promoter were similar, indicating that the repression mediated by the NRE was relieved when the entire first exon was included in the constructs. We also analyzed an equivalent CAT reporter plasmid pGvW(−496/+244) in BAECs, cultured between passage 5 and 8, and in CPAE cells, which express vWF. In our experiments (Fig 2), the construct pGvW(−496/+244) does not express CAT in either of those bovine cells.
Expression of CAT reporter gene constructs in bovine endothelial cells. BAECs and CPAE cells were transfected with 10 μg of the CAT reporter vectors pGvW(−89/+19) or pGvW(−496/+244) and 0.5 μg of pCMVβ plasmid. The level of CAT expression, normalized for β-galactosidase activity and expressed relative to the CAT activity of plasmid pGvW(−89/+19), is shown. (□) BAECs; (▧) CPAE. The results are the mean ± SD of 3 experiments.
Expression of CAT reporter gene constructs in bovine endothelial cells. BAECs and CPAE cells were transfected with 10 μg of the CAT reporter vectors pGvW(−89/+19) or pGvW(−496/+244) and 0.5 μg of pCMVβ plasmid. The level of CAT expression, normalized for β-galactosidase activity and expressed relative to the CAT activity of plasmid pGvW(−89/+19), is shown. (□) BAECs; (▧) CPAE. The results are the mean ± SD of 3 experiments.
The negative cis-regulatory regions of the vWF promoter inhibit the transcriptional activity of a heterologous promoter.
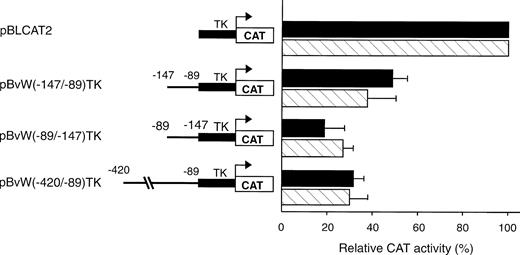
To test whether the NREs identified between −496 and −89 could inhibit the transcriptional activity of a heterologous promoter, two fragments spanning nt −420 to −89 and −147 to −89, respectively, were subcloned upstream of the ubiquitous TK promoter in plasmid pBLCAT2 to construct the plasmids pBvW(−420/−89)TK and pBvW(−147/−89)TK. The CAT activities of the constructs transfected into HUVECs and HeLa cells are presented in Fig 3. When the longer −420 to −89 fragment is inserted into the construct, the CAT activity of plasmid pBvW(−420/−89)TK decreases by more than 70% in HUVECs and HeLa cells. The fragment −147 to −89 retains the capacity to reduce TK promoter activity by more than 50% in both endothelial and nonendothelial cells. Furthermore, the DNA fragment spanning nt −147 to −89 inhibits the TK promoter when cloned in reverse orientation. These results confirm that the NRE situated between nt −142 and −89 is sufficient to inhibit the transcriptional activity of the heterologous TK promoter in both orientations.
Effect of negative regulatory elements in the vWF gene promoter on the transcriptional activity of the TK promoter. Blunt-ended fragments of the vWF gene 5′-flanking sequence were ligated in the sense or antisense direction into the BamHI site of pBLCAT2 upstream of the TK promoter. Transfections were performed in HUVECs and HeLa cells as described in the Materials and Methods. Forty-eight hours after transfection, cell extracts were prepared and aliquots, normalized for transfection efficiency, were used for the determination of CAT activity. The inserts are schematized on the left. The CAT activity of the vector pBLCAT2 with the TK promoter alone is set at 100%. The CAT activities associated with the other vectors are expressed relative to the activity of pBLCAT2 and are given on the right. (▪) HUVECs; () HeLa cells. The results are the mean ± SD of 4 to 6 experiments.
Effect of negative regulatory elements in the vWF gene promoter on the transcriptional activity of the TK promoter. Blunt-ended fragments of the vWF gene 5′-flanking sequence were ligated in the sense or antisense direction into the BamHI site of pBLCAT2 upstream of the TK promoter. Transfections were performed in HUVECs and HeLa cells as described in the Materials and Methods. Forty-eight hours after transfection, cell extracts were prepared and aliquots, normalized for transfection efficiency, were used for the determination of CAT activity. The inserts are schematized on the left. The CAT activity of the vector pBLCAT2 with the TK promoter alone is set at 100%. The CAT activities associated with the other vectors are expressed relative to the activity of pBLCAT2 and are given on the right. (▪) HUVECs; () HeLa cells. The results are the mean ± SD of 4 to 6 experiments.
HUVECs and HeLa nuclear proteins bind to the −142 to −89 NRE.
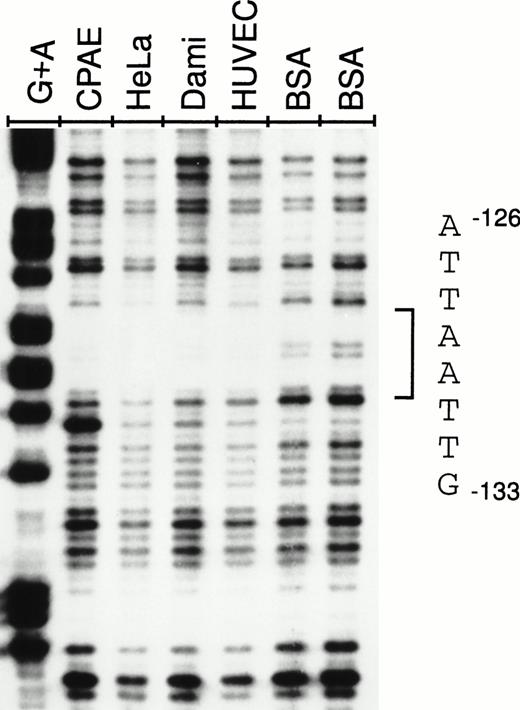
To determine whether the silencing activity of the −142 to −89 region correlates with the binding of nuclear factors from endothelial and nonendothelial cell nuclear extracts, DNase I footprintings were performed with a 32P-end-labeled restriction fragment spanning the vWF gene from −178 to +48. Analysis of the sense strand of the probe shows that a region between nt −126 and −133 is protected from DNase I digestion in the presence of endothelial (CPAE and HUVECs), megakaryoblastic (Dami), and HeLa nuclear extracts (Fig 4). This region includes the sequence TTAATTAA, an octamer-like AT-rich palindrome.
DNase I footprint of the −142 to −89 NRE. A −178 to +48 fragment was labeled at −178 on the antisense strand and incubated for 30 minutes with 90 μg of nuclear extract from CPAE cells, HeLa cells, Dami cells, or HUVECs or 90 μg of BSA and used in footprinting analysis as described in the Materials and Methods. The −126 to −133 protected sequence is shown on the right. G+A lane corresponds to cleavage at bases G and A by Maxam-Gilbert chemical sequencing of the probe.
DNase I footprint of the −142 to −89 NRE. A −178 to +48 fragment was labeled at −178 on the antisense strand and incubated for 30 minutes with 90 μg of nuclear extract from CPAE cells, HeLa cells, Dami cells, or HUVECs or 90 μg of BSA and used in footprinting analysis as described in the Materials and Methods. The −126 to −133 protected sequence is shown on the right. G+A lane corresponds to cleavage at bases G and A by Maxam-Gilbert chemical sequencing of the probe.
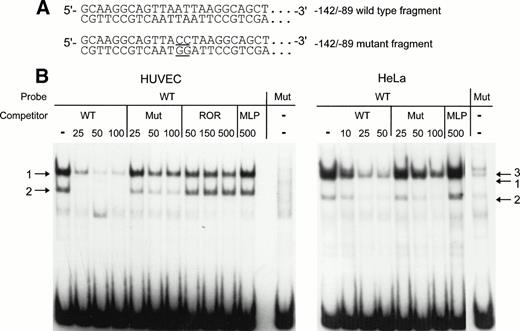
To confirm that the AT-rich sequence is part of a binding site for one or several nuclear factors, mobility shift assays were performed with labeled wild-type (WT) −142 to −89 or mutant (Mut) −142 to −89 fragment as probes. In the Mut probe, the AT-rich sequence TTAATTAA was mutated to TTACCTAA (Fig 5A). For competition studies, reactions were performed with an excess of cold WT or Mut fragment. After incubation with HUVEC nuclear extracts, the −142 to −89 probe forms two retarded DNA-protein complexes in the absence of competitor (arrows 1 and 2 in Fig 5B, left panel). Complexes in bands 1 and 2 are specific, because binding to the labeled WT probe is almost abolished with 25-fold molar excess of unlabeled WT fragment and binding is no longer observed with higher molar excess of competitor. The mutant competitor does not interfere to the same extent as the WT fragment with the formation of the major complex (band 1). Even at a 100-fold molar excess of Mut fragment, complex 1 can still be detected. Band 2 is weak, but the relative intensities of band 1 and band 2 appear to be unchanged in the presence or the absence of Mut competitor. Unrelated oligonucleotides were next used as competitors in the mobility shift assays with HUVEC nuclear extract to confirm that the −142 to −89 probe binds sequence-specific nuclear proteins. The oligonucleotides used were ROR, which contains an AT-rich binding site for the orphan nuclear receptor RORα, and MLP, containing the CCAAT sequence of the Adenovirus major late promoter. Complexes 1 and 2 are not affected by competition with a 500-fold molar excess of these oligonucleotides, except for a slight decrease of the intensity of band 1 in the presence of large quantities of ROR. The labeled mutant probe does not bind significant amounts of HUVEC nuclear proteins (Fig 5B, last lane in left panel), confirming that binding of the nuclear factors in complexes 1 and 2 is greatly reduced by mutation of the AT-rich sequence.
Detection of nuclear proteins interacting with the −142 to −89 NRE of the vWF gene by mobility shift assays. (A) 5′-end of the sequence of the −142 to −89 wild-type probe and of the −142 to −89 mutant probe. The mutated basepairs are underlined. (B) The restriction enzyme fragments were labeled and incubated with nuclear extracts from HUVECs or HeLa cells before electrophoresis as described in the Materials and Methods. The competitors were wild-type fragment (WT), mutant fragment (Mut), ROR, and MLP oligonucleotides at the indicated molar excess. Arrows designate the specific retarded complexes as described in the text.
Detection of nuclear proteins interacting with the −142 to −89 NRE of the vWF gene by mobility shift assays. (A) 5′-end of the sequence of the −142 to −89 wild-type probe and of the −142 to −89 mutant probe. The mutated basepairs are underlined. (B) The restriction enzyme fragments were labeled and incubated with nuclear extracts from HUVECs or HeLa cells before electrophoresis as described in the Materials and Methods. The competitors were wild-type fragment (WT), mutant fragment (Mut), ROR, and MLP oligonucleotides at the indicated molar excess. Arrows designate the specific retarded complexes as described in the text.
With nuclear extracts from HeLa cells (Fig 5B, right panel), two bands corresponding to complexes 1 and 2 are also detected. Again, a 25-fold molar excess of WT fragment competes more efficiently for the formation of the major complex 1 as compared with a 25-fold molar excess of Mut fragment. An additional band, indicated by arrow 3, is also visible without competitor or with a 10-fold excess of WT competitor. In the presence of 100-fold excess of Mut competitor, the complex in band 3 is completely abolished in contrast to the complex in band 1. Therefore, the formation of this complex seems to be independent of the mutated nucleotides in the AT-rich sequence.
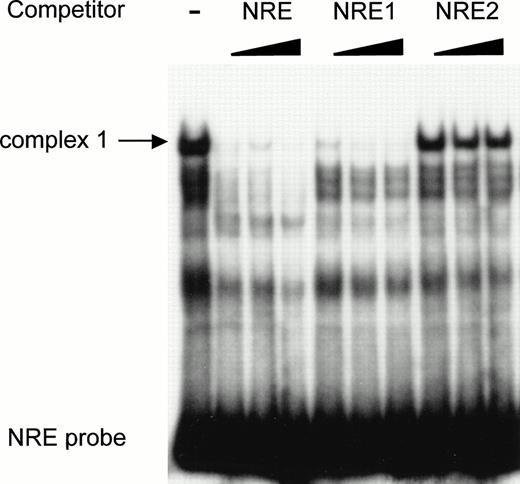
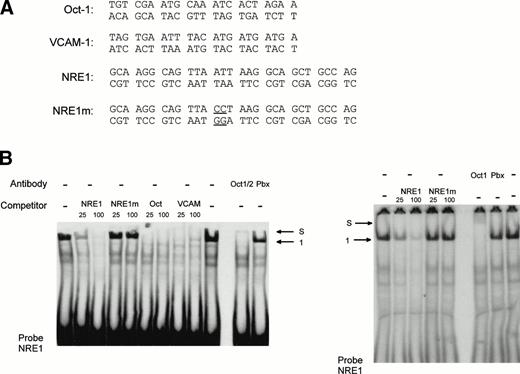
To further investigate the binding of the specific nuclear factors to the AT-rich sequence, a double-stranded oligonucleotide (NRE) covering the same −142 to −89 region as the restriction enzyme fragment (Fig 5) was synthesized and used as probe in a mobility shift assay. Two shorter fragments encompassing nt −142 to −114 (NRE1) and nt −113 to −85 (NRE2), respectively, were used as competitors. The full-length −142 to −89 NRE was end-labeled and incubated with HUVEC nuclear extracts in the absence and in the presence of NRE, NRE1, or NRE2 as competitors. Figure 6 shows that, in the absence of competitor, a major band corresponding to complex 1 is again observed with the NRE probe. However, additional bands are also present, among them the minor complex 2. The wild-type NRE sequence competes for the formation of complex 1. Complex 1 is completely abolished in the presence of NRE1 spanning nt −142 to −114 and including the AT-rich binding site. The downstream oligonucleotide NRE2 (nt −113 to −85) is not effective in preventing the formation of the DNA-protein complexes, indicating that it does not contain high-affinity nuclear protein binding sites. Results from Figs 5 and 6are consistent with the hypothesis that a nuclear protein interacts with the octamer-like AT-rich sequence spanning nt −133 to −126 to form the major complex 1.
The high-affinity nuclear protein binding site in the vWF gene promoter NRE is located inside the sequence NRE1 (−142 to −114). The double-stranded oligonucleotide NRE (−142 to −89) was labeled, incubated with nuclear extracts from HUVECs, and analyzed as described in the Materials and Methods. Unlabeled double-standed competitors NRE, NRE1 (−142 to −114) and NRE2 (−113 to −85) were included at 50-, 100-, and 200-fold molar excess. The major complex 1 is indicated by an arrow.
The high-affinity nuclear protein binding site in the vWF gene promoter NRE is located inside the sequence NRE1 (−142 to −114). The double-stranded oligonucleotide NRE (−142 to −89) was labeled, incubated with nuclear extracts from HUVECs, and analyzed as described in the Materials and Methods. Unlabeled double-standed competitors NRE, NRE1 (−142 to −114) and NRE2 (−113 to −85) were included at 50-, 100-, and 200-fold molar excess. The major complex 1 is indicated by an arrow.
The AT-rich binding site is a negative cis-acting regulatory element.
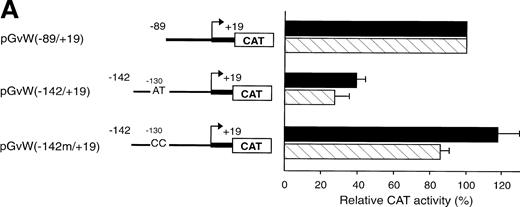
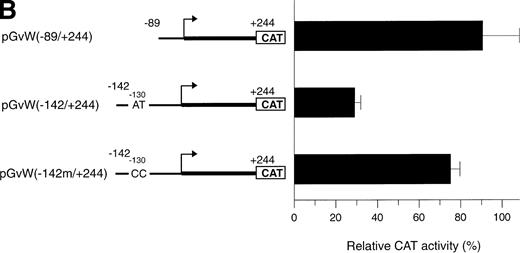
The octamer-like AT-rich sequence, located between nt −133 and −126 of the negative regulatory region of the vWF gene promoter, is a binding site for nuclear protein(s) present in nuclear extracts from HUVECs and HeLa cells. To examine whether this AT-rich motif is responsible for the silencing of the −142 to +19 and −142 to +244 promoter fragments, the same point mutations at position −130 and −129, which inhibit DNA-protein interaction (Fig5A), were introduced into the constructs pGvW(−142m/+19) and pGvW(−142m/+244). The reporter construct pGvW(−142/+19) exhibits reduced transcriptional activity in HUVECs and HeLa cells compared with pGvW(−89/+19) (Fig 7A). Mutation of the AT-rich sequence in plasmid pGvW(−142m/+19) restores the level of CAT activity to that obtained with the core (−89/+19) promoter. As shown in Fig 7B, the double base mutation in the AT-rich sequence also increases the transcriptional activity of construct pGvW(−142m/+244) to 75% of the activity of the −89 to +244 promoter in HUVECs. These results suggest that the nuclear protein(s) binding to the AT-rich sequence is involved in the negative regulation of the activity of the human vWF gene promoter.
Functional analysis of mutations in the AT-rich sequence. HUVECs and HeLa cells were transfected as described in the legend to Fig 1. (A) The vectors pGvW(−89/+19), pGvW(−142/+19), and pGvW(−142m/+19) are depicted on the left side of the figures. The construct pGvW(−142m/+19) is mutated at nt −130 and −129 inside the AT-rich sequence. The CAT activities associated with these vectors are expressed relative to the activity of pGvW(−89/+19), which is set at 100%, and are given on the right. (▪) HUVECs; () HeLa cells. (B) The vectors pGvW(−89/+244), pGvW(−142/+244), and pGvW(−142m/+244) are shown on the left side of the panel. The CAT activities in HUVECs are expressed relative to the activity of the plasmid pGvW(−89/+19). The results are the mean ± SD of 4 to 6 experiments.
Functional analysis of mutations in the AT-rich sequence. HUVECs and HeLa cells were transfected as described in the legend to Fig 1. (A) The vectors pGvW(−89/+19), pGvW(−142/+19), and pGvW(−142m/+19) are depicted on the left side of the figures. The construct pGvW(−142m/+19) is mutated at nt −130 and −129 inside the AT-rich sequence. The CAT activities associated with these vectors are expressed relative to the activity of pGvW(−89/+19), which is set at 100%, and are given on the right. (▪) HUVECs; () HeLa cells. (B) The vectors pGvW(−89/+244), pGvW(−142/+244), and pGvW(−142m/+244) are shown on the left side of the panel. The CAT activities in HUVECs are expressed relative to the activity of the plasmid pGvW(−89/+19). The results are the mean ± SD of 4 to 6 experiments.
Oct-1 binds to related NREs in the vWF and VCAM-1 promoters.
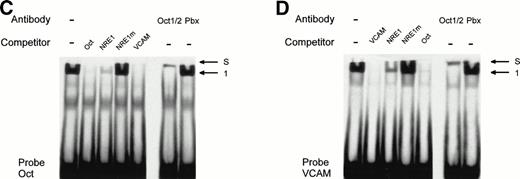
The AT-rich NRE of the vWF gene promoter conforms to the canonical homeodomain binding sequence TAATTA and exhibits limited homology with the octamer-1 (Oct-1) transcription factor binding site ATGCAAATAA. Octamer-like sequences in the human VCAM-1 promoter were shown to act as negative regulatory elements in endothelial and nonendothelial cells.17 To investigate whether both the vWF AT-rich and VCAM-1 octamer-like NREs are recognized by the same octamer binding factors, we used as competitors in mobility shift assays the oligonucleotides Oct, containing the cognate Oct-1 binding sequence, and VCAM, consisting of the VCAM-1 silencer sequence. The probe NRE1 was incubated with HeLa nuclear extracts and different amount of these competitors (Fig 8). At a 100-fold molar excess, Oct and VCAM compete as effectively as NRE1 for the binding of the nuclear factor in complex 1, suggesting that a transcription factor of the Oct family binds both to the vWF and VCAM NREs. To identify the protein involved in vWF and VCAM-1 transcriptional repression, a monoclonal antibody that recognizes the Oct-1 and Oct-2 POU-homeodomains was added to the binding reactions. In the presence of the antibody, complex 1 is abolished and partly supershifted (Fig 8B, left panel). This result identifies the vWF repressor as Oct-1, because Oct-2 is only expressed in lymphoid cells. A control antibody, directed against the homeodomain of the Pbx family of transcription factors, had no effect on the formation of the band. In Fig 8B (right panel), a second antibody directed specifically against the Oct-1 POU-domain also abolishes the formation of complex 1. Mobility shift assays with the labeled Oct-1 consensus sequence (probe Oct) gives rise to a nuclear protein-DNA complex migrating at the same level as complex 1 (Fig 8C). The vWF NRE1 and the VCAM octamer-like sequence, but not NRE1m, are able to compete for the binding of this nuclear protein to its cognate octamer binding site. This protein is also clearly supershifted by the Oct-1/2 antibody. Finally, the formation of complex 1 with the VCAM-1 probe is abolished (Fig 8D) by competition with NRE1 and Oct. Complex 1 is again supershifted in the presence of the Oct-1/2 antibody.
Identification and characterization of Oct-1 binding to the vWF AT-rich NRE, to the consensus Oct-1 binding site, and to the VCAM-1 promoter octamer-like sequence. Double-stranded oligonucleotides were labeled and incubated with nuclear extracts from HeLa cells before electrophoresis as described in the Materials and Methods. (A) Sequences of the probes used for MSA. These probes spanned the vWF AT-rich NRE between nt −142 and −114 (NRE1), the binding site for the bipartite POU domain of Oct-1 (Oct), and the octamer-like silencer element in the VCAM-1 promoter (VCAM). The competitors NRE1, NRE1m (the mutated base pairs in the AT-rich element are underlined), Oct, and VCAM were used at the indicated molar excess. (B, C, and D) Mobility shift assays with probes NRE, Oct, and VCAM. Arrows show the specific retarded complex 1 and the supershifted complexes between Oct-1 and the probes (S) as described in the text. Supershifts with probe NRE were performed with two different antibodies, an anti–Oct-1/2 antibody (B, left panel) and an antibody specific to Oct-1 (B, right panel). The supershifted complex S is situated just underneath the level of the wells of the polyacrylamide gel.
Identification and characterization of Oct-1 binding to the vWF AT-rich NRE, to the consensus Oct-1 binding site, and to the VCAM-1 promoter octamer-like sequence. Double-stranded oligonucleotides were labeled and incubated with nuclear extracts from HeLa cells before electrophoresis as described in the Materials and Methods. (A) Sequences of the probes used for MSA. These probes spanned the vWF AT-rich NRE between nt −142 and −114 (NRE1), the binding site for the bipartite POU domain of Oct-1 (Oct), and the octamer-like silencer element in the VCAM-1 promoter (VCAM). The competitors NRE1, NRE1m (the mutated base pairs in the AT-rich element are underlined), Oct, and VCAM were used at the indicated molar excess. (B, C, and D) Mobility shift assays with probes NRE, Oct, and VCAM. Arrows show the specific retarded complex 1 and the supershifted complexes between Oct-1 and the probes (S) as described in the text. Supershifts with probe NRE were performed with two different antibodies, an anti–Oct-1/2 antibody (B, left panel) and an antibody specific to Oct-1 (B, right panel). The supershifted complex S is situated just underneath the level of the wells of the polyacrylamide gel.
Taken together, these results establish that the protein that forms complex 1 with the vWF AT-rich NRE and the VCAM-1 octamer-like silencer is the ubiquitously expressed POU-domain transcription factor Oct-1 acting as a repressor.
DISCUSSION
To determine the molecular mechanisms contributing to the expression of vWF in human endothelial cells, we have studied the transcriptional regulation of the vWF gene. Using nested deletions of the 5′-region linked to a CAT reporter gene, we identified a core promoter, spanning nt −89 to +244, that is active in HUVECs but not in HeLa cells. This DNA fragment contains the first nontranslated exon of the vWF gene and was previously shown to be a functional promoter in BAEC.14 The transcriptional activity of this promoter region in HUVECs depends on the presence of an intact GATA binding site at position +221. Indeed, the mutation of the GATA binding site abolishes the activity of the core promoter in HUVECs and does not restore transcriptional activity in HeLa cells. These results show that the −89 +244 vWF gene promoter contains a silencer sequence, which is different from the GATA element. The silencer is offset by the positive GATA element in HUVECs only. It may thus be hypothesized that the GATA site contributes to the activity of the −89/+244 promoter fragment in endothelial cells by overcoming the effect of a repressor localized in the first exon. However, endogenous GATA-binding proteins (such as GATA-2) are expressed by HeLa cells.29Therefore, the −89 to +244 fragment may not be an effective promoter in HeLa cells, because the level of expression of the functional GATA-binding protein is too low. Another possibility is that a distinct (and perhaps endothelial cell-restricted) GATA-binding protein is responsible for cell-specific derepression in HUVECs.
vWF is expressed exclusively in two cell types, endothelial cells and megakaryocytes. The basal transcription of other genes, which are either expressed in endothelial cells (endothelin,29,30platelet/endothelial cell adhesion molecule-1,31endothelial nitric oxide synthase32) or which are expressed in megakaryocytes (glycoprotein IIb33), requires the presence of GATA cis-acting regulatory sequences. Several megakaryocytic gene promoters contain GATA sequences that are active in conjunction with Ets sequences to establish positive promoter activity.33-38 Recently, we have identified functional Ets binding sites in the core vWF promoter,39 and it could be hypothesized that GATA and Ets family members contribute in a coordinated fashion to the endothelial cell-restricted activity of the vWF promoter.
In this study, we have identified an NRE between nt −142 and −89 of the vWF gene promoter that represses the transcriptional activity of the −89 to +244 promoter fragment in HUVECs. The NRE also inhibits the −89 to +19 promoter fragment and the TK promoter in both HUVECs and HeLa cells. The core sequence of the NRE has been mapped to an AT-rich palindromic octamer (TTAATTAA) between nt −133 and −126. Site-directed mutagenesis in this core sequence sharply reduces protein-DNA interaction and abolishes the NRE-mediated repression in transfection assays.
Our experimental results confirm that the vWF NRE is bound by nuclear proteins related in their binding specificity to the Oct family of transcription factors. Indeed, the protein binding to the AT-rich NRE is recognized by antibodies directed against the POU domain of the widely expressed transcription factor Oct-1. Therefore, we conclude that the protein that binds to the NRE and that is involved in the negative regulation of vWF gene promoter is Oct-1. The AT-rich NRE of the vWF gene promoter conforms to the consensus homeodomain binding sequence, TAATTA.40,41 The homeodomain, a structural DNA binding module that contains the helix-turn-helix (HTH) motif, occurs in a large family of proteins that regulate transcription.42,43 Among transcription factors exhibiting the homeodomain motif are Oct-1 and Oct-2 (only expressed in lymphoid cells44), which contain the composite POU domain, a conserved DNA binding region of approximately 160 amino acids.45 The structure of the Oct-1-DNA complex has been determined by nuclear magnetic resonance and by crystallography, showing that the preferred binding site of Oct-1 is related to the vWF AT-rich element.46,47 The ability of Oct-1 and other POU domain factors to bind to a variety of degenerate octamer motifs is well established48 and could represent an important feature of their transcriptional regulatory function.
The transcriptional regulation of the VCAM-1 gene in endothelial cells has been extensively studied.17,49,50 Octamer-like sequences act as silencers to prevent VCAM-1 expression in unstimulated HUVECs and in other cell types.17,49 Experimental evidence presented in this report clearly shows that the extensively characterized VCAM-1 repressing element located at nt −1554 is related to the vWF AT-rich NRE and to the Oct-1 binding site. We demonstrate in this study for the first time that the VCAM-1 octamer-like silencer is bound by Oct-1. Therefore, Oct-1 may play an important role in the well-characterized promoter repression mechanism of the VCAM-1 promoter51 and in the negative regulation of the vWF promoter both in HUVECs and in other nonendothelial cell types.
The role of Oct-1 as a transcriptional repressor is further strengthened by recent studies that have implicated Oct-1 in the downregulation of the human pituitary-specific transcription factorpit1/ghf1 gene promoter52 and the repression of the human immunodeficiency virus (HIV) long terminal repeat.53 In addition, a detailed study54 of the human thyrotropin β subunit (hTSH β) gene promoter has shown that Oct-1 mediates silencing of this promoter through AT-rich, degenerate Oct-1 binding sites. As is the case for the vWF AT-rich NRE, the Oct-1–mediated silencing was acting in all cell types, including HeLa cells. Oct-1 was found to possess an intrinsic silencing activity in the carboxy-terminal domain.54
In response to TNFα, NF-κB transcription factor family members bind to the VCAM-1 promoter in HUVECs and activate transcription by overcoming the negative effect of the octamer-like silencers. In skeletal muscle cells, the inhibition of the VCAM-1 promoter can also be lifted by a cell-type–specific and position-specific enhancer located between the TATA box and the transcription initiation start site.17 A similar mechanism is found in the hTSHβ promoter, in which Oct-1–mediated silencing is overcome in thyrotrops by an upstream enhancer in the −10,000 to −1,200 region.54 By analogy, we would hypothesize that endothelial cell-specific enhancer sequences are located in the vWF promoter upstream or downstream of the sequences analyzed in this study. In this study, the extension of the upstream flanking sequence to nt −496 does not lift the repression of the −496 to +244 vWF promoter but leads to even stronger inhibition in HUVECs. The results from these deletion experiments therefore point to the presence of other NREs between nt −496 and −142 and suggest that putative enhancer sequences could be located in the introns and/or in more upstream sequences.
Jahroudi and Lynch14 have reported that the GATA site at position +221 was necessary and sufficient for cell-type–specific transcriptional activity in BAE cells of a human vWF promoter fragment spanning nt −487 to +247, which was active at the same level as a −90/+20 promoter. In this report, we show that an analogous −496 to +244 promoter region fused to the CAT reporter gene lacks the specific sequences required for high promoter activity in HUVECs, in CPAE cells, and in BAECs. These results suggest that this promoter fragment is dependent on the cell line used or that the different reporter vectors used in these two studies (growth hormone v CAT) influence cell-type–specific activity. However, our previous study on the transcriptional regulation of the human vWF promoter in CPAE cells13 and the results in human endothelial cells presented in this report show a mechanism of promoter repression involving Oct-1, which was not observed in the study in BAECs. Furthermore, when the same −487 to +246 vWF promoter fragment that shows transcriptional activity in BAECs was fused to theEscherichia colilacZ gene and was used to generate transgenic mice,16 only a limited subpopulation of embryonic yolk sac endothelial cells and adult brain endothelial cells expressed the LacZ construct. No expression was detected in adult mouse endothelial cells outside the brain. Recently, Aird et al55 demonstrated that a longer human vWF gene fragment, spanning 2,182 bp of 5′ flanking sequence, the first exon, and first intron, is active in the endothelial lining of blood vessels in the brain, heart, and skeletal muscle, contrasting again with the much more widespread expression of the endogenous vWF gene. The expression of the transgene was dependent on the tissue environment. Taken together, the results of studies in bovine endothelial cells,13-15 in HUVECs, and in transgenic mice suggest that transcriptional repression of the human vWF gene promoter by POU domain proteins and other trans-acting factors is overcome in a tissue-specific manner through positive cis-acting elements that may be different in endothelial cells of various origins and depend on the tissue environment.
POU domain proteins play an important role in embryonic development and the establishment of distinct cell lineages.56 As we have shown in this study, Oct-1, a member of the POU domain transcription factor family, may be involved in the transcriptional regulation of two important genes expressed by endothelial cells.
ACKNOWLEDGMENT
The authors thank Lydie Barek and Annick Ganieux for excellent technical assistance. We thank Dr Philippe Huber for stimulating discussions.
J.-L.S. was supported by a grant from the Ministère de l’Education Nationale, Luxembourg. N.J. was supported by the Ministère de l’Enseignement Supérieur et de la Recherche, France. J.E.R. and D.H. were supported by funds from the Research Council of the University of Leuven (OT/94/22).
Address reprint requests to Jean-Luc Schwachtgen, PhD, Vascular Diseases Unit, Glaxo Wellcome, Medecines Research Centre, Gunnels Wood Road, Stevenage, Herts., SG1 2NY, UK.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal