Abstract
Current prognostic indicators such as age, sex, and white blood cell count (WBC) fail to identify all children with more aggressive forms of B-precursor acute lymphoblastic leukemia (ALL), and a proportion of patients without poor prognostic indicators still relapse. Results obtained from an analysis of 65 pediatic B-precursor ALL patients indicated that subclone formation leading to clonal diversity, as detected by Ig and T-cell receptor (TCR) gene rearrangements, may represent a very useful prognostic indicator, independent of age, sex, and WBC. Disease-free survival was significantly shorter in those patients showing clonal diversity at presentation. Furthermore, clonal diversity was detected not only in the majority of high-risk patients who relapsed but was also associated with a high probability of relapse in standard-risk patients. Sixty-five percent (13/20) of standard-risk patients who also showed clonal diversity subsequently relapsed, whereas the percentage of relapses among standard-risk patients without clonal diversity was much lower at 19% (7/36). Continued clonal evolution during disease progression is an important feature of aggressive B-precursor ALL. All 5 patients with clonal diversity who were followed up in our study showed a change in the pattern of clonality between presentation and relapse. This implies an important role for clonal diversity as a mechanism of disease progression through the process of clonal variation and clonal selection.
© 1998 by The American Society of Hematology.
SIGNIFICANT IMPROVEMENTS have been made over the past 2 decades in the treatment of childhood acute lymphoblastic leukemia (ALL).1 In particular the most common subtype, B-precursor ALL, generally has a good prognosis, with a survival rate of approximately 70%.1-3 An important contribution to this improvement has been the recognition of a subgroup of patients, at high risk of relapse, who require more aggressive treatment. These high-risk patients may be identified on the basis of classical clinical parameters such as age, sex, white blood count, immunophenotype, cytogenetics, and DNA index.3 4
For the remaining 30% of patients, improvement in survival rates will require progress in two areas. First, further stratification of standard-risk patients, a proportion of whom show unpredictable relapse on current treatment protocols despite the absence of conventional poor prognostic indicators (approximately 32% in our series), and second, the development of more effective treatment strategies based on a better understanding of the biological/genetic factors influencing disease progression.
Lymphoid malignancies offer a unique opportunity for the investigation of disease evolution and progression at the genetic level as a consequence of the occurrence of recombination within the immune system gene loci. During normal B- and T-cell development, recombination in Ig and T-cell receptor (TCR) genes gives rise to unique combinations of variable (V), diversity (D), and joining (J) gene segments. The junctional regions of each of the gene segments are different in each lymphocyte or lymphocyte clone, and hence may be regarded as tumor-specific markers.5-8
It is becoming increasingly clear, however, that leukemogenesis is a dynamic process, with continued evolution of Ig and TCR rearrangements.9,10 This is also likely to reflect more general genetic change giving rise to the outgrowth of more aggressive, treatment-resistant tumor subclones. Several studies have shown, either by Southern blotting or polymerase chain reaction (PCR) amplification, multiple rearrangements of Ig and/or TCR genes at presentation in up to 40% of patients with B-precursor ALL.11-17 There is some evidence to suggest that the resulting clonal diversity may be associated with a poor prognosis in children with B-precursor ALL.11,14 16 In this study, we investigate whether clonal diversity can be used to identify patients with poor prognosis, particularly among those who are classified as standard risk using current prognostic indicators.
MATERIALS AND METHODS
Patient material.
Bone marrow samples were collected from 65 patients who presented with B-lineage ALL at Birmingham Children's Hospital between 1989 and 1996. Patients were selected only on the basis of availability of material for study. A total of 207 patients were treated in this period according to UK Medical Research Council UKALL Trial protocols (UKALL X, XI) or on the MRC Infant Leukaemia Trial. The 5-year disease-free survival (DFS) of 142 cases that were not included in the study, due to unavailibility of material, was 65% (95% confidence interval = 55.2 − 74.8).
For the purposes of the analyses performed in this study, patients were classified as being at high risk of treatment failure on the basis of the presence of one or more of the following parameters: age less than 1 year, Ph chromosome, translocation t(4;11), or a hazard score of greater than or equal to 0.8 (calculated retrospectively). The Oxford hazard score,4 as implemented in the MRC UKALL 1997 Trial, relates risk of treatment failure for children over 1 year to age, gender, and leucocyte count as follows: Hazard Score = 0.22 × loge (WBC + 1.0) + 0.0043 × Age2 − 0.39 × Sex (male = 1; female = 2). The boundary value of 0.8 defines a group of patients with 5-year DFS of less than 40%.
Bone marrow mononuclear cells were isolated by centrifugation on Ficoll density gradients, and DNA extracted by standard methods.18 Chromosome analyses of bone marrow samples were performed using standard G-banding techniques.
Southern blot analysis.
The following probes were used for Southern blot hybridization: a 2.7-kb BglII/Pst I fragment specific for the JH region (M13CTGR51A), a 0.8-kbHindIII/EcoRI fragment for the Cκ region (pRH10), an 8-kb EcoRI fragment for the Cλ region, a 1.5-kb Sac I fragment for the Jδ region (JdS16), a 5-kb EcoRI fragment for Jα region (JαRR), a 0.7-kb HindIII/EcoRI fragment for the Jγ region (M13H60), and a 0.7kb HindIII/EcoRI fragment for the Cβ region, all kindly provided by Dr T.H. Rabbitts (MRC Laboratory of Molecular Biology, Cambridge, UK).
IgH gene amplification.
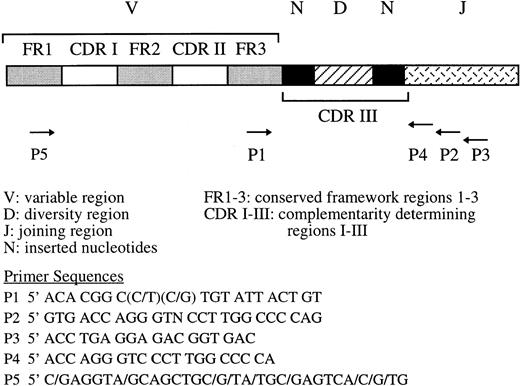
PCR amplification of the third complementarity determining region (CDR-III) was performed using either primers P1 and P2 for direct amplification or primers P1/P3 followed by P1/P419 for seminested PCR as shown in Fig 1. The whole of the VNDNJ region was amplified using seminested PCR and primers P5/P3 for the first round followed by the primers P5/P2 for the second round of amplification (Fig 1).
Position and sequence of primers for single round (P1/P2) and seminested (P1/P3, P1/P4, or P5/P3, P5/P2) PCR amplification of the third complementarity region (CDR-III) of the IgH gene.
Position and sequence of primers for single round (P1/P2) and seminested (P1/P3, P1/P4, or P5/P3, P5/P2) PCR amplification of the third complementarity region (CDR-III) of the IgH gene.
TCRδ and γ amplification.
The Vδ2-Dδ3 rearrangement was amplified using primers 5′GAG TCA TGT CAG CCA TG AG (forward), and 5′AGG GAA ATG GCA CTT TTG CC (reverse). The primers used for TCRγ amplification were 5′CTG TGA CAA CAA GTG TTG TTC CAC and 5′GTG CTT CTA GCT TTC CTG TCT C,20 with internal primers for nested PCR: 5′GAG TAC GCT GCC TAC AGA GAG G and 5′CCA CTG CCA AAG AGT TTC TT. PCR amplifications were performed using 100 ng of genomic DNA. The amplified products were resolved on 8% acrylamide or 3.5% Metaphor (FMC) agarose gels, and bands of interest were excised and eluted in water for sequencing.
CDR-III sequencing.
Gel-purified PCR products were either sequenced directly using the automated chain termination sequencing system (Applied Biosystems Inc, Warrington, UK) or following subcloning into pUC18. Sequenced rearrangements were assigned to their component variable (V), diversity (D), and joining (J) region sequences by comparison with published sequences.20-27
Criteria for detection of clonal diversity.
The criteria for the identification of clonal diversity of leukemic blast cells by either Southern blotting or PCR amplification of immune system gene rearrangements were: (1) the presence of an excess of bands on Southerns or number of PCR products in relation to the number of alleles, ie, more than 2 bands on Southerns or more than 2 PCR products when only 2 copies of the relevant chromosome were present, and (2) the existence of 2 bands of unequal density on Southern blots.
Statistical methods.
DFS was defined as the time between diagnosis and the first relapse, or if no relapse occurred, between diagnosis and the censor date of August 31, 1997. For patients who did not enter remission, the DFS was defined as zero; patients who died without relapse were censored at the date of death. Differences in the prognostic cofactors (white cell count, age, sex, high-/standard-risk category) between the oligoclonal and normal group were analyzed using the Chi-squared and Fisher's exact test and Mann-Whitney U-test. Differences in DFS between oligoclonal and nonoligoclonal patients were evaluated using Kaplan-Meier survival curves and the log-rank test. The effects of prognostic cofactors were investigated using stepwise Cox's proportional hazard regression.
RESULTS
Detection of clonal diversity.
To ensure optimal detection of clonal diversity we analyzed immune system gene rearrangements by both Southern blotting, using seven immune system gene probes (see Materials and Methods) and JH, TCR Jγ, and TCR Jδ PCR assays whenever the amount of material allowed such extensive study. Complete analysis was performed in 33 patients with B-precursor ALL, whereas in a further 32 patients, for whom only small samples were available, the PCR method was used. The results obtained indicated that while clonal diversity was detected in the majority of cases by both methods, a number of cases was detected only by Southern blotting or only by PCR. Six of 33 fully analyzed patients (18%), for example, were detected only by Southern blotting; the negative PCR result in these cases was probably a consequence of loss of primer binding site(s) or incomplete DJH rearrangement. PCR methods did, however, show greater sensitivity in the detection of minor subclones, which may account for the observation of clonal diversity in 3 patients using IgH PCR amplification but not with Southern blotting. The most informative assays, in terms of subclone detection, were Southern blotting with IgJH, IgCκ, and TCR Jγ probes and PCR amplification across the CDR-III region of the JH locus. Using a combination of these assays we identified oligoclonality in 40% (26/65) of patients. However, as the Southern blotting showed a higher frequency of subclone detection than PCR analysis, we cannot rule out the possibility that we may have underestimated the frequency of clonal diversity in the 32 patients analyzed by PCR only.
Clonal diversity and DFS in pediatric B-precursor ALL.
In all patients the presence of clonal diversity was investigated at diagnosis and, in some cases, at intervals during the course of their treatment. The results obtained from a total of 65 patients (Table 1), (29 of whom have been reported previously16) indicated that approximately 40% of patients (26/65) showed clonal diversity at the presentation of their disease.
Clinical and Laboratory Characteristics of Pediatric ALL Patients
| Patient No. . | Sex . | Age (yr) . | Diagnosis . | WBC ×109/L . | Hazard Score* . | Oligoclonal . | Time to Relapse (mo) . | Site of Relapse-164 . |
|---|---|---|---|---|---|---|---|---|
| 1 | M | 2.2 | cALL | 5.2 | 0.0 | Yes | ||
| 2 | F | 1.9 | cALL | 32.4 | 0.0 | Yes | 19 | CNS |
| 3 | M | 2.4 | cALL | 7.1 | 0.1 | Yes | ||
| 4 | F | 0.9 | pre B | 144.0 | NA-152 | Yes | ||
| 5 | F | 4.4 | cALL | 25.8 | 0.0 | Yes | 0# | |
| 6 | M | 1.1 | cALL | 24.7 | 0.3 | Yes | 41 | BM |
| 7 | F | 4.1 | pre B | 94.0 | 0.3 | Yes | ||
| 8 | M | 3.4 | cALL | 116.0 | 0.7 | Yes | 18 | BM |
| 9 | M | 3.3 | cALL | 138.0 | 0.7 | Yes | 19 | CNS, BM |
| 10 | M | 4.8 | cALL | 51.0 | 0.6 | Yes | 25 | testes |
| 11 | F | 9.4 | ALL-151 | 7.1 | 0.1 | Yes | 17 | intrathoracic |
| 12 | M | 7.8 | cALL | 204.0 | 1.0 | Yes | 23 | BM |
| 13 | F | 6.2 | cALL | 3.2 | −0.3 | No | ||
| 14 | M | 1.5 | cALL | 7.5 | 0.1 | No | ||
| 15 | M | 4.6 | cALL | 40.0 | 0.5 | No | ||
| 16 | F | 7.2 | cALL | 5.2 | −0.2 | No | ||
| 17 | M | 3.6 | cALL | 44.0 | 0.5 | No | 29 | BM, CNS |
| 18 | F | 8.6 | cALL | 7.7 | 0.0 | No | ||
| 19 | M | 7.4 | pre B | 9.6 | 0.4 | No | ||
| 20 | M | 11.8 | ALL-151 | 1.6 | 0.4 | No | ||
| 21 | M | 2.9 | cALL | 6.3 | 0.1 | No | 29 | BM, testes |
| 22 | M | 2.3 | cALL | 3.1 | −0.1 | No | ||
| 23 | M | 3.3 | cALL | 31.9 | 0.4 | No | ||
| 24 | M | 3.4 | cALL | 80.5 | 0.6 | Yes | 29 | testes |
| 25 | M | 4.1 | cALL | 35.8 | 0.5 | No | ||
| 26 | M | 3.8 | cALL | 15.8 | 0.3 | No | 0# | |
| 27 | M | 2.2 | cALL | 8.6 | 0.1 | No | ||
| 28 | M | 3.2 | cALL | 11.4 | 0.2 | No | ||
| 29 | F | 4.5 | cALL | 5.5 | −0.3 | No | ||
| 30 | M | 10.9 | cALL | 19.7 | 0.7 | No | ||
| 31 | M | 9.2 | cALL | 55.1 | 0.9 | No | ||
| 32 | M | 6.1 | cALL | 5.6 | 0.2 | No | 25 | BM, CNS |
| 33 | M | 9.1 | cALL | 3.3 | 0.3 | No amp-160 | ||
| 34 | F | 0.4 | null-155 | 1200.0 | NA-152 | Yes | 0¶ | |
| 35 | M | 2.6 | pre B | 41.2 | 0.5 | No | ||
| 36 | F | 5.6 | cALL | 7.5 | −0.2 | No amp-160 | ||
| 37 | F | 3.5 | cALL | 4.3 | −0.4 | No | ||
| 38 | M | 5.5 | pre B | 19.0 | 0.4 | No | 27 | BM |
| 39 | M | 13.1 | cALL | 1.6 | 0.6 | No | 28 | BM, testes |
| 40 | F | 2.6 | cALL | 24.2 | 0.0 | No | ||
| 41 | M | 1.2 | cALL | 3.2 | −0.1 | No | ||
| 42 | M | 13.3 | cALL | 3.2 | 0.7 | No | ||
| 43 | F | 2.2 | cALL | 84.1 | 0.2 | No | ||
| 44 | M | 4.1 | cALL | 6.7 | 0.1 | No | 39 | BM |
| 45 | M | 2.0 | cALL | 34.3 | 0.4 | Yes | ||
| 46 | M | 9.8 | cALL | 243.0 | 1.2 | No amp-160 | 41 | testes |
| 47 | M | 1.8 | cALL | 50.9 | 0.5 | Yes | 31 | BM, CNS |
| 48 | F | 2.0 | cALL | 7.4 | −0.3 | Yes | 54 | BM |
| 49 | M | 5.9 | pre B | 65.0 | 0.7 | Yes | ||
| 50 | M | 4.6 | cALL | 22.2 | 0.4 | Yes | 23 | testes, BM |
| 51 | M | 2.2 | pre B | 24.0 | 0.3 | No | ||
| 52 | M | 3.2 | cALL | 7.1 | 0.1 | No | 19 | BM |
| 53 | F | 13.3 | pre B | 5.6 | 0.4 | No | ||
| 54 | F | 3.1 | cALL | 67.8 | 0.2 | Yes | 32 | BM, CNS |
| 55 | M | 0.4 | null-155 | 295.0 | NA-152 | No | 26 | BM |
| 56 | F | 3.3 | cALL | 43.0 | 0.1 | No | ||
| 57 | M | 2.6 | cALL | 17.7 | 0.3 | No | ||
| 58 | M | 3.5 | pre B | 23.3 | 0.4 | Yes | ||
| 59 | M | 9.7 | cALL | 110.0 | 1.0 | Yes | ||
| 60 | F | 3.7 | cALL | 8.0 | −0.2 | No | ||
| 61 | M | 1.9 | cALL | 2.5 | −0.1 | No | ||
| 62 | M | 9.6 | cALL | 60.7 | 0.9 | Yes | 28 | CNS |
| 63 | M | 4.1 | cALL-153 | 58.9 | 0.6 | Yes | 35 | testes |
| 64 | M | 4.7 | cALL | 15.1 | 0.3 | Yes | 12 | CNS |
| 65 | F | 7.9 | cALL | 5.1 | −0.1 | Yes | 61 | BM |
| Patient No. . | Sex . | Age (yr) . | Diagnosis . | WBC ×109/L . | Hazard Score* . | Oligoclonal . | Time to Relapse (mo) . | Site of Relapse-164 . |
|---|---|---|---|---|---|---|---|---|
| 1 | M | 2.2 | cALL | 5.2 | 0.0 | Yes | ||
| 2 | F | 1.9 | cALL | 32.4 | 0.0 | Yes | 19 | CNS |
| 3 | M | 2.4 | cALL | 7.1 | 0.1 | Yes | ||
| 4 | F | 0.9 | pre B | 144.0 | NA-152 | Yes | ||
| 5 | F | 4.4 | cALL | 25.8 | 0.0 | Yes | 0# | |
| 6 | M | 1.1 | cALL | 24.7 | 0.3 | Yes | 41 | BM |
| 7 | F | 4.1 | pre B | 94.0 | 0.3 | Yes | ||
| 8 | M | 3.4 | cALL | 116.0 | 0.7 | Yes | 18 | BM |
| 9 | M | 3.3 | cALL | 138.0 | 0.7 | Yes | 19 | CNS, BM |
| 10 | M | 4.8 | cALL | 51.0 | 0.6 | Yes | 25 | testes |
| 11 | F | 9.4 | ALL-151 | 7.1 | 0.1 | Yes | 17 | intrathoracic |
| 12 | M | 7.8 | cALL | 204.0 | 1.0 | Yes | 23 | BM |
| 13 | F | 6.2 | cALL | 3.2 | −0.3 | No | ||
| 14 | M | 1.5 | cALL | 7.5 | 0.1 | No | ||
| 15 | M | 4.6 | cALL | 40.0 | 0.5 | No | ||
| 16 | F | 7.2 | cALL | 5.2 | −0.2 | No | ||
| 17 | M | 3.6 | cALL | 44.0 | 0.5 | No | 29 | BM, CNS |
| 18 | F | 8.6 | cALL | 7.7 | 0.0 | No | ||
| 19 | M | 7.4 | pre B | 9.6 | 0.4 | No | ||
| 20 | M | 11.8 | ALL-151 | 1.6 | 0.4 | No | ||
| 21 | M | 2.9 | cALL | 6.3 | 0.1 | No | 29 | BM, testes |
| 22 | M | 2.3 | cALL | 3.1 | −0.1 | No | ||
| 23 | M | 3.3 | cALL | 31.9 | 0.4 | No | ||
| 24 | M | 3.4 | cALL | 80.5 | 0.6 | Yes | 29 | testes |
| 25 | M | 4.1 | cALL | 35.8 | 0.5 | No | ||
| 26 | M | 3.8 | cALL | 15.8 | 0.3 | No | 0# | |
| 27 | M | 2.2 | cALL | 8.6 | 0.1 | No | ||
| 28 | M | 3.2 | cALL | 11.4 | 0.2 | No | ||
| 29 | F | 4.5 | cALL | 5.5 | −0.3 | No | ||
| 30 | M | 10.9 | cALL | 19.7 | 0.7 | No | ||
| 31 | M | 9.2 | cALL | 55.1 | 0.9 | No | ||
| 32 | M | 6.1 | cALL | 5.6 | 0.2 | No | 25 | BM, CNS |
| 33 | M | 9.1 | cALL | 3.3 | 0.3 | No amp-160 | ||
| 34 | F | 0.4 | null-155 | 1200.0 | NA-152 | Yes | 0¶ | |
| 35 | M | 2.6 | pre B | 41.2 | 0.5 | No | ||
| 36 | F | 5.6 | cALL | 7.5 | −0.2 | No amp-160 | ||
| 37 | F | 3.5 | cALL | 4.3 | −0.4 | No | ||
| 38 | M | 5.5 | pre B | 19.0 | 0.4 | No | 27 | BM |
| 39 | M | 13.1 | cALL | 1.6 | 0.6 | No | 28 | BM, testes |
| 40 | F | 2.6 | cALL | 24.2 | 0.0 | No | ||
| 41 | M | 1.2 | cALL | 3.2 | −0.1 | No | ||
| 42 | M | 13.3 | cALL | 3.2 | 0.7 | No | ||
| 43 | F | 2.2 | cALL | 84.1 | 0.2 | No | ||
| 44 | M | 4.1 | cALL | 6.7 | 0.1 | No | 39 | BM |
| 45 | M | 2.0 | cALL | 34.3 | 0.4 | Yes | ||
| 46 | M | 9.8 | cALL | 243.0 | 1.2 | No amp-160 | 41 | testes |
| 47 | M | 1.8 | cALL | 50.9 | 0.5 | Yes | 31 | BM, CNS |
| 48 | F | 2.0 | cALL | 7.4 | −0.3 | Yes | 54 | BM |
| 49 | M | 5.9 | pre B | 65.0 | 0.7 | Yes | ||
| 50 | M | 4.6 | cALL | 22.2 | 0.4 | Yes | 23 | testes, BM |
| 51 | M | 2.2 | pre B | 24.0 | 0.3 | No | ||
| 52 | M | 3.2 | cALL | 7.1 | 0.1 | No | 19 | BM |
| 53 | F | 13.3 | pre B | 5.6 | 0.4 | No | ||
| 54 | F | 3.1 | cALL | 67.8 | 0.2 | Yes | 32 | BM, CNS |
| 55 | M | 0.4 | null-155 | 295.0 | NA-152 | No | 26 | BM |
| 56 | F | 3.3 | cALL | 43.0 | 0.1 | No | ||
| 57 | M | 2.6 | cALL | 17.7 | 0.3 | No | ||
| 58 | M | 3.5 | pre B | 23.3 | 0.4 | Yes | ||
| 59 | M | 9.7 | cALL | 110.0 | 1.0 | Yes | ||
| 60 | F | 3.7 | cALL | 8.0 | −0.2 | No | ||
| 61 | M | 1.9 | cALL | 2.5 | −0.1 | No | ||
| 62 | M | 9.6 | cALL | 60.7 | 0.9 | Yes | 28 | CNS |
| 63 | M | 4.1 | cALL-153 | 58.9 | 0.6 | Yes | 35 | testes |
| 64 | M | 4.7 | cALL | 15.1 | 0.3 | Yes | 12 | CNS |
| 65 | F | 7.9 | cALL | 5.1 | −0.1 | Yes | 61 | BM |
*Oxford Hazard Score as defined in Materials and Methods; ≥0.8 = patient at high risk of treatment failure.
Acute leukemia with lymphoid and myeloid markers.
NA: hazard score not applicable if age <1 yr.
Ph+.
t(4;11).
¶Patient died of disease without achieving remission.
#Patient died of infection without achieving remission.
DNA amplified with control but not with JHprimers.
CNS, central nervous system; BM, bone marrow.
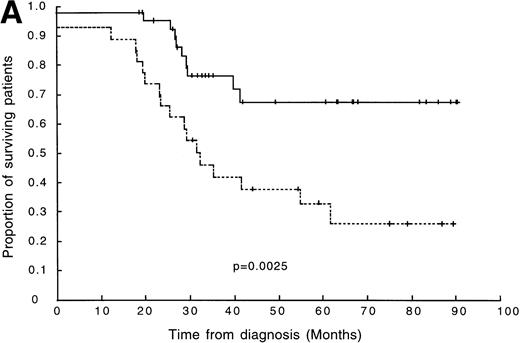
Eighteen out of 26 (69%) oligoclonal patients relapsed or died without entering remission compared with only 10 of the remaining 39 patients (26%) without clonal diversity. Patients were followed up for between 18 and 90 months from diagnosis, (median follow-up was 59 months; 80.4% of surviving patients had ≥30 months follow-up). The association between clonal diversity at presentation and shorter DFS was shown to be significant by a log-rank test (P = .0025; Fig 2A).
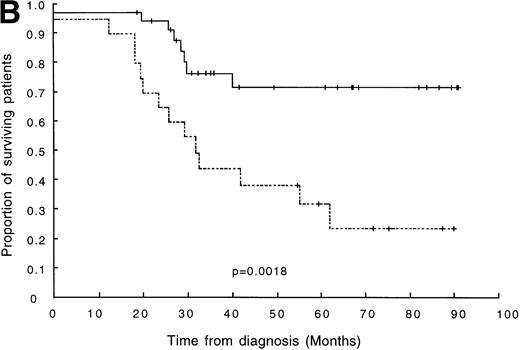
(A) DFS in 65 children with ALL. The broken line represents survival in patients with clonal diversity (n = 26), and the solid line represents survival in patients without clonal diversity (n = 39). (B) DFS in 56 patients with standard-risk ALL. The broken line represents survival in standard-risk patients with clonal diversity (n = 20), and the solid line represents survival in standard-risk patients without clonal diversity (n = 36).
(A) DFS in 65 children with ALL. The broken line represents survival in patients with clonal diversity (n = 26), and the solid line represents survival in patients without clonal diversity (n = 39). (B) DFS in 56 patients with standard-risk ALL. The broken line represents survival in standard-risk patients with clonal diversity (n = 20), and the solid line represents survival in standard-risk patients without clonal diversity (n = 36).
Prognostic significance of clonal diversity in patients with standard-risk B-precursor ALL.
In view of these results it was important to determine whether prognostic information obtained from the investigation of clonal diversity could be used to improve the stratification of patients as compared with currently used indicators.
Initially we compared the relationship between DFS and five prognostic indicators: clonal diversity, age, sex, white blood cell count, and high-risk status (as defined in Materials and Methods) in a stepwise Cox's regression analysis. Both white blood cell count and clonal diversity were independently predictive of outcome (clonal diversity,P = .013; white blood cell count, P = .005), whereas the effects of age, sex, and high-risk status were not significant. The relative risk of treatment failure in patients showing clonal diversity, independent of white blood cell count, was 2.73 (95% confidence interval = 1.24 − 6.00).
We next looked in more detail at patients classified as standard risk (ie, age >1 year, absence of t(4;11) and t(9;22), hazard score < 0.8). Again, DFS was shown to be significantly shorter in patients with clonal diversity (P = .002; 5-year DFS = 32% and 72% for patients with and without clonal diversity, respectively; Fig2B). In our series of 56 patients with standard-risk ALL, 65% (13/20) of oligoclonal patients subsequently relapsed without achieving remission. This was in contrast to a relapse rate of only 19% (7/36) observed among the patients with a standard-risk and monoclonal disease.
A Cox's regression analysis of patients with standard risk indicated that clonal diversity was more predictive of outcome than any other parameter (clonal diversity P = .002;white blood cell countP = .028; sex P = .560; age P = .921). In the presence of clonal diversity no other factor was significantly prognostic. The relative risk of treatment failure in clonal diversity versus monoclonal patients was 3.66.
In summary, an important feature of these results was the fact that the improved prognostic information contributed by the assessment of clonal diversity appeared to relate largely to a group of patients currently classified as standard risk. Consequently, the use of this indicator may make a significant contribution to the identification of patients within this group who have a greater risk of treatment failure.
Clonal diversity and disease progression.
Having shown that clonal diversity was closely associated with aggressive disease, we next looked in more detail at the pattern of clonal evolution between presentation and relapse. We sought to investigate whether clonal diversity is always generated from (1) the major clone, (2) could be generated from a tumor cell at an earlier stage of development, and (3) the extent to which clonal diversity contributed to clonal selection and emergence of the final highly resistant clone.
The pattern of clonality was followed by IgH CDR-III PCR through various stages of disease progression in six oligoclonal patients, five with oligoclonality at presentation and one with oligoclonality at the end of treatment. In all analyzed patients the number and/or size of IgH CDR-III fragments differed between presentation and relapse, although in four out of six cases at least one fragment of identical size was observed (Table2).
Clonal Evolution Between Presentation and Relapse in cALL Patients
| Patient No. . | Time of Relapse* (mo) . | Site of Relapse† . | No. of IgH PCR Products . | IgH Fragment Size (bp) . | ||
|---|---|---|---|---|---|---|
| Presentation . | Relapse . | Presentation . | Relapse . | |||
| 47 | 33 | BM, CNS | Multiple | 2 | 80 | |
| 90 | ||||||
| 48 | 54 | BM | 4 | 2 | 75 | 100 |
| 100 | 110 | |||||
| 120 | ||||||
| 130 | ||||||
| 50 | 28 | BM | 3‡ | 1 | 75 | 65 |
| 85 | ||||||
| 105 | ||||||
| 64 | 11 | CNS | 3 | 3 | 60 | 80 |
| 70 | 100 | |||||
| 110 | 110 | |||||
| 65 | 61 | BM | 11-153 | 2 | 110 | 110 |
| 125 | ||||||
| 44 | 39 | BM | 11-155 | 2 | 100 | 100 |
| 115 | ||||||
| Patient No. . | Time of Relapse* (mo) . | Site of Relapse† . | No. of IgH PCR Products . | IgH Fragment Size (bp) . | ||
|---|---|---|---|---|---|---|
| Presentation . | Relapse . | Presentation . | Relapse . | |||
| 47 | 33 | BM, CNS | Multiple | 2 | 80 | |
| 90 | ||||||
| 48 | 54 | BM | 4 | 2 | 75 | 100 |
| 100 | 110 | |||||
| 120 | ||||||
| 130 | ||||||
| 50 | 28 | BM | 3‡ | 1 | 75 | 65 |
| 85 | ||||||
| 105 | ||||||
| 64 | 11 | CNS | 3 | 3 | 60 | 80 |
| 70 | 100 | |||||
| 110 | 110 | |||||
| 65 | 61 | BM | 11-153 | 2 | 110 | 110 |
| 125 | ||||||
| 44 | 39 | BM | 11-155 | 2 | 100 | 100 |
| 115 | ||||||
*Time interval between presentation and relapse.
BM, bone marrow; CNS, central nervous system.
Oligoclonal at presentation and at end of treatment.
Oligoclonal at presentation by Southern blotting.
Oligoclonal at end of treatment.
To determine the relationship between individual subclones, JH rearrangements from various stages of disease were sequenced in three patients and the sequences aligned to V, D, and J regions. Sequencing of the major clones at different stages of the disease in patient 48 revealed that all detectable subclones appeared to be related, since they all shared common D (DLR4) and J (J6) sequences (Table 3). However, the greater preservation of the D sequences in one of the relapse clones suggested that this was not derived directly from a presentation clone but rather was present at an earlier stage of leukemogenesis. Interestingly, this patient was initially classified as a standard-risk ALL, with no other recognizable features of aggressive disease. The pattern of clonality started to change 2 weeks after presentation, during induction treatment (Fig 3), and the patient eventually relapsed 54 months later, showing an entirely different pattern of bands to that observed previously (Fig 3).
Clonal Evolution During Disease Progression
| Patient . | Stage . | FR3A Sequence . | N Sequence . | D Sequence . | N Sequence . | J Sequence . |
|---|---|---|---|---|---|---|
| 48 | Presentation | GCGAGG | tagttaggatgg | TGTAGGAGTA (DLR4) | ttgtgtgtagaa | GGTA (J6) |
| GCGA | aagatccccatgtggggt | GTAGTAACCAGCTG (DLR4) | ttatagtata | TACTAC (J6) | ||
| Week 2 | GGAG | — | TAGTAG (DLR4) | gttgt | TACT (J6) | |
| GGAG | taggttag | GTAG (DLR4) | ttgggatcg | TACT (J6) | ||
| Relapse | GCGAG | gccccccg | AGTAGTA (DLR4) | taaggggaaccgc | CTACTACTACTA (J6) | |
| GCGAGA | gatcgagagagattgcc | TATTGTAGTAGTACCAGCTGCTAT (DLR4) | agtg | TACTACT (J6) | ||
| 65 | Presentation | ACCTGT | acctgcccaacctttctccc | CTATGGTTCGGGGAGTTAT (DXP′1) | gcacgttg | ATACT (J1) |
| Relapse | ACCTGT | acctgcccaacctttctccc | CTATGGTTCGGGGAGTTAT (DXP′1) | gcacgttg | ATACT (J1) | |
| GCAAGA | gacgaaa | TATGATAGTAGTGG (D21/9) | ggcctattggg | CTACT (J6) | ||
| 44 | Presentation | GCGAGA | ggg | AGGACTGGAACTA (DLR1) | cccccggga | TTCGACC (J5) |
| Relapse | GCGAGA | ggg | AGGACTGGAACTA (DLR1) | cccccggga | TTCGACC (J5) | |
| ACCAAGA | tcgagaaggagaagacccca | TATAGCAGCTCGT (DN4/M4) | – | ACTACTTTGACTA (J4) |
| Patient . | Stage . | FR3A Sequence . | N Sequence . | D Sequence . | N Sequence . | J Sequence . |
|---|---|---|---|---|---|---|
| 48 | Presentation | GCGAGG | tagttaggatgg | TGTAGGAGTA (DLR4) | ttgtgtgtagaa | GGTA (J6) |
| GCGA | aagatccccatgtggggt | GTAGTAACCAGCTG (DLR4) | ttatagtata | TACTAC (J6) | ||
| Week 2 | GGAG | — | TAGTAG (DLR4) | gttgt | TACT (J6) | |
| GGAG | taggttag | GTAG (DLR4) | ttgggatcg | TACT (J6) | ||
| Relapse | GCGAG | gccccccg | AGTAGTA (DLR4) | taaggggaaccgc | CTACTACTACTA (J6) | |
| GCGAGA | gatcgagagagattgcc | TATTGTAGTAGTACCAGCTGCTAT (DLR4) | agtg | TACTACT (J6) | ||
| 65 | Presentation | ACCTGT | acctgcccaacctttctccc | CTATGGTTCGGGGAGTTAT (DXP′1) | gcacgttg | ATACT (J1) |
| Relapse | ACCTGT | acctgcccaacctttctccc | CTATGGTTCGGGGAGTTAT (DXP′1) | gcacgttg | ATACT (J1) | |
| GCAAGA | gacgaaa | TATGATAGTAGTGG (D21/9) | ggcctattggg | CTACT (J6) | ||
| 44 | Presentation | GCGAGA | ggg | AGGACTGGAACTA (DLR1) | cccccggga | TTCGACC (J5) |
| Relapse | GCGAGA | ggg | AGGACTGGAACTA (DLR1) | cccccggga | TTCGACC (J5) | |
| ACCAAGA | tcgagaaggagaagacccca | TATAGCAGCTCGT (DN4/M4) | – | ACTACTTTGACTA (J4) |
Patient 65 had 1 PCR-amplified clone at presentation and 2 at relapse; patient 48 had 4 clones at presentation and 2 at relapse; only the major clones were sequenced. Patient 44 had 1 PCR-amplified clone at presentation and 2 at relapse.
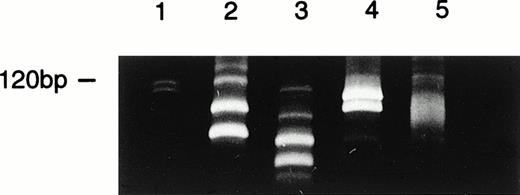
Clonal evolution identified by FRA3/JH PCR at various stages of disease in patient 48. Lane 1, DNA size marker; lane 2, four clones identified at presentation; lane 3, two major clones together with 2 minor subclones identified at week 2; lane 4, two clones identified at relapse; lane 5, a smear of PCR products in control sample from normal bone marrow.
Clonal evolution identified by FRA3/JH PCR at various stages of disease in patient 48. Lane 1, DNA size marker; lane 2, four clones identified at presentation; lane 3, two major clones together with 2 minor subclones identified at week 2; lane 4, two clones identified at relapse; lane 5, a smear of PCR products in control sample from normal bone marrow.
In the second patient, patient 65, the clone from presentation was accompanied at the time of relapse, 61 months later, by a new, independent subclone with the involvement of different D and J regions (Table 3). The presence of two independent subclones indicated the derivation of clonal diversity from an immature, nonrearranged tumor progenitor. Bearing in mind the time of the relapse in both patients, 54 and 61 months, respectively, the possibility of a secondary leukemia was considered. However, secondary leukemia was unlikely in either of these patients because in patient 48 all identified subclones from presentation to relapse were related, and in patient 65 the presentation clone was still detectable at the time of relapse.
In a further two patients, 44 and 50, an increase in the number of amplified JH rearrangements was observed at the end of treatment. Interestingly, patient 44 presented with a single leukemic clone, and clonal diversity was identified only later at the end of treatment. Subsequently, both children relapsed within 1 year. Sequencing of the terminal clones in patient 44 revealed coexistence of the initial clone with a new independent subclone (Table 3).
DISCUSSION
Current approaches to the treatment of pediatric ALL involve the stratification of patients into different risk groups so that treatment regimens may be tailored to match the specific needs of each group. Those patients at highest risk of relapse, for example, will require more aggressive treatment to improve their chances of long-term survival. Equally important, however, is the need to avoid over-treating those children in the better prognostic groups in order to minimize long-term side-effects.28-30 Discrimination between the different risk groups is not straightforward, and several different systems have been proposed.4,31 32 None of these appear to be entirely satisfactory, however, since a proportion of children currently classified as low or standard risk still relapse.
In a follow-up to previous reports11,14,16 that suggested that the detection of clonal diversity in pediatric B-precursor ALL patients might provide a new indicator of poor prognosis, we have sought to investigate this association further and to develop a straightforward strategy that would allow the routine assessment of clonal diversity in large numbers of patient samples. Results obtained indicate that combination of a PCR-based approach, involving the amplification of the CDR-III region of IgH (using consensus FR1, FR3, and JH primers), and Southern blotting using IgH, Igκ, and TCRγ allowed a high level of detection of clonal diversity comparable to previously published reports.11 13
A study of 65 pediatric B-precursor ALL patients confirmed that clonal diversity was significantly associated with a poor outcome. A reduction in long-term survival (>5 years) of almost 50% was observed as compared with patients who did not show clonal diversity (Fig 2A). Statistical analyses confirmed that clonal diversity was independant of, and more predictive of poor outcome, than any other prognostic indicator tested.
It was apparent from this study however, that the prognostic information obtained differed in some ways from that provided by other prognostic indicators. Of particular interest was the observation that a proportion of patients regarded as standard risk showed clonal diversity and also had significantly shorter DFS; 5-year survival for these patients was only 32% as compared with 72% in standard-risk patients without clonal diversity.
It is interesting to speculate whether clonal diversity represented only a marker of aggressive disease or whether there was a biological role for subclone formation during tumor progression. The observations from this study, particularly with respect to the change in the pattern of clonality between presentation and relapse support the hypothesis that clonal diversity, indeed, represents a mechanism of disease progression. It is possible that this mechanism operates through the process of clonal variation and clonal selection providing a highly proliferative, resistant malignant clone in the terminal stage of the disease.33 The circumstances that promote both continued or de novo immunoglobulin gene rearrangements in clonally unstable cells are not clear. The process of VHDJHrecombination is regulated at several levels and, therefore, many factors may be involved. At one level, the appearance in a normal cell of a productive rearrangement leads to the subsequent inhibition of recombinase activity. Continued, multiple VHDJHgene rearrangements, which are nonfunctional, are common in ALL tumor cells,10 suggesting that inhibition of the recombinase activity might not take place in leukemic blasts. At another level, it is possible that factors affecting accessibility of chromosomal regions containing the signal sequences34 contribute to the continuously active recombinations during leukemic proliferation. The same mechanism could cause both subclone formation and a wider genomic instability, involving genome wide recognition signal–like sequences, with consequent involvement of other genes important for tumor proliferation.35 There is a need, therefore, for the identification of factors promoting both clonal diversity and the aggressive phenotype that could be potentially used in an appropriate assay in clinical studies. In this respect, the expression of recombinase (RAG) proteins that are essential for B-cell differentiation might be one of the factors affected in patients with clonal diversity.
In two patients, 44 and 65, in whom subclones were sequenced, the source of clonal diversity might have been an early B nonrearranged progenitor cell. One can speculate that differences in the time before relapse (39 and 61 months) was related to the requirement for different additional molecular change and further clonal selection to produce a more aggressive, treatment-resistant subclone. However, an alternative explanation for the presence of the further, independent VHDJH rearrangement at the time of relapse in these patients is that a second, previously nonrearranged or DJ-rearranged IgH allele underwent a complete VHDJH rearrangement in the original leukemic clone. Such a possibility cannot be excluded, particularly in the light of the fact that both the original and the new independent VHDJH rearrangement were present at the time of relapse in both patients. In contrast to patients 44 and 65, all the identified subclones in patient 48 were related and showed utilization of the same DH and JH region. The CDR-III sequences of the subclones showed substantial variability at the VHJH junctions suggesting that V-V replacement was an unlikely mechanism for the generation of clonal diversity in this patient. The variability of the DJH joints was also remarkable in patient 48 and, therefore, de novo V rearrangements to a preexisting DJH rearrangement, as an alternative mechanism, seems doubtful. A further possibility is that all these subclones were independently derived from a nonrearranged tumor progenitor cell in an environment that directed rearrangement to the same VH, D and J segments as has been previously postulated for another patient.36
The present observations are restricted to a small number of patients that were available for the study. To clarify the origin of clonal diversity and its biological significance it would be informative, in a further study, to extend sequence analysis of the identified VHDJH rearrangements to all patients with clonal diversity irrespective of relapse. It would be important to determine whether the relationship between subclones and the mechanism of their generation, in patients who do not relapse, differs from that observed in patients who do relapse. Such a distinction would provide further stratification of patients with standard risk and improve the power of clonal diversity as a predictor of relapse. The conclusions from this study, therefore, have potential clinical implications, particularly with respect to the future treatment of standard-risk ALL patients. Clearly, it is neccessary to perform both larger scale retrospective and prospective studies to confirm our findings and to provide a firm basis on which to make any subsequent changes in the clinical management of pediatric B-precursor ALL.
ACKNOWLEDGMENT
We thank the Department of Haematology, Birmingham Children's Hospital and the Regional Cytogenetics Unit, Birmingham Heartlands Hospital for immunophenotype and cytogenetics data. Patient data were compiled from the West Midlands Regional Children's Tumour Research Group Registry. We also thank Dr C.G. Steward for helpful comments on the manuscript, and S. Rees and K. Willis for technical assistance.
Supported in part by The British Council and the Cancer Research Campaign. E.G. was a Price Hall Fellow (Former United Birmingham Hospitals Endowment Fund).
Address reprint requests to Tatjana Stankovic, MD, PhD, CRC Institute for Cancer Studies, University of Birmingham, Birmingham, UK B15 2TT.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal