Abstract
Although the majority of patients with acute promyelocytic leukemia (APL) are potentially cured by treatments combining all-trans retinoic acid (ATRA) and chemotherapy (CHT), a sizable proportion (around 30%) will relapse during follow-up. Retrospective molecular monitoring studies using reverse transcriptase-polymerase chain reaction (RT-PCR) for the specific PML/RARα fusion gene, have shown that a positive test usually precedes the occurrence of hematologic relapse. Prospective RT-PCR analyses were performed since 1993 at diagnosis and at preestablished time intervals during follow-up in bone marrow (BM) samples of 163 patients with PML/RARα+ APL enrolled in the multicenter Gruppo Italiano Malattie Ematologiche Maligne dell' Adulto (GIMEMA) trial AIDA (All-trans retinoic acid plus Idarubicin). Treatment consisted of ATRA and idarubicin for induction followed by three polychemotherapy courses as consolidation. The sensitivity level of the RT-PCR assay for PML/RARα, as assessed by serial dilution experiments, was 10−4. All patients were in hematologic remission and tested PCR− at the end of consolidation. Of 21 who converted to PCR-positive thereafter, 20 underwent hematologic relapse at a median time of 3 months (range, 1 to 14) from the first PCR+ result. Seventeen of these 21 (81%) PCR+ conversions were recorded within the first 6 months postconsolidation. Of 142 who tested persistently PCR− in ≥2 tests after consolidation, 8 had hematologic relapse and 134 remained in complete remission (CR) after a median follow-up of 18 months (range, 6 to 38) postconsolidation. Using a time-dependent Cox model, the relative risk of hematologic relapse of patients who converted to PCR+ was 31.8 (confidence limits 95%, 12.9 to 78.3). Our results indicate that conversion to PCR positivity for PML/RARα during remission is highly predictive of subsequent hematologic relapse and highlight the prognostic value of stringent molecular monitoring during the early postconsolidation phase in APL. As a result of the present study, salvage treatment in patients enrolled in the GIMEMA trial AIDA is now anticipated at the time of molecular relapse, defined as the conversion to PCR positivity in two successive BM samplings during follow-up.
© 1998 by The American Society of Hematology.
THE MAJORITY (up to 70%) of patients with acute promyelocytic leukemia (APL) are currently induced into long-term remission and potential cure with modern treatment approaches combining all-trans retinoic acid (ATRA) and chemotherapy. However, despite this progress, relapse occurs in 20% to 30% of patients receiving such therapy and still represents a major obstacle to final cure.1-8
Although patients with relapsed APL have a good chance of achieving second remission, disease recurrence is associated with higher frequency of refractory leukemia and with shorter survival.1,2 Novel treatment approaches, including AM80 (a synthetic retinoid) or arsenic trioxide seem effective in APL relapse,9 10 but their use is still investigational.
The identification during hematologic remission of patients at highest risk of relapse is relevant to adjust treatment choices and may allow us to anticipate salvage therapy. We and others have shown that reverse transcriptase-polymerase chain reaction (RT-PCR) analysis of the PML/RARα fusion gene derived from the t(15;17) may enable the detection of residual leukemic cells during remission in APL patients.11-21 Using PCR assays with a sensitivity threshold ranging between 10−3 and 10−4, several retrospective studies reported that patients who remain (or convert to) PCR+ after consolidation are very likely to relapse within a few months, whereas patients in long-term remission and potentially cured show no PCR-detectable residual disease.11-21
Following these observations, the Italian Gruppo Italiano Malattie Ematologiche Maligne dell' Adulto (GIMEMA) and Associazione Italiana Ematologia e Oncologia Pediatrica (AIEOP) groups initiated in 1993 a trial for newly diagnosed APL (“AIDA” [All-trans retinoic acid plus Idarubicin] 0493), which included serial prospective PCR evaluations. The recently reported interim results indicate that 60% and 96% of patients convert to PCR− after induction and consolidation, respectively.5 However, because ≈20% of patients achieving molecular remission relapsed during follow-up, it appears that PCR tests performed at the end of consolidation fail to identify all patients at risk.5
We report here the results of a prospective PCR monitoring study in 163 PML/RARα+ patients enrolled in the AIDA trial. Our findings, which indicate that conversion to PCR positivity after consolidation is almost uniformly followed by hematologic relapse, prompted us to anticipate salvage therapy at the time of minimal disease recurrence.
MATERIALS AND METHODS
Patients and sample collection.
As of July 1997, 519 patients with a genetically confirmed diagnosis of APL were eligible to enter the Italian GIMEMA “AIDA” trial. Of these, 479 (92%) were characterized by RT-PCR as PML/RARα+. After the end of consolidation treatment, 312 of 324 (96%) tested PCR− and 12 (4%) PCR+. These latter 12 cases received allogeneic transplant from HLA-identical or unrelated donor, or alternative treatments at the physician's discretion and are not included in this monitoring analysis. Of the 312 patients who tested PCR−, 163 with ≥6 months postconsolidation follow-up, and in which at least two bone marrow (BM) samples were collected at the scheduled time intervals and sent to referral laboratories, are included in the present study.
All investigators of participating institutions (see Appendix) were requested to prepare locally Ficoll-Hypaque (Nycomed Pharma AS, Oslo, Norway) isolated mononuclear cells collected from BM aspirates, wash cells twice in sterile phosphate-buffered saline (PBS), and then store samples at −20°C in a 4-mol/L guanidium thiocyanate (GTC) solution. According to the study design, BM aspirates for molecular monitoring analyses were called at diagnosis, after induction, at the end of consolidation, every 3 months during the first and second year, and then every 6 months during the third and fourth year after consolidation. GTC aliquots were provided to peripheral institutions by the two central laboratories. Specific recommendations were made for the use of RNAase-free disposable materials during cell manipulation. Cryopreserved GTC samples were sent in dry ice to two referral molecular biology laboratories (Hematology, University “La Sapienza” of Rome and Clinica Pediatrica, University of Milano-Monza) where RT-PCR analyses were performed.
RT-PCR of PML/RARα.
Total RNA was extracted by the method of Chomczynsky and Sacchi.22 To assess the integrity of RNA, samples were run after extraction on a formaldehyde minigel. Cases showing partial or total RNA degradation were not processed further and a new sample was requested in such cases to the peripheral Center. RT-PCR amplification of the PML/RARα hybrid gene was performed as reported elsewhere,12 13 with two modifications: the annealing temperature was raised to 56°C and, starting from April 1996, the Amplitaq Gold enzyme (Roche Molecular Systems Inc, Branchburg, NJ) was used as Taq polymerase in both laboratories. In addition to the amplification of the hybrid gene in patient RNA, each RT-PCR experiment included the coamplification of: (1) RNA from the NB4 cell line as positive control; (2) all reagents plus water and no RNA as negative control; and (3) a cDNA fragment containing RARα exons 2 and 3 obtained from the same patient under analysis (internal control) to further verify RNA integrity and to assess the efficiency of the RT step.
The sensitivity of the RT-PCR assay was determined by amplifying serially diluted RNA mixtures of a diagnostic sample with 100% of blasts and the t(15;17)-negative myeloid cell line GF-D8, as reported.12 The PML/RARα transcript was still detectable in the presence of 0.1 ng total RNA, that is a final dilution of 10−4. Such detection level was repeatedly obtained in several new experiments performed using Amplitaq Gold as DNA polymerase. The use of the latter apparently increased either the specificity or the sensitivity of the assay. However, in only two of nine experiments performed using Amplitaq Gold, were we able to visualize on the ethidium bromide gel a 10−5dilution, whereas a 10−4 dilution was constantly detected.
To verify the reproducibility of results between the two reference laboratories, 66 RNA samples including diagnostic and remission specimens, were divided in two aliquots and blindly analyzed in parallel. The results showed 100% concordance as regarding both the presence or absence of the fusion mRNA and the type of transcript in positive cases.
Treatment plan.
The AIDA protocol consists of an induction phase combining oral ATRA given daily until complete remission (CR) and four doses of intravenous idarubicin, followed by three polychemotherapy consolidation courses. At the end of consolidation, PCR− patients were randomized into four arms including chemotherapy alone with 6-mercaptopurine and methotrexate (arm 1), ATRA alone (arm 2), alternating chemotherapy and ATRA (arm 3), or observation (arm 4). Patients who tested PCR+ after consolidation underwent, if eligible, allogeneic BM transplantation in first CR or received alternative therapy at the physician's discretion. Detailed drug doses and schedule of the protocol have been reported elsewhere.5 23
Criteria for hematologic and molecular response.
Hematologic remission was defined as a normal BM cellularity with less than 5% leukemic promyelocytes and normalization of peripheral blood counts. PCR negativity was defined as the absence, on ethidium bromide-stained electrophoresis gel, of the specific PML/RARα amplification band detected at diagnosis, in the presence of RNA integrity as evaluated by minigel visualization, and successful amplification of the internal control. PCR positivity in follow-up studies was defined as the reappearance on ethidium bromide-stained gel of the same amplification band detected at diagnosis. A second BM sample was requested in all cases who converted to PCR+ and the assay repeated to avoid false positivity due to contamination or amplification of nonspecific PCR products. Hematologic relapse was defined as the reappearance of ≥5% leukemic promyelocytes in the BM.
Statistical analysis.
Relapse probability was calculated from the end of consolidation to the time of hematologic relapse or last follow-up, according to the Kaplan-Meier method.24 Patients PCR− in the BM at the time of isolated extrahematologic relapse were considered as censored in the relapse curves. The relative risk and 95% confidence limits were estimated using a time-dependent proportional hazard Cox model.25
RESULTS
A total of 163 patients who tested PCR− at the end of consolidation are included in this molecular monitoring analysis. A minimum potential follow-up of 6 months after consolidation and at least two scheduled and evaluable PCR tests were considered for patient inclusion in the present evaluation. The remaining 149 of the 312 patients who tested PCR− after consolidation were excluded due to protocol violation, because it was too early for evaluation, or, as a main cause (115 cases), because less than two scheduled and evaluable BM samples were received by referral laboratories.
A mean of 5 (range, 2 to 11) BM samples per patient collected during postconsolidation follow-up were analyzed. In the vast majority of cases, specimens were processed and the results obtained within 3 weeks from receipt. The appropriate oligonucleotide set, using the PML external primer (M4, located on PML exon 3) for patients initially characterized as having the short or bcr3 isoform or, alternatively, the PML internal primer (M2, for patients initially characterized as having the long type or bcr1-2 isoform) was adapted to each case during monitoring analyses to visualize a single PML/RARα amplification band and to avoid the resolution of multiple bands due to PML alternative splicing. Morphologic examinations of BM smears and PCR tests were always done independently avoiding any exchange of information between cytology reviewers and molecular biologists to minimize interpretation bias. Clinical updates, including written reports of marrow appearance and PCR data, were timely and independently sent to the GIMEMA data center by local clinical institutions and by referral molecular biology laboratories.
Twenty-one of the 163 patients tested PCR+ at least once during follow-up. BM morphologic reports documented hematologic remission in all 21 cases at the time of conversion to PCR+. Of these 21, 20 (95%) underwent subsequent hematologic relapse within a median time of 3 months (range, 1 to 14) from the time of first conversion to PCR+. In 8 of these 20 patients, at least one further BM sample was analyzed and found positive before overt relapse. Seventeen of the 21 (81%) PCR+ conversions were recorded within the first 6 months, ie, on the first or second scheduled sampling collected postconsolidation. Detailed characteristics of these 20 patients who converted to PCR+ and underwent hematologic relapse are reported in Table 1. One patient showed a transient PCR-positivity 16 months after the end of consolidation, with a subsequent analysis performed 3 weeks later showing again PCR-negativity. This patient remained in remission after 2 months.
Characteristics of 20 Patients Who Showed Conversion to PCR+ and Subsequent Hematologic Relapse
| Patient No. . | Age/Sex . | PML Break Type . | Maintenance . | First Molecular CR Duration (mo)-150 . | Time From PCR+ to Hematologic Relapse (mo) . |
|---|---|---|---|---|---|
| 1. | 53/F | L | None | 3 | 3 |
| 2. | 36/F | S | None | 3 | 6 |
| 3. | 38/M | L | CHT-151 | 5 | 3 |
| 4. | 35/F | S | CHT + ATRA | 5 | 2 |
| 5. | 43/M | L | None | 4 | 2 |
| 6. | 54/M | L | CHT + ATRA | 2 | 14 |
| 7. | 44/M | S | CHT + ATRA | 8 | 9 |
| 8. | 43/M | L | ATRA | 5 | 1 |
| 9. | 25/M | S | None | 3 | 1 |
| 10. | 35/M | S | CHT + ATRA | 3 | 4 |
| 11. | 62/M | L | CHT | 10 | 4 |
| 12. | 20/M | L | None | 1 | 4 |
| 13. | 10/M | L | CHT | 9 | 2 |
| 14. | 26/F | L | CHT | 4 | 3 |
| 15. | 46/F | S | ATRA | 9 | 8 |
| 16. | 29/M | L | ATRA | 2 | 3 |
| 17. | 13/M | L | ATRA | 4 | 1 |
| 18. | 13/F | L | None | 5 | 3 |
| 19. | 44/F | S | None | 4 | 1 |
| 20. | 37/M | S | None | 4 | 1 |
| Patient No. . | Age/Sex . | PML Break Type . | Maintenance . | First Molecular CR Duration (mo)-150 . | Time From PCR+ to Hematologic Relapse (mo) . |
|---|---|---|---|---|---|
| 1. | 53/F | L | None | 3 | 3 |
| 2. | 36/F | S | None | 3 | 6 |
| 3. | 38/M | L | CHT-151 | 5 | 3 |
| 4. | 35/F | S | CHT + ATRA | 5 | 2 |
| 5. | 43/M | L | None | 4 | 2 |
| 6. | 54/M | L | CHT + ATRA | 2 | 14 |
| 7. | 44/M | S | CHT + ATRA | 8 | 9 |
| 8. | 43/M | L | ATRA | 5 | 1 |
| 9. | 25/M | S | None | 3 | 1 |
| 10. | 35/M | S | CHT + ATRA | 3 | 4 |
| 11. | 62/M | L | CHT | 10 | 4 |
| 12. | 20/M | L | None | 1 | 4 |
| 13. | 10/M | L | CHT | 9 | 2 |
| 14. | 26/F | L | CHT | 4 | 3 |
| 15. | 46/F | S | ATRA | 9 | 8 |
| 16. | 29/M | L | ATRA | 2 | 3 |
| 17. | 13/M | L | ATRA | 4 | 1 |
| 18. | 13/F | L | None | 5 | 3 |
| 19. | 44/F | S | None | 4 | 1 |
| 20. | 37/M | S | None | 4 | 1 |
Abbreviations: L, long type; S, short type.
Months after consolidation.
CHT, chemotherapy including methotrexate and 6-mercaptopurine.
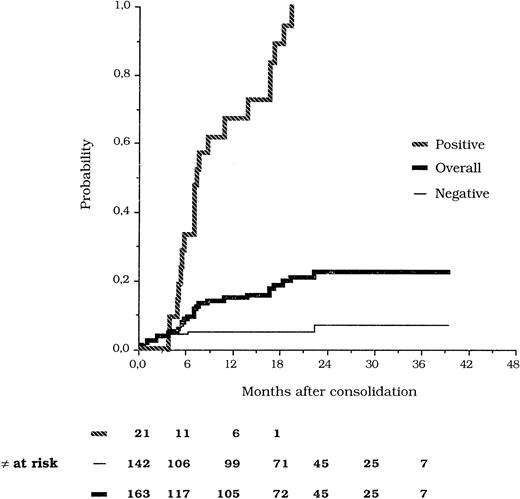
Of the 142 patients who tested persistently PCR− in ≥2 postconsolidation tests, eight (5.6%) underwent hematologic relapse at a median time of 1.5 months (range, 1 to 23). In seven of these eight cases, a BM specimen collected within 3 months before hematologic relapse could be analyzed and tested negative, whereas no sample was sent at such scheduled time in the other patient. The remaining 134 patients in this group are in hematologic remission at a median postconsolidation follow-up of 18 months (range, 6 to 38). Figure 1 shows the probability of hematologic relapse according to the results of molecular monitoring. Two patients who tested PCR− in the BM at the time of isolated extramedullary relapse were censored in the relapse curves.
Kaplan-Meier estimates of the relapse risk according to PCR analysis of PML/RARα.
Kaplan-Meier estimates of the relapse risk according to PCR analysis of PML/RARα.
Considering that conversion to PCR+ is observed at variable times during follow-up, the relapse risk analysis to compare positive and negative patients was performed using a time-dependent Cox model. The relative risk of developing hematologic relapse for patients who converted to PCR+ at any time during follow-up compared with patients who remained persistently PCR− was 31.8 (confidence limits 95%, 12.9 to 78.3).
DISCUSSION
In recent years, a number of studies have highlighted the relevance of PML/RARα detection in the clinical management of APL.1,2,11-21 At diagnosis, such importance relies on the possibility of identifying virtually 100% of patients responsive to a specific therapy including ATRA. After treatment, longitudinal retrospective studies of minimal residual disease (MRD) monitoring have shown that PCR amplification of this hybrid gene is prognostically informative because overt relapse is usually (but not uniformly) preceded by a positive test and, conversely, long-term survivors test PCR−.11-21 These findings further distinguish APL from all other acute leukemia subsets in which associations with a specific genetic lesion are never equally consistent and where MRD studies have yielded more controversial results.26 On the other hand, several investigators have pointed out the technical difficulties of the RT-PCR assay for PML/RARα, which are mainly related to the low amount and instability of the fusion transcript, resulting in a less sensitive test compared with the amplification of other well-characterized leukemia associated fusion genes such as, for example, BCR/ABL and AML1/ETO.27,28 It seems, however, that this relatively poor sensitivity (most studies report detection levels between 10−3 and 10−4) results in a clinically useful threshold, whereby patients who test positive during remission have been reported to be at highest risk of developing hematologic relapse.11-21
The present study was performed prospectively in a large number of patients characterized at diagnosis as having PML/RARα-positive APL and enrolled in a single treatment protocol. According to the design of our trial,5 one of the aims of molecular monitoring was to identify at the end of consolidation patients at risk of relapse to adjust further treatment choices (including allogeneic transplantation in patients still positive at such time). The exceedingly high fraction of PCR− cases observed after consolidation (96%) compared with the hematologic relapse rate at 2 years (≈20%, data not shown), indicate that a sizable proportion of patients at risk are not identified at this time. By contrast, we found that the results of subsequent postconsolidation analyses are extremely relevant in terms of outcome prediction. In fact, 20 of 21 patients who converted to PCR+ during follow-up underwent successively overt relapse, whereas only 8 of 142 who tested ≥2 times negative after consolidation relapsed thereafter. Because they are obtained prospectively, these data provide compelling and definite evidence that PCR positivity during hematologic remission predicts relapse in APL.
With respect to the sensitivity of the assay, we observe that although our method allowed us to reach clinically useful conclusions, a greater degree of standardization is needed to enable better reproducibility in large clinical trials. Future studies should be aimed at determining whether a given amount of the PML/RARα transcript correlates with a clinical remission status and, conversely, which copy number detection would anticipate the occurrence of impending relapse in individual patients. It is expected that newly developed automated methods, such as Taqman quantitative PCR, will be of considerable help for this purpose, ensuring more reliable comparison among different studies.
As to the time elapsed between the first molecular evidence of MRD and hematologic relapse, the majority of patients in the present study had overt disease recurrence within a few months from the first PCR+ test, although a rather heterogeneous behavior was observed considering the whole group (range, 1 to 14 months). It is presumed that intrinsic biologic diversity related to the clonogenic capacity of leukemia cells may account for such variability. Alternatively, this heterogeneous clinical evolution may simply be related to differences in the amount of MRD, which are undetected using a nonquantitative assay. With respect to a potential influence of maintenance treatment, we found no significant correlations between treatment type and time elapse from PCR positivity to disease recurrence (Table 1).
Because this study was performed in the context of a multicenter clinical trial involving more than 70 institutions (listed in the Appendix), some considerations on the logistic and technical difficulties encountered may be of interest. Molecular monitoring data could be obtained in approximately half of the patients with an evaluable follow-up. The main reasons for failure to analyze the remaining patients were related to poor compliance from some local institutions as regarding scheduled samplings or inadequate shipment and late receipt of samples resulting in poor RNA yield after extraction. We remark, however, that the vast majority of PCR+ conversions were recorded in our study within the first 6 months after the end of consolidation. This observation emphasizes the prognostic value of a stringent molecular assessment of MRD performed in the early posttherapy period. Thus, despite the logistic and technical difficulties, we strongly recommend that all efforts are made to warrant this monitoring analysis during the first 6 months after consolidation treatment.
Based on the findings reported here, the GIMEMA and AIEOP groups have recently adopted an amendment in the AIDA protocol. According to this, patients who convert to PCR+ at any time after consolidation are tested again in a new BM sample collected within 2 to 4 weeks after the first one, and therapy of relapse is anticipated after confirmation of PCR positivity in the second BM specimen. Such repeated sampling certainly provides the most adequate experimental condition to rule out false positivity due to PCR contamination.
To the best of our knowledge, this is the first report on acute leukemia showing that MRD results are translated into operationally active medical decisions. Whether the anticipation of treatment at the time of MRD recurrence will improve the outcome of APL relapsed patients remains to be established. However, we presume that at least some significant advantages will be obtained, such as the disappearance or minimization of early deaths due to hemorrhage or ATRA syndrome, and the possibility of outpatient-based treatment. Together with the important recent advances in the front-line therapy of APL, early identification and treatment of relapse might represent a further step towards the final cure of all patients with this disease.
ACKNOWLEDGMENT
We are grateful to Dr M. L. Vegna for statistical analysis.
APPENDIX
The following clinical departments participated in the AIDA 0493 trial: Ematologia, Università “La Sapienza”, Roma, F. Mandelli, G. Avvisati, D. Diverio, A.M. Testi, F. Lo Coco, M.C. Petti, M.L. Vegna; Divisione di Ematologia, Ospedale S. Martino, Genova, E. Damasio, R Cerri; Istituto di Ematologia L. e A.Seragnoli, Università, Bologna, S. Tura, G. Visani, G. Martinelli; Divisione di Ematologia, Ospedale S. Bortolo, Vicenza, F. Rodeghiero, E. Di Bona; Divisione di Ematologia, Policlinico S. Matteo, Pavia. C. Bernasconi, M. Lazzarino; Divisione di Medicina E, Opedale S. Giovanni, Torino, L. Resegotti, M. Falda; Divisione di Ematologia, Policlinico Careggi, Firenze, P. Rossi Ferrini, F. Leoni; Divisione di Ematologia, Ospedali Riuniti, Bergamo, T. Barbui, A. Rambaldi; Divisione di Ematologia, Ospedale Civile, Pescara, G. Fioritoni, A. Recchia; Servizio di Ematologia, Policlinico, Bari, V. Liso, G. Specchia; Divisione di Ematologia, Ospedale A. Businco, Cagliari, G. Broccia, W. Deplano; Servizio di Ematologia, Ospedale Civile, Avellino, E. Volpe, N. Cantore; Divisione di Ematologia, Ospedale A. Pugliese, Catanzaro, A. Peta, F. Iuliano; Divisione di Ematologia, Ospedale S. Gerado, Monza, E. Pogliani, G. Corneo; Ematologia, Ospedale Generale e Regionale, Bolzano, P. Coser, P. Fabris; Sezione di Ematologia Spedali Civili, Brescia, T. Izzi, G. Rossi; Cattedra di Ematologia, Università, Catania, E. Cacciola, F. Di Raimondo; Cattedra di Ematologia, Università, Parma, V. Rizzoli, C. Almici; Cattedra di Ematologia, Università, Verona, G. Perona, D. Veneri; Cattedra di Ematologia, Università, Genova, M. Gobbi, M. Clavio; Divisione di Ematologia, Ospedale Cardarelli, Napoli, R. Cimino, F. Ferrara; Divisione di Ematologia, Osp. Nuovo Pellegrini, Napoli, R. De Biasi, E. Miraglia; Divisione di Ematologia, T.E.R.E., Napoli, L. De Rosa, V. Mettivier; Cattedra di Ematologia, Università Tor Vergata, Roma, S. Amadori, G. Aronica; Clinica Pediatrica, Ospedale S. Gerardo, Monza, G. Masera, A. Biondi, A. Luciano; Divisione di Ematologia, UniversitàCattolica, Roma, G. Leone, S. Sica; Divisione di Ematolgia, Ospedali Riuniti, Reggio Calabria, F. Nobile, B. Martino; Sezione di Ematolgia, Ospedale S. Croce, Cuneo, E. Gallo, A. Gallamini; Divisione di Ematologia, Ospedale S. Maria Goretti, Latina, L. Deriu, A. Cherichini; Sezione di Ematologia, CTMO, Cremona, A. Porcellini, S. Morandi; Divisione di Ematologia, Nuovo Policlinico, Napoli, B. Rotoli, C. Selleri; Cattedra di Ematologia, Università, Perugia, M.F. Martelli, A. Tabilio; Clinica Medica, Università, Palermo, G. Mariani, M. Musso; Divisione di Ematologia, Ospedale V. Cervello, Palermo, F. Caronia, S. Mirto, A. Santoro; Divisione di Ematologia, Ospedale B. Gesù, Roma, G. De Rossi, M. Caniggia; Istituto di Ematologia, Nuovo Ospedale Torrette, Ancona, P. Leoni, M. Montillo; Centro di Riferimento Oncologico, Aviano, S. Monfardini, V. Zagonel; Patologia Medica, Università, Genova, R. Ghio, E. Balleari; Clinica Medica, Policlinico S. Matteo, Pavia, E. Ascari, R. Invernizzi; Divisione di Ematologia, Università, Pisa, B. Grassi, M. Petrini; Ematologia, Ospedale S.S. Annunziata, Taranto, P. Mazza, G. Lazzari; Cattedra di Ematologia, Università, Udine, M. Baccarani, A. Candoni; Ematologia Pediatrica, Università, Catania, G. Schilirò, A.M. Ippolito; Ematologia, IV Divisione Pediatrica, Genova, L. Massimo, C. Micalizzi; Cinica Pediatrica, Università, Pavia, F. Severi, F. Locatelli; Ematologia, Ospedale Regionale A. Di Summa, Brindisi, G. Quarta, A. Melpignano; Cattedra di Ematologia, Università, Ferrara, G. Castoldi, F. Lanza; Semeiotica Medica, Università, Genova, F. Patrone, M. Sessarego; Divisione di Ematologia, Ospedale Niguarda, Milano, E. Morra, A.M. Nosari; Ematologia, Ospedale S. Raffaele, Milano, C. Bordignon, L. Camba; Ematologia ed Autotrapianto Ospedale S. Martino, Genova, A.M. Carella, F. Frassoni; Sezione di Ematologia, Ospedale S. Francesco, Nuoro, A. Gabbas, G. Latte; Cattedra di Ematologia, Policlinico, Palermo, P. Citarella, S. Grisanti; Divisione di Ematologia, Ospedale S. Salvatore, Pesaro, G. Lucarelli, G. Sparaventi; Sezione di Ematologia, Ospedale S. Carlo, Potenza, F. Ricciuti, M. Pizzuti; Divisione di Ematologia, Ospedale S. Camillo, Roma, A. De Laurenzi, L. Pacilli; Div. di Ematologia, Casa Sollievo della Sofferenza, S.G. Rotondo, M. Carotenuto, L. Melillo; Divisione di Ematologia, Ospedale A. Sclavo, Siena, E. Dispensa, A. Bucalossi; Clinica Pediatrica, Ospedale G. Salesi, Ancona, P. Giorgi, L. Felici; Clinica Pediatrica I, Policlinico, Bari, F. Schettini, N. Santoro; Onco-Ematologia Pediatrica, Ospedale Regionale, Cagliari, P. Biddau; II Divisione Pediatrica, Ospedale Pausilipon, Napoli, V. Poggi, M.F. Pintà; Clinica Pediatrica I, Università, Napoli, M.T. Di Tullio, M. Giuliano; Clinica Pediatrica II, Università, Padova, L. Zanesco, M. Pilon; Clinica Pediatrica III, Università, Pisa, P. Macchia, C. Favre; Clinica Pediatrica, Università, Torino, E. Madon, R. Miniero; Department of Hematology, University Nijmegen (NL), T.de Witte, P. Muus; Medizinische Klinik III, University Munich (D), U. Jehn; Department of Hematology, University Leiden (NL), R. Willemze; Department of Hematology, University Ankara (TK), M. Beksac; Az Middelheim, Afdeling Hemato-Oncologie, Antwerpen (B), R. De Bock.
Supported by ROMAIL (Associazione Italiana contro le Leucemie, Sezione di Roma), CNR Project “Biotecnologie” and Fondazione Tettamanti, Monza.
Presented in part at the 39th Annual Meeting of the American Society of Hematology, held in San Diego, CA, December 5-9, 1997.
Address reprint requests to Francesco Lo Coco, MD, Dipartimento di Biotecnologie Cellulari ed Ematologia, Università“La Sapienza”, Via Benevento 6, 00161 Rome, Italy; e-mail:lococo@bce.med.uniroma1.it.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal