Abstract
The SH2-containing inositol phosphatase, SHIP, often appears as multiple bands in anti-SHIP immunoblots. To characterize these bands, antisera were generated against the N-terminal (anti-N), mid-region (anti-M), and C-terminal (anti-C) portions of SHIP. Immunoprecipitation and immunoblotting studies showed that 145-, 135-, 125-, and 110-kD bands were detected in lysates from the murine hematopoietic cell line, DA-ER, with either anti-N or anti-M antisera, whereas only the 145- and 135-kD bands were recognized by the anti-C antiserum. This finding suggested that the smaller proteins might be C-terminal truncations of the full-length SHIP. To confirm this and determine if these proteins arose through alternate splicing or posttranslational cleavage, a 5′-hemagglutin (HA)-tagged full-length SHIP cDNA was expressed in these cells. We observed, via Western analysis with anti-HA antibodies, the same 4 bands with either anti-N or anti-M and only the 145- and 135-kD bands with anti-C immunoprecipitation. After interleukin-3 stimulation of HA-SHIP–expressing DA-ER cells, only the 145-kD form coprecipitated with Shc, raising the possibility that different forms of SHIP may have distinct intracellular sites. This was confirmed by subcellular fractionation, which showed that only the 110-kD form is present in the cytoskeleton of DA-ER cells. This 110-kD form possesses the same PIP3 5-ptase activity as the 145-kD form and can be generated from the latter in vitro by digestion with calpain. It is therefore possible that the different forms of SHIP are generated in vivo by calpain-mediated C-terminal truncations and perform distinct functions within hematopoietic cells.
© 1998 by The American Society of Hematology.
RECENTLY, WE1 AND Lioubin et al2 cloned the cDNA for a 145-kD protein that becomes tyrosine phosphorylated and associated with Shc in response to multiple cytokines in hematopoietic cell lines. Based on its predicted amino acid sequence, which contains an amino terminal Src homology (SH)1 2 domain, two centrally located motifs highly conserved among inositol polyphosphate 5-phosphatases (5-ptases), two phosphotyrosine binding (PTB) consensus sequences (ie, NPXY sequences),3 and several C-terminal proline-rich SH3 binding regions (see Fig 1A), we agreed to call this protein SHIP for SH2-containing inositol phosphatase.1,2 Unlike most 5-ptases,4 SHIP selectively hydrolyzes the 5′-phosphate from inositol-1,3,4,5-tetraphosphate and phosphatidylinositol-3,4,5-trisphosphate (PI-3,4,5-P3),1 two inositol polyphosphates recently implicated in growth factor-mediated signaling.5-8SHIP is also unique in that it is the only 5-ptase cloned to date that possesses an SH2 domain, and we recently demonstrated that this domain is critical for SHIP’s tyrosine phosphorylation, for its association with either Shc9 or SHP-2,10 and for SHIP-mediated apoptosis.9
During our purification of SHIP from the murine hematopoietic cell line, B6SUtA1, we found that, in addition to the 145-kD SHIP protein, 3 other major proteins with relative molecular masses of 135, 125, and 110 kD bound consistently to both Grb2-C-terminal-SH3 beads1 and to pYXN beads, suggesting that these proteins also possessed both proline-rich and SHIP-like SH2 domains. Subsequently, we found that antiserum generated against the SH2 domain of SHIP9,10 recognized 145-, 135-, 125-, and 110-kD proteins, or subsets of these proteins, in many hematopoietic cell lines, suggesting that SHIP-related proteins may exist. Interestingly, both Kavanaugh et al11 and Geier et al12 have also reported that multiple forms of SHIP may exist. Specifically, Kavanaugh et al11 demonstrated the presence of 145-, 130-, and 110-kD immunologically related forms of SHIP and suggested that the 130-kD form results from an internal translation initiation site within the mRNA of the 145-kD form and that the 110-kD form is an mRNA splice variant that lacks SHIP’s SH2 domain. Geier et al12 also reported multiple forms of SHIP in hematopoietic progenitors, with molecular masses ranging from 100 to 165 kD, and showed that the pattern of expression of the different SHIP proteins changes with hematopoietic differentiation. They also hypothesized that the different forms might be the result of different mRNA species. In this study, we have further characterized these SHIP-related proteins and conclude that at least some of them are derived via C-terminal truncations of the full-length SHIP protein.
MATERIALS AND METHODS
Reagents.
Pure COS cell-derived murine interleukin-3 (IL-3) was obtained as described previously.13 Glutathione S-transferase (GST) fusion proteins consisting of the 27-kD amino terminal of GST linked to the SH2 domain of murine SHIP (amino acids 7-133),1 the middle region of murine SHIP (amino acids 930 to 1042), the C-terminal SH3 domain of murine Grb2 (amino acids 163 to 215),14 and the PTB domain of Shc (amino acids 1 to 200)15 were expressed in Escherichia coli in pGEX-2T plasmids (Pharmacia LKB Biotechnology, Inc, Baie d’Urfe, Quebec, Canada) and purified from the sonicated bacteria using glutathione-agarose (Pharmacia), as described previously.1 Rabbit antisera to SHIP were generated by immunization with the GST-SHIP SH2 fusion protein (anti-N), with the GST-SHIP middle region (anti-M), or with a 22mer corresponding to amino acids 1157 to 1178, which is near the C-terminus of the full-length 1190 amino acid murine SHIP protein (anti-C). Affinity-purified polyclonal antibodies to Shc were purchased from Transduction Laboratories (Lexington, KY). Anti-hemagglutinin (HA) monoclonal antibodies were from BAbCO (Richmond, CA). Horseradish peroxidase-conjugated second antibodies were from Jackson Immunoresearch (West Grove, PA). Protein-grade Nonidet P-40 was from Calbiochem (San Diego, CA). The enhanced chemiluminescence Western blotting reagents were obtained from Pierce (Rockford, IL).
Generation of DA-ER cells expressing HA-tagged SHIP.
HA-tagged SHIP was generated by fusing the HA-tag in frame at the 5′ end to the murine SHIP cDNA in a murine stem cell virus (MSCV) vector containing the puromycin resistance gene, pac,16 as described earlier.9 The construct was calcium phosphate transfected into the producer cell line BOSC 23 and 48-hour retroviral supernates were used to infect DA-ER cells (DA-3 cells expressing retrovirally infected erythropoietin [Epo] receptors17). Infected DA-ER cells were selected for 2 weeks in puromycin and clones were analyzed for SHIP expression by anti-HA Western analysis.
Immunoprecipitations and Western blotting.
Murine DA-ER cells infected or not with HA-tagged SHIP and maintained in RPMI 1640 with 10% fetal calf serum (FCS) and 5 ng/mL IL-3 were growth factor deprived for 4 to 6 hours at 37°C in RPMI 1640 containing 0.1% bovine serum albumin (BSA) and then stimulated at 37°C for 5 minutes with IL-3 (400 ng/mL). The cells were then washed with phosphate-buffered saline (PBS), solubilized at 5 × 107 cells/mL with 0.5% NP-40 in 4°C phosphorylation solubilization buffer (PSB), ie, 50 mmol/L HEPES, pH 7.4, 100 mmol/L NaF, 10 mmol/L NaPPi, 2 mmol/L Na3VO4, 4 mmol/L EDTA, 2 mmol/L phenylmethyl sulfonyl fluoride (PMSF), 10 mg/mL leupeptin, and 2 mg/mL aprotinin and subjected to immunoprecipitation and Western blotting as described previously.18
35S-methionine labeling studies.
Logarithmically growing HA-SHIP–expressing DA-ER cells were washed with methionine-free Dulbecco’s modified Eagle’s medium (DMEM), resuspended at 1.5 × 107cells/mL in the same medium for 30 minutes at 37°C, and then labeled for 30 minutes with 100 μCi/mL of EASYTAG L-[35S]-methionine (1,175 Ci/mmol; NEN, Guelph, ON). The cells were washed twice with methionine-containing medium, resuspended at 1.5 × 106 cells/mL in RPMI, 10% FCS, 25 U/mL Epo, and, at the indicated times, lysed and subjected to anti-HA immunoprecipitation and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Gels were fixed, treated with EN3HANCE (NEN), dried, and exposed to REFLECTION autoradiography film (NEN) at −70°C.
Calpain digestion studies.
HA-SHIP–expressing DA-ER cells were lysed and immunoprecipitated with anti-C and protein-A Sepharose. The beads were washed 3 times with 0.1% NP-40-100 in PSB, 3 times with PBS, and suspended in 75 μL of 0.15 mol/L NaCl, 20 mmol/L Tris-Cl, pH 7.4, 100 μg/mL BSA, 1 mmol/L dithiothreitol (DTT), and 1 mmol/L CaCl2. Pure rabbit m-calpain (10 mU/mL; Sigma) was then added and, after incubation at 37°C for various times, the reactions were stopped by boiling for 2 minutes with SDS-sample buffer.
Subcellular fractionation.
Subcellular fractionation was performed as described by Thomas et al.19 Briefly, HA-SHIP–expressing DA-ER cells were lysed at 5 × 107 cells/450 mL with 2% TX-100 in PSB for 10 minutes at 4°C and centrifuged at 10,000g for 10 minutes. The pellet was washed twice at 10,000g for 10 minutes with 1% TX-100 in PSB and the TX-100–insoluble fraction was extracted from this pellet with 450 mL PSB containing 1% TX-100, 0.3% deoxycholate, and 1 mol/L NaCl for 30 minutes at 4°C. This extract was diluted twofold with PSB containing 1% TX-100 for anti-N immunoprecipitations. The three 10,000g supernatants were combined and centrifuged at 100,000g to yield the TX-100 soluble fraction (the supernatant) and the pellet was washed once with 1% TX-100 in PSB and extracted as described above to yield the submembranous cytoskeletal fraction.
RESULTS AND DISCUSSION
Multiple forms of SHIP can be generated by C-terminal truncations of the full-length SHIP protein and have long half lives in vivo.
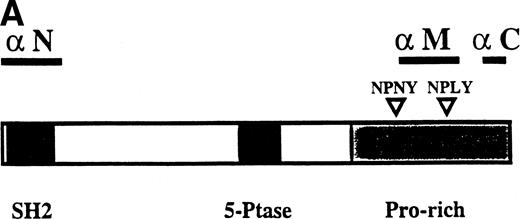
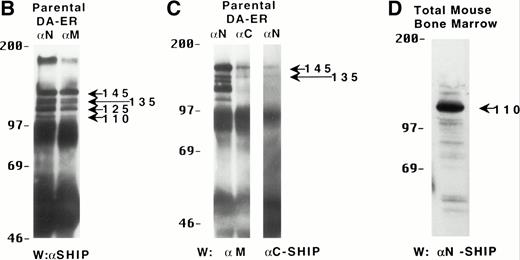
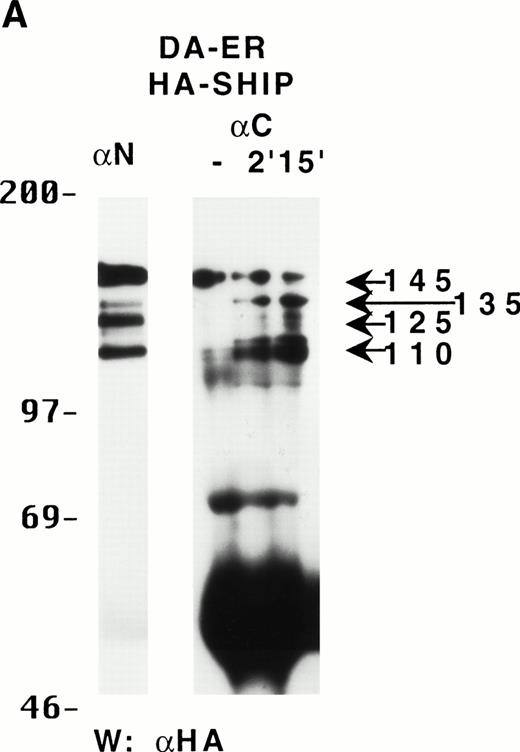
To characterize the SHIP-related proteins that we1 and others11,12,20 have observed, we generated antisera to 3 distinct regions of SHIP, ie, the SH2 domain (anti-N), the region spanning the NPXY motifs (anti-M), and the C-terminal region (anti-C; see Fig 1A). Immunoprecipitation of lysates from the murine hematopoietic cell line DA-ER with either anti-N or anti-M demonstrated, upon Western analysis with anti-M (or anti-N; data not shown), that 145-, 135- (doublet), 125-, and 110-kD bands were recognized by both antibodies (Fig 1B). However, anti-C immunoprecipitated only the 145- and 135-kD forms (Fig 1C, middle lane). Moreover, reprobing the anti-N immunoprecipitated sample (Fig1C, left lane) with anti-C showed that anti-C recognized only the 145- and 135-kD proteins in immunoblots as well. This suggested that the smaller SHIP-like proteins might be C-terminal truncations of the full-length protein. Interestingly, both the 135- and 145-kD proteins were immunoprecipitated by our anti-C antibodies. Because our anti–C-terminal antiserum was generated against a 22mercorresponding to amino acids 1157 to 1178 of the full-length 1190 amino acid SHIP, it is conceivable that the 135-kD protein is a clipped form that migrates aberrantly at 135 kD. Alternatively, the full-length SHIP protein might actually be the 135-kD species (the predicted molecular mass of full-length SHIP is 133 kD) and the larger 145-kD form a posttranslationally modified form. This is consistent with previous 2D-SDS gel studies in which we demonstrated that SHIP is phosphorylated at multiple sites.18 This phosphorylation may also be responsible for our seeing, in some blots, up to 7 SHIP-like bands (ie, doublets at 135, 125, and 110 kD).
Multiple forms of SHIP are present in the murine hematopoietic cell line, DA-ER. (A) A diagrammatic representation of SHIP and an indication of the regions used to generate anti–N-, anti–M-, and anti–C-antisera. (B) An anti-M immunoblot of anti-N (lane 1) and anti-M (lane 2) immunoprecipitates from IL-3–stimulated DA-ER cells. (C) The left panel is an anti-M immunoblot of anti-N or anti-C immunoprecipitates from IL-3–stimulated DA-ER cells and the right panel is a reprobing of the anti-N immunoprecipitated sample with anti-C. (D) An anti-N immunoblot of a total cell lysate from 1 × 107 nucleated mouse bone marrow cells.
Multiple forms of SHIP are present in the murine hematopoietic cell line, DA-ER. (A) A diagrammatic representation of SHIP and an indication of the regions used to generate anti–N-, anti–M-, and anti–C-antisera. (B) An anti-M immunoblot of anti-N (lane 1) and anti-M (lane 2) immunoprecipitates from IL-3–stimulated DA-ER cells. (C) The left panel is an anti-M immunoblot of anti-N or anti-C immunoprecipitates from IL-3–stimulated DA-ER cells and the right panel is a reprobing of the anti-N immunoprecipitated sample with anti-C. (D) An anti-N immunoblot of a total cell lysate from 1 × 107 nucleated mouse bone marrow cells.
Recently, Geier et al12 demonstrated by Western analysis with antibodies generated to amino acids 670-868 of murine SHIP that the most prominent SHIP band in normal human bone marrow is approximately 100 kD. To determine if this represented the 110-kD C-terminal truncated SHIP we observed in our hematopoietic cell lines or the 110-kD SHIP protein lacking an N-terminal SH2 domain reported by Kavanaugh et al,11 we performed Western analysis with normal mouse bone marrow using anti-N. As can be seen in Fig 1D, a prominent 110-kD band was detected, consistent with the normal mouse bone marrow form of SHIP being a C-terminally truncated derivative of the full-length SHIP protein.
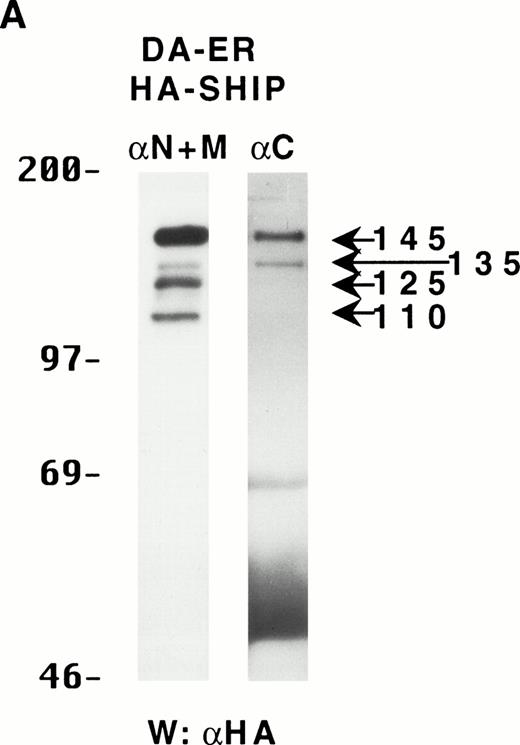
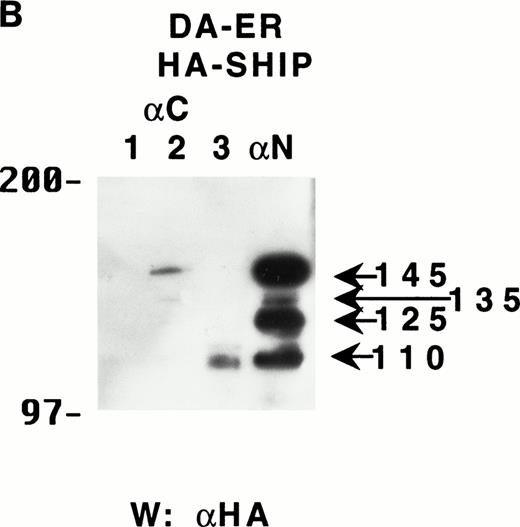
To determine if these proteins arose from distinct mRNAs, alternate splice forms of a single mRNA, or posttranslational C-terminal cleavages of a single protein, the full-length murine SHIP cDNA was tagged with HA at its 5′-terminus and retrovirally infected into DA-ER cells. Immunoprecipitation of these cell lysates with anti-M plus anti-N (or either alone; data not shown) and anti-HA Western analysis showed the presence of the same sized bands as those observed for endogenously expressed SHIP (Fig 2A). Moreover, anti-C immunoprecipitates from the same lysates contained only the 145- and 135-kD bands, as assessed by anti-HA immunoblotting (Fig 2A). Based on the epitopes recognized by the 3 antibodies and the fact that the HA-tag was at the 5′ end of the SHIP cDNA, we concluded from these studies that the different SHIP-like proteins were most likely the result of C-terminal cleavages of the full-length protein and not different mRNAs or alternate splice forms of a single mRNA.
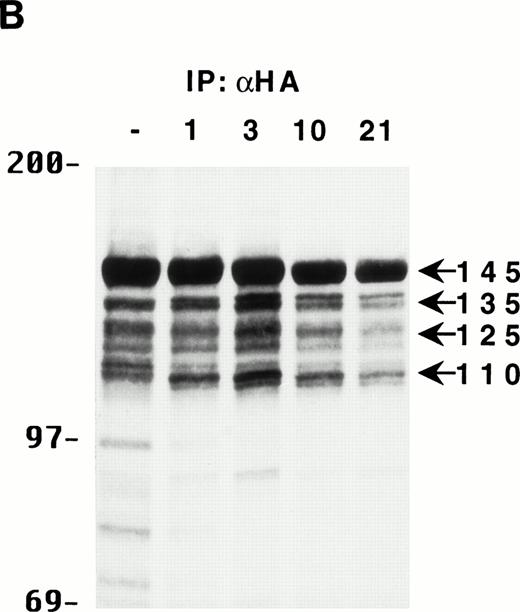
The multiple forms of SHIP can be generated from a single cDNA and are all long lived in vivo. (A) An anti-HA Western blot of anti-N + anti-M (left panel) or anti-C immunoprecipitates (right panel) from IL-3–stimulated DA-ER cells expressing HA-tagged SHIP. (B) An autoradiogram of anti-HA immunoprecipitates from 35S-methionine pulse-labeled HA-SHIP–expressing DA-ER cells taken at 0 (−), 1, 3, 10, and 21 hours after the 30-minute labeling period. Similar results were obtained in 3 separate experiments.
The multiple forms of SHIP can be generated from a single cDNA and are all long lived in vivo. (A) An anti-HA Western blot of anti-N + anti-M (left panel) or anti-C immunoprecipitates (right panel) from IL-3–stimulated DA-ER cells expressing HA-tagged SHIP. (B) An autoradiogram of anti-HA immunoprecipitates from 35S-methionine pulse-labeled HA-SHIP–expressing DA-ER cells taken at 0 (−), 1, 3, 10, and 21 hours after the 30-minute labeling period. Similar results were obtained in 3 separate experiments.
To gain some insight into the generation kinetics of the multiple forms of SHIP, HA-SHIP–expressing DA-ER cells were pulse-labeled for 30 minutes at 37°C with 35S-methionine and lysates were immunoprecipitated with anti-HA at different times. As can be seen in Fig 2B, all forms appear to be generated rapidly and the relative intensities of the bands do not change with time, indicating that they all have similar, long half lives (ie, ∼10 hours). Similar results were obtained when the cells were pulse-labeled for only 15 minutes at 23°C, suggesting that the smaller forms may actually be generated during translation of the full-length protein (data not shown). Identical results were observed with B6SUtA1 cells expressing only endogenous SHIP. To rule out that the truncated proteins were a lysis artifact, HA-SHIP–expressing DA-ER cells were lysed with 0.5% NP-40 in PSB for 5 or 60 minutes in the presence and absence of various inhibitor cocktails or were lysed directly by boiling in SDS-sample buffer. Anti-HA immunoblots showed that there was no significant difference in the relative intensities of the SHIP bands under these conditions (data not shown).
Calpain digests the 145-kD species of SHIP to a stable 110-kD species in vitro.
In an attempt to identify the protease(s) responsible for the cleavage of SHIP, DA-ER cells were incubated with lysosome, proteosome, caspase, and calpain inhibitors (ie, methylamine,21lactacystin,22 BD-FMK,23 and calpeptin24 or calpastatin,25 respectively), starting 15 minutes before pulse-labeling with35S-methionine, and then the cells were lysed and subjected to anti-HA immunoprecipitation and autoradiography. None of the inhibitors, either alone or in combination, prevented the appearance of the smaller forms (data not shown). Nonetheless, because we had previously identified two areas within murine SHIP with unusually high concentrations of prolines, glutamic/aspartic acids, serines, and threonines, ie, PEST sequences26 that might be targets for calpain cleavage,27 we postulated that DA-ER cells might contain a tissue-specific calpain28 that is resistant to calpeptin and performed in vitro digestions with calpain to see if the shorter forms could be produced from the full-length SHIP. Specifically, HA-SHIP–expressing DA-ER cells were lysed and immunoprecipitated with anti-C to selectively isolate the 145/135-kD species and aliquots were incubated with m-calpain for various times. As can be seen in Fig 3A, within 2 minutes of digestion, a prominent 110-kD band appeared that comigrated with the 110-kD band observed in anti-N immunoprecipitates. Although intermediates between 135 and 110 kD (that might correspond to the in vivo 135- and 125-kD SHIP forms) were observed in these digests, they were short lived. Control cultures incubated for 15 minutes in the absence of m-calpain yielded no detectable breakdown products (Fig 3A, lane 2). As shown in Fig 3B, after 4 hours of digestion with calpain, the 110-kD species is the only form present. Interestingly, when DA-ER cell lysates are prepared in the absence of inhibitors (lanes 1 and 3), SHIP is broken down to fragments that migrate with the dye front (lane 1), unless calpain is present (lane 3), suggesting that the 110-kD protein is more resistant to proteases than its full-length counterpart. Although we do not as yet know the cleavage sites within SHIP, it is noteworthy that one of the calpain cleavage sites mapped within the integrin β3 subunit is carboxyl to the Y within the sequence NNPLY,29 a site similar to an ENPLY sequence within SHIP. Cleavage of SHIP at this site would theoretically result in a truncated protein of approximately 110 kD.
The multiple forms of SHIP can be generated in vitro by digestion with calpain. (A) Lysates from IL-3–stimulated DA-ER cells expressing HA-SHIP were immunoprecipitated with anti-N (lane 1) or anti-C (next 3 lanes), and the latter was subjected to digestion with 10 mU of pure rabbit m-calpain for the times indicated. The control sample (lane 2) was incubated for 15 minutes in the absence of m-calpain. The samples were then subjected to anti-HA Western analysis. (B) Lysates, prepared in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of the standard protease inhibitors (see the Materials and Methods) from IL-3–stimulated DA-ER cells expressing HA-SHIP, were immunoprecipitated with anti-C (lanes 1, 2, and 3) or anti-N (lane 4) and the lane 3 sample was digested with 10 mU of pure rabbit m-calpain for 4 hours. The samples were then subjected to anti-HA Western analysis.
The multiple forms of SHIP can be generated in vitro by digestion with calpain. (A) Lysates from IL-3–stimulated DA-ER cells expressing HA-SHIP were immunoprecipitated with anti-N (lane 1) or anti-C (next 3 lanes), and the latter was subjected to digestion with 10 mU of pure rabbit m-calpain for the times indicated. The control sample (lane 2) was incubated for 15 minutes in the absence of m-calpain. The samples were then subjected to anti-HA Western analysis. (B) Lysates, prepared in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of the standard protease inhibitors (see the Materials and Methods) from IL-3–stimulated DA-ER cells expressing HA-SHIP, were immunoprecipitated with anti-C (lanes 1, 2, and 3) or anti-N (lane 4) and the lane 3 sample was digested with 10 mU of pure rabbit m-calpain for 4 hours. The samples were then subjected to anti-HA Western analysis.
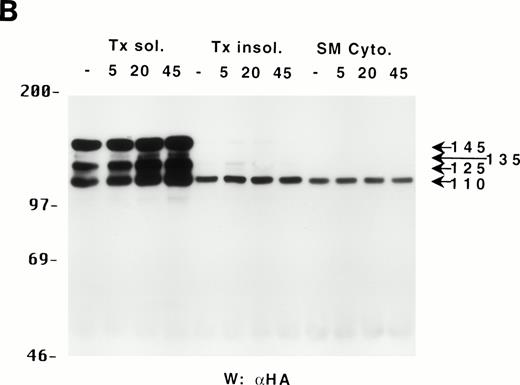
The different forms of SHIP bind to different proteins and only p110 is present in the cytoskeletal fraction of DA-ER cells.
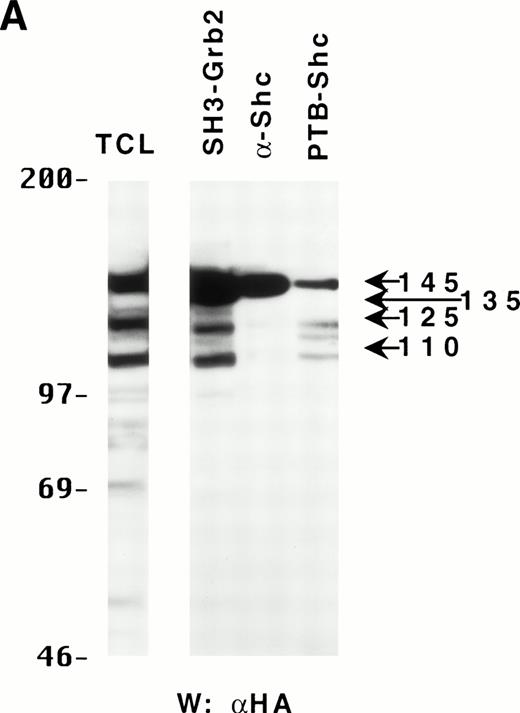
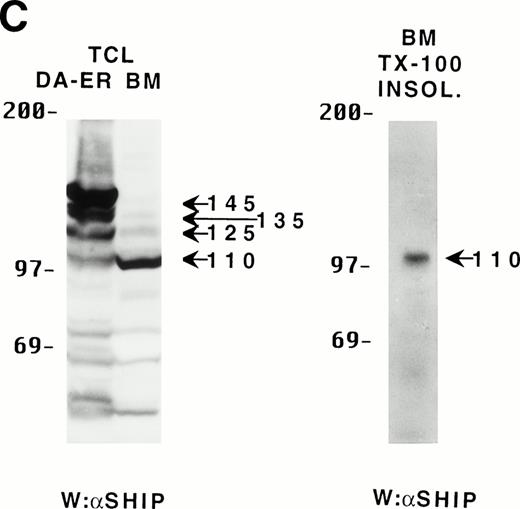
Because the C-terminus of SHIP contains many proline-rich regions, it is conceivable that the various truncated forms of SHIP bind to different SH3-containing proteins30 and thus may be translocated to different intracellular sites. As a preliminary test to see which proteins the various forms of SHIP were capable of binding, we incubated lysates from HA-SHIP–expressing, IL-3–stimulated DA-ER cells with anti-Shc antibodies or with beads containing either the C-terminal SH3 domain of Grb2 or the PTB domain of Shc. As can be seen in Fig 4A, all forms were capable of binding to the C-terminal SH3 domain of Grb2. Moreover, all forms bound to the PTB domain of Shc, indicating they are all tyrosine phosphorylated on one or both of the two NPXY motifs in response to IL-3. However, only the 145-kD form coprecipitated with Shc, raising the possibility that different forms of SHIP may have different partners and therefore distinct intracellular sites. To test this, HA-SHIP–expressing DA-ER cells were stimulated with IL-3 for various times and then subfractionated into TX-100–soluble, TX-100–insoluble, and submembranous cytoskeletal fractions. Anti-N immunoprecipitations were then performed with these fractions and anti-HA Western analysis showed that all forms of SHIP were present in the TX-100–soluble fraction, but only the 110-kD form was detectable in the two cytoskeletal fractions (Fig 4B). Interestingly, there was no apparent redistribution of any of the SHIP forms upon cytokine stimulation. In addition, as expected from our in vitro affinity studies (Fig 4A), IL-3 stimulated the coprecipitation of Shc with SHIP only in the TX-100–soluble fractions (data not shown). We then asked if the prominant 110-kD form of SHIP that is present in normal bone marrow cells also associates with the cytoskeleton. As shown in the left panel of Fig 4C, if total cell lysates from DA-ER and normal mouse bone marrow cells are coelectrophoresed and coblotted with anti-N+M antisera, the 110-kD forms from the two sources comigrate (as do the higher molecular weight species). More importantly, as shown in the right panel of Fig 4C, this bone marrow cell-derived 110-kD SHIP is present in the TX-100–insoluble fraction from these cells. Together, these data suggested the possibility of a distinct function for the 110-kD form of SHIP, and we sought to characterize it further by comparing the PIP3-5 ptase activity of this 110-kD form (isolated from anti-N immunoprecipitates of the submembranous cytosketal fraction of HA-SHIP–expressing DA-ER cells) with an equivalent amount (as determined by anti-HA immunoblots) of full-length SHIP (isolated from anti-Shc immunoprecipitates or from the TX-100–soluble fraction of these cells). Using assay conditions described previously,31 we found no significant difference in the catalytic activity of the 2 species (data not shown). Of note in this regard is that Norris et al32 recently showed that another inositol polyphosphatase, 4-ptase, is a substrate for calpain in platelets and that cleavage markedly reduces its ptase activity.
Binding properties and subcellular location of the various SHIP proteins. (A) Cell lysates from HA-SHIP–expressing DA-ER cells, treated with IL-3 for 5 minutes at 37°C, were either subjected directly to SDS-PAGE (right panel) or were incubated with beads containing the C-terminal SH3 domain of Grb2 or the PTB domain of Shc for 1 hour at 4°C, or immunoprecipitated with anti-Shc antibodies. The beads were then washed 3 times with PSB containing 0.5% NP40 and boiled in SDS-sample buffer and all of the samples were subjected to anti-HA Western analysis. (B) DA-ER cells expressing HA-SHIP were stimulated with IL-3 for the indicated times and fractionated into TX-100–soluble (Tx sol), TX-100–insoluble (Tx insol), and submembranous cytoskeletal (SM Cyto) fractions and subjected to anti-N immunoprecipitation and anti-HA Western analysis. (C) (Left panel) Total cell lysates from DA-ER and murine bone marrow cells were subjected to anti-N+anti-M Western analysis. (Right panel) Murine bone marrow cells (2 × 107 cells) were lysed in 100 μL 2% TX-100 in PSB for 10 minutes at 4°C and centrifuged at 10,000g for 10 minutes. The TX-100–insoluble fraction was extracted from the pellet with 100 μL 1% TX-100, 0.3% deoxycholate, and 1 mol/L NaCl in PSB for 30 minutes at 4°C and, after centrifuging at 10,000g for 10 minutes, it (the supernatant) was subjected to anti-N+anti-M Western analysis.
Binding properties and subcellular location of the various SHIP proteins. (A) Cell lysates from HA-SHIP–expressing DA-ER cells, treated with IL-3 for 5 minutes at 37°C, were either subjected directly to SDS-PAGE (right panel) or were incubated with beads containing the C-terminal SH3 domain of Grb2 or the PTB domain of Shc for 1 hour at 4°C, or immunoprecipitated with anti-Shc antibodies. The beads were then washed 3 times with PSB containing 0.5% NP40 and boiled in SDS-sample buffer and all of the samples were subjected to anti-HA Western analysis. (B) DA-ER cells expressing HA-SHIP were stimulated with IL-3 for the indicated times and fractionated into TX-100–soluble (Tx sol), TX-100–insoluble (Tx insol), and submembranous cytoskeletal (SM Cyto) fractions and subjected to anti-N immunoprecipitation and anti-HA Western analysis. (C) (Left panel) Total cell lysates from DA-ER and murine bone marrow cells were subjected to anti-N+anti-M Western analysis. (Right panel) Murine bone marrow cells (2 × 107 cells) were lysed in 100 μL 2% TX-100 in PSB for 10 minutes at 4°C and centrifuged at 10,000g for 10 minutes. The TX-100–insoluble fraction was extracted from the pellet with 100 μL 1% TX-100, 0.3% deoxycholate, and 1 mol/L NaCl in PSB for 30 minutes at 4°C and, after centrifuging at 10,000g for 10 minutes, it (the supernatant) was subjected to anti-N+anti-M Western analysis.
Interestingly, the levels of the various forms of SHIP have been shown recently to change both during hematopoietic differentiation12 and with leukemogenesis20 and it is tempting to speculate that this may occur in part as a result of changes in the activation state of calpain in these cells, either by changes in the level of calpain itself, as has been reported during megakaryopoiesis,33 or via changes in the levels of intracellular calcium and/or phospholipids, two well-established activators of calpain.27
Very recently, we generated a SHIP knock out mouse by targeting the first exon (the SH2-containing region) of this gene. Hematopoietic cells from these mice lack all 4 SHIP bands, whereas their normal littermates possess all 4. Moreover, bone marrow cells from these knock out mice do not have the prominant 110-kD SHIP band (as assessed by anti-N or anti-M antiserum), whereas normal littermates do,34 further establishing that these proteins are derived from the same gene.
In summary, we demonstrate for the first time that different forms of SHIP can be generated via C-terminal truncations of the full-length protein. However, this does not rule out the possibility that some forms of SHIP are generated by alternate splicing at the mRNA level. We also show that these truncations can be generated in vitro with calpain and that the 110-kD protein (which is the predominant species in total mouse bone marrow) has a distinct intracellular distribution.
ACKNOWLEDGMENT
The authors thank Vivian Lam for excellent technical assistance and Christine Kelly for typing the manuscript.
Supported by the NCI-C and the MRC-C with core support from the B.C. Cancer Foundation and the B.C. Cancer Agency. G.K. is a Terry Fox Cancer Research Scientist of the NCI-C supported by funds from the Canadian Cancer Society.
Address reprint requests to Gerald Krystal, PhD, Terry Fox Laboratory, B.C. Cancer Research Centre, 601 W 10th Ave, Vancouver, British Columbia, Canada V5Z 1L3.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal