Abstract
In bone marrow transplantation, the detection of chimerism is an important adjunct to the repertoire of tests available for determining acceptance of the graft. In solid organ transplantation, there is currently intense interest in the role that chimerism plays in both short- and long-term host reactivity to the graft. Allogeneic blood transfusion has been associated with a subtle immunosuppressive effect in renal transplantation and chimerism is implicated as a possible mechanism for this effect. To assess the survival of allogeneic cells after blood transfusion or transplantation, we have developed a technique based on molecular typing for HLA class II alleles, which enables the detection of donor-derived cells in patients receiving blood transfusions. While developing this technology, we investigated why we and others observe false amplification. Sequencing of false products has shown that they arise from amplification of both pseudogenes and non-pseudogenes present in the DNA under test. Elucidation of this phenomenon allows the amplification of these false products to be predicted in any given combination and hence avoided by the judicious selection of primers. Validation has been achieved by following donor alleles after transfusion of blood containing defined numbers of leukocytes expressing selected mismatched antigens.
THE DEVELOPMENT OF chimerism after allogeneic bone marrow transplantation is paramount to its success. However, the connection between the development of chimerism after solid organ transplantation and graft outcome is tenuous. In solid organ transplantation, microchimerism is commonly used to define the presence of donor cells/DNA outside of the graft environment. It has been hypothesized that spontaneous hematopoietic microchimerism may be essential for the development and maintenance of immunologic unresponsiveness to organ allografts.1,2 Hematopoietic microchimerism has been demonstrated after transplantation of livers,3-5 kidneys,6,7 hearts,8 and lungs9,10 using methods based on polymerase chain reaction (PCR) amplification of the Y-chromosome3 and the HLA-DR region of the major histocompatibility complex (MHC).6-8 10 An accurate assessment of the level of donor-derived cells surviving within recipients may lead to an improved understanding of the relative importance of microchimerism after either blood transfusion or solid organ transplantation.
The transfusion of allogeneic blood is in many ways analogous to a bone marrow transplant without the accompanying immunosuppression and for this reason is often termed mononuclear cell transplantation.11 It has been associated with an improvement in subsequent renal allograft survival,12increased susceptibility to viral or bacterial infection,13,14 and to the possible increased risk of cancer recurrence.15,16 Experimental evidence suggests that the transfusion effect depends on recipient exposure to viable allogeneic donor cells.17,18 Although several studies have documented the survival of donor cells within recipients after blood transfusion in animal models,19-21 there has been limited investigation into the development of such microchimerism after allogeneic blood transfusion in humans. Human studies of donor leukocyte survival after HLA-unrelated transfusions have shown little evidence for long-term microchimerism, showing the presence of donor cells only in the first few days after transfusion.22-26These assays were restricted, as they were based on the detection of male donor cells in female recipients. In addition, the techniques may not have been sensitive enough to detect the low levels of donor leukocytes that may be present after allogeneic transfusions.
There are many factors, both immunologic and nonimmunologic, that may influence the survival of donor cells after allogeneic blood transfusion including the length and method of storage of the blood, HLA matching, and natural killer cell activity. To be able to assess the relative influence of such factors, a reproducible and accurate assay system for the identification of donor cells in the recipient is required. We have now developed a technique based on published methods,27-29 which enables the detection of donor-derived cells in patients receiving blood transfusions. While investigating the commonly used techniques for the detection of microchimerism after solid organ transplantation, we frequently observed the presence of what are euphemistically termed “false positives”. In this report, we describe the conditions under which these false positives occur and how to compensate for their presence.
MATERIALS AND METHODS
DNA Isolation
Genomic DNA was isolated from leukocytes obtained from anticoagulated blood using the salting out procedure,30 precipitated with ethanol, and resuspended in sterile water at a concentration of 200 ng/μL.
HLA DR Typing
PCR-SSP typing (PCR amplification with sequence-specific primers).
Allele specific primers (0.5 μmol/L), designed on the basis of published sequences,29,31 were used in multiple amplification reactions (Table 1) consisting of 200 ng genomic DNA, 67 mmol/L Tris pH 8.8, 16.6 mmol/L NH4SO4 (ammonium sulphate), 200 μmol/L of each deoxyadenosine triphosphate (dATP), deoxycytidine triphosphate (dCTP), deoxyguanosine triphosphate (dGTP), deoxythymidine triphosphate (dTTP), 2.0 mmol/L MgCl2(magnesium chloride), and 0.25 U BioTaq polymerase (Bioline, London, UK). PCR amplifications were performed in a PTC200-96v thermal cycler (Genetic Research Instrumentation, Essex, UK) according to the method of Bunce et al.29 All products were analyzed by agarose gel electrophoresis.
Primer Mix Composition for PCR-SSP Typing
| PM . | Primers . | Size (bp) . | Locus . | HLA Alleles Amplified . |
|---|---|---|---|---|
| 113 | 36 + 39 | 194 | DRB1 | *0101, *01021/2, *0104 |
| 114 | 36 + 40 | 195 | DRB1 | *0103 |
| 115 | 41 + 252 | 206 | DRB1 | *15011/2, *15021/2, *1503-6 |
| 185 | 41 + 491 | 136 | DRB1 | *16011/2, *16021/2,*1603-8 |
| 162 | 68 + 255 | 211 | DRB1 | *03011/2,*03021/2, *0303-8, *1107 |
| 118 | 46 + 38 | 216 | DRB1 | *03011/2, *0304/6/8/11, *1327 |
| 119 | 44 + 37 | 188 | DRB1 | *03021/2, *0305, *1109/20, *1302/5/26/29/31,*1402/3/9/13/19/24/27/30, *1608, DRB3*0206 |
| 120 | 47 + 37 + 38 | 259 | DRB1 | *0401-22, *0424, *1122,*1410 |
| 121 | 48 + 49 | 231 | DRB1 | *0701 |
| 122 | 51 | 213 | DRB1 | *0801-4, *0806-13, *0815-17,*1415 |
| 227 | 46 + 49 | 192 | DRB1 | *09012 |
| 124 | 53 + 37 | 203 | DRB1 | *1001 |
| 125 | 68 + 273 + 54 | 175 | DRB1 | *1101-29, *0308, *1204,*1411 |
| 126 | 50 + 256 | 162 | DRB1 | *1201-5 |
| 127 | 263 + 252 + 259 | 152 | DRB1 | *1301/2/8/9/16/20/27-29/31/32, *1116/20, *1416 |
| 183 | 68 + 273 + 259 | 210 | DRB1 | *1301/2/4/8/15-17/19/20/22-24/27/28/31/32, *1102/3/11/14/16/20/21,*1416 |
| 170 | 68 + 102 | 170 | DRB1 | *1303/4/12/13/21/30/32, *1413 |
| 169 | 44 + 258 | 136 | DRB1 | *1305/6, *1109 |
| 171 | 263 + 264 + 51 + 58 | 165 | DRB1 | *1401/3-5/7-8/10-12/14-15/18/23/26-28, *0809,*1117, *1318 |
| 135 | 44 + 39 + 51 | 128/151 | DRB1 | *1402/3/6/9/12/13/17/27/29/30, *1318 |
| 134 | 68 + 107 | 171 | DRB1 | *1401/7/16/22/25/26 |
| 136 | 150 + 151 | 179 | DRB3 | *0101-2, *0201-6, *0301 |
| 186 | 61 + 492 | 172 | DRB5 | *0101-5, *0202-4 |
| 140 | 283 + 314 | 151 | DRB4 | *0101-5, *0201N |
| PM . | Primers . | Size (bp) . | Locus . | HLA Alleles Amplified . |
|---|---|---|---|---|
| 113 | 36 + 39 | 194 | DRB1 | *0101, *01021/2, *0104 |
| 114 | 36 + 40 | 195 | DRB1 | *0103 |
| 115 | 41 + 252 | 206 | DRB1 | *15011/2, *15021/2, *1503-6 |
| 185 | 41 + 491 | 136 | DRB1 | *16011/2, *16021/2,*1603-8 |
| 162 | 68 + 255 | 211 | DRB1 | *03011/2,*03021/2, *0303-8, *1107 |
| 118 | 46 + 38 | 216 | DRB1 | *03011/2, *0304/6/8/11, *1327 |
| 119 | 44 + 37 | 188 | DRB1 | *03021/2, *0305, *1109/20, *1302/5/26/29/31,*1402/3/9/13/19/24/27/30, *1608, DRB3*0206 |
| 120 | 47 + 37 + 38 | 259 | DRB1 | *0401-22, *0424, *1122,*1410 |
| 121 | 48 + 49 | 231 | DRB1 | *0701 |
| 122 | 51 | 213 | DRB1 | *0801-4, *0806-13, *0815-17,*1415 |
| 227 | 46 + 49 | 192 | DRB1 | *09012 |
| 124 | 53 + 37 | 203 | DRB1 | *1001 |
| 125 | 68 + 273 + 54 | 175 | DRB1 | *1101-29, *0308, *1204,*1411 |
| 126 | 50 + 256 | 162 | DRB1 | *1201-5 |
| 127 | 263 + 252 + 259 | 152 | DRB1 | *1301/2/8/9/16/20/27-29/31/32, *1116/20, *1416 |
| 183 | 68 + 273 + 259 | 210 | DRB1 | *1301/2/4/8/15-17/19/20/22-24/27/28/31/32, *1102/3/11/14/16/20/21,*1416 |
| 170 | 68 + 102 | 170 | DRB1 | *1303/4/12/13/21/30/32, *1413 |
| 169 | 44 + 258 | 136 | DRB1 | *1305/6, *1109 |
| 171 | 263 + 264 + 51 + 58 | 165 | DRB1 | *1401/3-5/7-8/10-12/14-15/18/23/26-28, *0809,*1117, *1318 |
| 135 | 44 + 39 + 51 | 128/151 | DRB1 | *1402/3/6/9/12/13/17/27/29/30, *1318 |
| 134 | 68 + 107 | 171 | DRB1 | *1401/7/16/22/25/26 |
| 136 | 150 + 151 | 179 | DRB3 | *0101-2, *0201-6, *0301 |
| 186 | 61 + 492 | 172 | DRB5 | *0101-5, *0202-4 |
| 140 | 283 + 314 | 151 | DRB4 | *0101-5, *0201N |
All primer mixes have been previously described in Bunce et al29 with the exception of primer mix 136. Primer mix 136 consists of primer 150 (5′-CCACgTTTCTTggAgCTgC) and primer 151 (5′-CCgTAgTTgTgTCTgCAgTAA), which amplify all DRB3 alleles (PM, primer mix number; Size, size of PCR product).
Nested PCR-SSP typing.
Forward primers DRFR1 CCCCACAgCACgTTTCTTg and DRFR2 CCCCACAgCACgTTTCCTg and reverse primer DRFR3 CCgCTgCACTgTGAAgCTCT amplify a 280-bp region of exon 2 of DRB1-9.32 The combination of primers is necessary to amplify all of the currently known HLA-DR alleles.
Amplification.
A total of 200 ng DNA was initially amplified in a buffer containing 67 mmol/L Tris pH 8.8, 16.6 mmol/L NH4SO4 , 200 μmol/L of each dATP, dCTP, dGTP, dTTP, 1.0 mmol/L MgCl2and 0.25 U BioTaq polymerase for 35 cycles (95°C for 30 seconds, 65°C for 45 seconds, and 72°C for 45 seconds) in a PTC200-96v thermal cycler. The resultant PCR product was diluted 1:200 in water before PCR-SSP typing.
Determination of Sensitivity and Specificity of Typing Methods
Sensitivity of PCR-SSP typing.
Decreasing amounts of DNA (200 ng/μL) from heterozygous individuals were mixed into DNA of known HLA type (200 ng/μL) to give final concentrations of 5%, 1%, 0.5%, and 0.1% (vol/vol). The resulting DNA mixtures were subsequently typed using equimolar HLA-DR sequence-specific primers and analyzed by agarose gel electrophoresis.
Sensitivity of nested PCR-SSP typing.
As for PCR-SSP typing, the sensitivity of nested PCR-SSP typing was analyzed by mixing DNA from healthy volunteers (200 ng/μL) with DNA of known type (200 ng/μL) at the following relative concentrations: 1%, 0.1%, 0.01%, 0.001%. The mixtures were then subjected to nested PCR-SSP typing and analyzed by agarose gel electrophoresis.
Specificity.
Specificity of nested PCR-SSP was determined by sequencing seven PCR products resulting from the amplification by primer mix 118 (Table 1). Amplicons were isolated from a low melting temperature agarose gel using the Wizard PCR Preps Purification System (Promega, Hampshire, UK). 33P cycle sequencing was performed with the Thermo Sequenase cycle sequencing kit (Amersham Life Science, Buckinghamshire, UK) using either [α-33P]-dATP or [α-33P]–dCTP and the specific forward or reverse primer. The products were run on a 7.5% polyacrylamide gel at 65 W for 2.5 hours. The gel was dried and autoradiography performed.
RESULTS
Determination of the Sensitivity of PCR-SSP Typing
The results summarized in Table 2 show that there is immense variation in the efficiency of amplification of HLA-DR alleles when using this system. While certain primer mixes (115 and 119) were capable of detecting DNA to a level of 0.1%, other primer mixes (120, 125, 126, and 127) only detected DNA to a level of between 1% and 5%. These results demonstrated that a greater sensitivity was required to detect donor-derived DNA after blood transfusion.
Comparison of the Sensitivities of PCR-SSP Typing and Nested PCR-SSP Typing
| Primer Mix . | PCR-SSP . | Nested PCR-SSP . | ||||||
|---|---|---|---|---|---|---|---|---|
| 5.0% . | 1.0% . | 0.5% . | 0.1% . | 1.0% . | 0.1% . | 0.01% . | 0.001% . | |
| 113 | ||||||||
| 114 | ||||||||
| 115 | ||||||||
| 185 | ||||||||
| 118 | NT | NT | NT | NT | ||||
| 119 | ||||||||
| 120 | ||||||||
| 125 | ||||||||
| 126 | ||||||||
| 127 | ||||||||
| 171 | ||||||||
| 121 | ||||||||
| 122 | ||||||||
| 227 | ||||||||
| 124 | ||||||||
| Primer Mix . | PCR-SSP . | Nested PCR-SSP . | ||||||
|---|---|---|---|---|---|---|---|---|
| 5.0% . | 1.0% . | 0.5% . | 0.1% . | 1.0% . | 0.1% . | 0.01% . | 0.001% . | |
| 113 | ||||||||
| 114 | ||||||||
| 115 | ||||||||
| 185 | ||||||||
| 118 | NT | NT | NT | NT | ||||
| 119 | ||||||||
| 120 | ||||||||
| 125 | ||||||||
| 126 | ||||||||
| 127 | ||||||||
| 171 | ||||||||
| 121 | ||||||||
| 122 | ||||||||
| 227 | ||||||||
| 124 | ||||||||
Decreasing amounts of DNA, representing all major DR alleles, were mixed with DNA of known type and HLA-DR typed using PCR-SSP and nested PCR-SSP to assess the sensitivity of both methods (shaded areas represent where alleles were detected; NT, not tested due to false amplification when this primer mix is used).
Nested PCR-SSP Typing
Sensitivity.
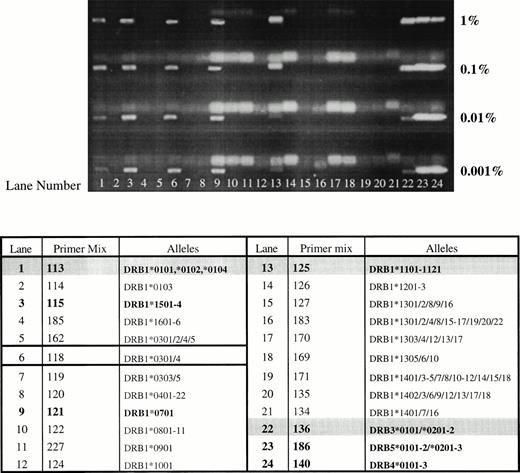
The results summarized in Table 2 demonstrate that the sensitivity of this nested PCR-SSP typing system is approximately 100-fold higher than that of PCR-SSP typing, and that 11 of 14 primer mixes tested could detect DNA to a level of 0.001%. However, primer mixes 114, 126, and 122 could only detect DNA to a level of between 0.01% and 0.001%. Figure 1 shows a typical set of results where all HLA-DR alleles from one donor (DRB1*0102, *1101/4, DRB3*0202/3) can be detected to a level of 0.001% in a recipient of HLA type DRB1*1501, *0701, DRB5*01, DRB4*0101. There is also an extra band that appears on the gel, in each case, which is neither donor nor recipient in origin (band detected with primer mix 118).
Sensitivity and specificity of nested PCR-SSP typing. DNA from one individual (HLA-DRB1*0102, *1101/4; DRB3*0202/3) was mixed with DNA of known type (HLA-DRB1*1501, *0701; DRB4*0101; DRB5*01) at relative concentrations of 1%, 0.1%, 0.01%, and 0.001%. DNA mixtures were typed using nested PCR-SSP and products analyzed by agarose gel electrophoresis. All HLA alleles from the donor can be detected to a level of 0.001% (donor and recipient lanes are in bold type, with donor lanes also shaded. The lane yielding the false amplification is outlined).
Sensitivity and specificity of nested PCR-SSP typing. DNA from one individual (HLA-DRB1*0102, *1101/4; DRB3*0202/3) was mixed with DNA of known type (HLA-DRB1*1501, *0701; DRB4*0101; DRB5*01) at relative concentrations of 1%, 0.1%, 0.01%, and 0.001%. DNA mixtures were typed using nested PCR-SSP and products analyzed by agarose gel electrophoresis. All HLA alleles from the donor can be detected to a level of 0.001% (donor and recipient lanes are in bold type, with donor lanes also shaded. The lane yielding the false amplification is outlined).
Specificity.
When nested PCR-SSP typing was used, extra bands appeared on the gel, one of which appeared every time, regardless of donor/recipient HLA type. Primer mix 118 yielded a band almost every time nested PCR-SSP typing was used (Fig 1). The only instance when this primer mix failed to yield a band was when DNA homozygous for DRB1*1001 was used (data not shown). To investigate the nature of the products amplified by this primer mix, DNA samples of known HLA type were amplified using this primer mix in nested PCR-SSP typing and sequenced by 33P Cycle sequencing. In conventional PCR-SSP typing systems, primer mix 118 amplifies the alleles DRB1*0301-8, with the exception of DRB1*0302 and DRB1*0305. Seven individuals, two of whom were DRB1*0301 (Table 3) and five who were not DRB1*0301-8 and, hence, should not be amplified by this primer mix, were sequenced and the results are summarized in Fig 2. As predicted, the two DNA samples typed as DRB1*0301 (A and B) had a sequence that corresponded exactly to that of the published sequence for DRB1*0301.31 When the remaining samples were compared with known HLA class II sequences, five of six corresponded to one or other of the HLA-DRB alleles present in the reaction. Interestingly, one of the samples (F) showed a number of differences to the sequence of DRB1*0301, which were only present in the sequence of HLA-DRB7*0101 (previously HLA-DRBψ), a pseudogene associated with the DR4, 7 and 9 haplotypes.33 The sequencing results show that the extra products seen when nested PCR-SSP typing is used are the result of mispriming events leading to the nonspecific amplification of HLA alleles or pseudogenes present in the PCR reaction.
Allelic HLA Types of DNA Samples Used for Specificity Determination
| DNA . | DRB1 . | DRB3 . | DRB4 . | DRB5 . | Pseudogenes . |
|---|---|---|---|---|---|
| A | *0301, *0401 | *0101 | *0101 | — | DRB2, DRB7, DRB8, DRB9 |
| B | *0701, *0301 | *0101 | *0101 | — | DRB2, DRB6, DRB9 |
| C | *0101 | — | — | — | DRB6, DRB9 |
| D | *1201 | *02/0301 | — | — | DRB2, DRB9 |
| E | *0102, *1101/4 | *02/0301 | — | — | DRB2, DRB6, DRB9 |
| F | *0401,*0701 | — | *01011, *0101 | — | DRB7, DRB8, DRB9 |
| G | *1501, *0701 | — | *0101 | *01 | DRB6, DRB7, DRB8, DRB9 |
| DNA . | DRB1 . | DRB3 . | DRB4 . | DRB5 . | Pseudogenes . |
|---|---|---|---|---|---|
| A | *0301, *0401 | *0101 | *0101 | — | DRB2, DRB7, DRB8, DRB9 |
| B | *0701, *0301 | *0101 | *0101 | — | DRB2, DRB6, DRB9 |
| C | *0101 | — | — | — | DRB6, DRB9 |
| D | *1201 | *02/0301 | — | — | DRB2, DRB9 |
| E | *0102, *1101/4 | *02/0301 | — | — | DRB2, DRB6, DRB9 |
| F | *0401,*0701 | — | *01011, *0101 | — | DRB7, DRB8, DRB9 |
| G | *1501, *0701 | — | *0101 | *01 | DRB6, DRB7, DRB8, DRB9 |
Pseudogenes are listed for each DNA sample.
Investigation into products detected when performing nested PCR-SSP typing with primer mix 118. Seven DNA samples (A to G) were subject to nested PCR-SSP typing and the resultant amplicons were purified and sequenced using 33P cycle sequencing. The resultant amplicon sequences were aligned to known HLA-DRB sequences31 and compared with HLA DRB1*0301 (redundancies are identified using the International Union of Biochemistry Group Codes where K = G/T, M = A/C, R = A/G, S = C/G, V = A/C/G, W = A/T, Y = C/T; * indicates base unknown).
Investigation into products detected when performing nested PCR-SSP typing with primer mix 118. Seven DNA samples (A to G) were subject to nested PCR-SSP typing and the resultant amplicons were purified and sequenced using 33P cycle sequencing. The resultant amplicon sequences were aligned to known HLA-DRB sequences31 and compared with HLA DRB1*0301 (redundancies are identified using the International Union of Biochemistry Group Codes where K = G/T, M = A/C, R = A/G, S = C/G, V = A/C/G, W = A/T, Y = C/T; * indicates base unknown).
DISCUSSION
The use of PCR-SSP typing alone for detection of HLA alleles was found not to be applicable to the detection of microchimerism as the level of sensitivity is poor for many alleles. The sensitivity experiments performed using deliberate mixing of DNA showed significant differences in the efficiency of amplification of the individual HLA-DR primer mixes (Table 2). These experiments suggested that a more sensitive method was required to enable donor-derived HLA alleles to be detected after allogeneic transfusion. A number of investigators attempting to assess the levels of microchimerism after solid organ transplantation have used a nested PCR approach,8,10,34,35primarily amplifying exon 2 of HLA DRB1, followed by a second PCR based on standard HLA typing procedures.27 28 We have used a similar technique and have found, in accordance with other groups, an increased sensitivity in the detection of donor-derived alleles in our mixing experiments (Table 2). We were able to detect HLA-DR alleles tested to a level of 0.001%, equivalent to a sensitivity of 1:100,000. However, we have shown that this nested PCR technique results in the presence of extra bands on the gel, bands that can easily be misinterpreted to indicate the presence of donor alleles.
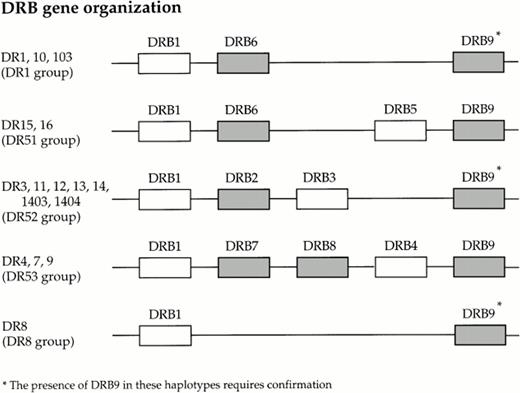
On sequencing a number of these nonspecific products, using primers that amplify DRB1*0301 in PCR-SSP typing, we have determined that one product is solely the result of a mispriming event which leads to the amplification of the HLA class II-associated pseudogene, HLA-DRB7, present on the DR4, 7 and 9 haplotypes. There are an additional four known pseudogenes associated with the class II region of the MHC; (Fig 3). The primers used in the first round amplification will amplify exon 2 of HLA-DRB1-9, with the exception of DRB2 and DRB8, as these pseudogenes do not contain exon 2.36 37 Although all alleles of HLA-DRB6, and HLA-DRB9 will be amplified by the first round primers, the forward and reverse primers from mix 118 are sufficiently mismatched to prevent any amplification of these alleles in the second round of amplification. On analysis of the other nested PCR-SSP amplicons, we have found that most of the nonspecific products are the result of mispriming events leading to the amplification of one or both of the HLA alleles present in the reaction.
Organization of the known HLA DRB loci within the class II region of the human MHC (known pseudogenes are shaded).
Organization of the known HLA DRB loci within the class II region of the human MHC (known pseudogenes are shaded).
The specificity of PCR-SSP typing relies on a mismatch at the 3′ residue of one of the primers being sufficient to prevent misamplification of alleles under carefully established conditions.38 However, we have shown that the addition of the preliminary amplification, so necessary for increased sensitivity, alters the stringency required for the subsequent PCR-SSP reactions. In many cases, the single base pair mismatch at the 3′ end of either primer is not then sufficient to prevent mispriming events, this in turn resulting in nonspecific amplification of many alleles. We have analyzed the sequences of the HLA alleles (including all pseudogenes) used in determining the specificity of nested PCR-SSP typing for their ability to bind to the individual primers in mix 118 (Table 4). The allele DRB1*0101, present in sample C, has a single base pair mismatch with the forward primer and two with the reverse, all being located at the 3′ end of the primers. Sequencing showed that this allele was amplified, hence the 3′ mismatches are not sufficient in this case to prevent amplification. All of the alleles present in sample D are also amplified with this primer mix, as shown by the sequencing result (Fig2). We believe this is because there is complete homology between the sequences of DRB1*1201 and DRB3*0201/4 and the reverse primer with only a single residue mismatched with the forward primer in both cases. There is obviously a hierarchy in terms of which alleles are amplified. This is illustrated by samples E, F, and G where only those alleles with one mismatch with the forward or reverse primers are amplified. The remaining alleles are not detected by sequencing and one can assume that they are not amplified by this primer set in these samples. Interestingly, the pseudogene DRB7*0101 was amplified in samples F and G. This is due to the fact that the forward primer will bind, possibly with complete homology to the DRB7*0101 sequence (there are still two bases unknown) and the reverse primer merely has one residue mismatched with DRB7*0101, the mismatch being located internally. This analysis shows that we can now predict which HLA alleles will be amplified in any given donor-recipient combination and we can control for their presence by the judicious selection of primers.
Degree of Mismatching of Forward and Reverse Primers Included in Primer Mix 118 With the HLA Alleles Used for Specificity Determination
| DNA Sample . | HLA-DR Allele . | Forward Primer . | Reverse Primer . | Amplified by DR17 Primers . | ||||
|---|---|---|---|---|---|---|---|---|
| Matched . | No. of Mismatches . | 3′ mm . | Matched . | No. of Mismatches . | 3′ mm . | |||
| C | DRB13-1500101 | No | 1 | Yes | No | 2 | Yes | √ |
| D | DRB13-1501201 | No | 1 | Yes | Yes | 0 | No | √ |
| DRB33-1500201/4 | No | 1 | Yes | Yes | 0 | No | √ | |
| E | DRB13-1500102 | No | 1 | Yes | Yes | 0 | No | √ |
| DRB13-1501104 | No | 1 | Yes | Yes | 0 | No | √ | |
| DRB33-1500202/3 | No | 1 | Yes | No | 2 | Yes | x | |
| F | DRB13-1500401 | No | 1 | Yes | No | 2 | Yes | x |
| DRB13-1500701 | No | 2 | Yes | No | 2 | Yes | x | |
| DRB43-1500101 | No | 3 | No | Yes | 0 | No | x | |
| DRB73-1500101 | Yes | 03-150 | No | No | 1 | No | √ | |
| G | DRB13-1501501 | No | 1 | Yes | Yes | 0 | No | √ |
| DRB13-1500701 | No | 2 | Yes | No | 2 | Yes | x | |
| DRB53-15001 | No | 1 | Yes | No | 2 | Yes | x | |
| DRB43-1500101 | No | 3 | No | Yes | 0 | No | x | |
| DRB73-1500101 | Yes | 03-150 | No | No | 1 | No | √ | |
| DNA Sample . | HLA-DR Allele . | Forward Primer . | Reverse Primer . | Amplified by DR17 Primers . | ||||
|---|---|---|---|---|---|---|---|---|
| Matched . | No. of Mismatches . | 3′ mm . | Matched . | No. of Mismatches . | 3′ mm . | |||
| C | DRB13-1500101 | No | 1 | Yes | No | 2 | Yes | √ |
| D | DRB13-1501201 | No | 1 | Yes | Yes | 0 | No | √ |
| DRB33-1500201/4 | No | 1 | Yes | Yes | 0 | No | √ | |
| E | DRB13-1500102 | No | 1 | Yes | Yes | 0 | No | √ |
| DRB13-1501104 | No | 1 | Yes | Yes | 0 | No | √ | |
| DRB33-1500202/3 | No | 1 | Yes | No | 2 | Yes | x | |
| F | DRB13-1500401 | No | 1 | Yes | No | 2 | Yes | x |
| DRB13-1500701 | No | 2 | Yes | No | 2 | Yes | x | |
| DRB43-1500101 | No | 3 | No | Yes | 0 | No | x | |
| DRB73-1500101 | Yes | 03-150 | No | No | 1 | No | √ | |
| G | DRB13-1501501 | No | 1 | Yes | Yes | 0 | No | √ |
| DRB13-1500701 | No | 2 | Yes | No | 2 | Yes | x | |
| DRB53-15001 | No | 1 | Yes | No | 2 | Yes | x | |
| DRB43-1500101 | No | 3 | No | Yes | 0 | No | x | |
| DRB73-1500101 | Yes | 03-150 | No | No | 1 | No | √ | |
The right hand column (√) and shading indicates alleles which were amplified by this primer mix.
There are two bases unknown in the sequence of DRB7*0101, which lie in the forward primer of mix 118.
Our data suggests that before using these techniques for the detection of microchimerism, adequate controls are mandatory. In addition to a negative or water control, a specificity control (pretransfusion/pretransplant recipient DNA) must be used to validate the technique for each recipient/donor combination, otherwise a band that is present after transfusion/transplant may be wrongly assumed to be indicative of the presence of a donor allele. Indeed, this has been demonstrated recently in a study evaluating microchimerism after solid organ transplantation using nested PCR for HLA genes, where 17% of patients would have yielded false positive results had a recipient pretransplant DNA sample not been included in the analysis.35
To validate our nested PCR-SSP technique, we have followed a number of different donor MHC class II alleles after transfusion of blood containing defined numbers of leukocytes bearing selected mismatches. Preliminary results indicate that donor-derived alleles are present in the early period after transfusion, and we are now in the process of analyzing further samples to give a clearer understanding of the patterns of survival of donor cells after HLA matched or mismatched transfusions.
Supported by a grant from the National Kidney Research Fund (Huntingdon, Cambs, UK).
Address reprint requests to Anthony Carter, Nuffield Department of Surgery, John Radcliffe Hospital, Headington, Oxford, OX3 9DU, UK; e-mail: anthony.carter@nds.ox.ac.uk.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.
© 1998 by the American Society of Hematology.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal