Abstract
The binding of all-trans retinoic acid (ATRA) to the ligand-binding region in the E-domain of retinoic acid receptor-α (RARα) modifies the transcriptional activity of RARα protein. ATRA probably induces differentiation of acute promyelocytic leukemia (APL) cells by binding to the E-domain of the RARα portion (RARα/E-domain) of PML/RARα chimeric protein. Therefore, molecular alteration in the RARα/E-domain of the chimeric gene is one mechanism by which patients with APL may acquire resistance to ATRA therapy. In this study using reverse transcription-polymerase chain reaction and single-strand conformation polymorphism, DNA segments amplified from the RARα/E-domain in fresh APL cells of 23 APL patients (8 males and 15 females from 4 to 76 years of age) were screened for mutations. Of those patients, 3 patients (1 with de novo and 2 with relapse) had clinical resistance to ATRA therapy. We found mutations in the RARα/E-domain of PML/RARα chimeric gene exclusively in the 2 patients who exhibited ATRA-resistance at relapse, whereas the mutations were not detected at their initial onset. Interestingly, these patients received a prolonged or intermittent administration of ATRA before relapse with ATRA-resistance. The mutations lead to the change of amino acid in the ligand-binding region of RARα/E-domain, Arg272Gln, or Met297Leu according to the amino acid sequence of RARα, respectively. Further study demonstrated that the in vitro ligand-dependent transcriptional activity of the mutant PML/RARα protein was significantly decreased as compared with that of wild-type PML/RARα. These findings suggest that mutations in the RARα/E-domain of the PML/RARα chimeric gene may confer clinical resistance to ATRA therapy in patients with APL.
ACUTE PROMYELOCYTIC leukemia (APL) is a unique subtype of leukemia characterized by a distinct chromosomal t(15;17) translocation with breakpoints within the retinoic acid receptor-α (RARα) gene on chromosome 17 and the PML gene, a putative transcription factor, on chromosome 15.1-3 The translocation generates a PML/RARα chimeric gene and, thereby, a PML/RARα fusion protein.4,5 Because the chimeric gene is present invariably and specifically in APL, the formation of PML/RARα is thought to be involved in the primary pathogenesis.3Recently, it was shown that the PML/RARα fusion protein inhibits differentiation or promotes the survival of myeloid precursor cells in vitro, suggesting a role of the fusion protein in the pathogenesis of APL.6 7
In the genomic structure of the PML/RARα chimeric gene, the breakpoints in the RARα gene localize exclusively within the second intron of the gene, and thereby the chimeric transcripts consistently retain not only the C-domain of the RARα for DNA-binding, but also the E-domain of the RARα that is required for ligand-binding and receptor-dimerization.8,9 Because of this structural property, the function of the PML/RARα fusion protein may be dependent on the binding of a specific ligand, all-transretinoic acid (ATRA), to the E-domain of the RARα portion (RARα/E-domain), as is the nuclear RARα.9-12Furthermore, it appears that this ligand-dependent transcription mechanism may underlie the cytodifferentiation of APL cells induced by pharmacological concentrations of ATRA.11
APL cells are induced by ATRA to differentiate in vitro and in vivo to mature myeloid cells.13-17 ATRA therapy induces complete remission in a high percentage of patients without severe complications associated with marrow hypoplasia or hemostatic impairment.17,18 However, most patients who relapse after ATRA therapy exhibit an apparent acquired ATRA-resistance.19-22 This acquired ATRA-resistance may be associated with several mechanisms such as insufficient plasma levels of ATRA concentration primarily caused by an increased oxidative catabolism of ATRA by cytochrome P-450 enzyme activity,23-26 increases in cellular retinoic acid binding protein (CRABP),27 or multidrug-resistance (MDR) gene product.28 More importantly, APL cells of patients with in vivo ATRA-resistance also exhibit frequently an ATRA-resistance or a decreased ATRA-sensitivity in vitro.20 27 These results indicate that ATRA-resistance could be due to an acquired cellular mechanism by which APL subclones survive and proliferate in vivo under the condition with pharmacological concentrations of ATRA.
As a model for an acquired resistance to ATRA, recent studies using ATRA-resistant myeloid leukemia cell lines showed the presence of point mutations in the E-domain of the RARα gene in HL-60 subclones that acquired ATRA-resistance in vitro.29-31 More recently, a NB4 subclone with ATRA-resistance had missense mutation also in the RARα/E-domain of PML/RARα chimeric gene, which abolished the ATRA-binding activity of the chimeric protein.32 Therefore, molecular alteration in the RARα/E-domain of the chimeric protein may impair ATRA binding, leading to an acquired ATRA-resistance of APL cells. On the basis of these findings, alteration of the RARα/E-domain of the chimeric gene has been anticipated, but not yet demonstrated, as a possible mechanism for ATRA-resistance in patients with APL.
In this study, we investigated molecular alteration of the RARα/E-domain in 23 patients with APL, including 3 with ATRA-resistance. We identified missense mutations in the RARα/E-domain of the PML/RARα chimeric gene at relapse in 2 patients who had ATRA-resistance after a prolonged or intermittent ATRA therapy. Our study may be useful to the understanding of the mechanism(s) by which patients with APL become ATRA-resistance during or after ATRA therapy.
PATIENTS
Clinical, hematological, and cytogenetic features of 23 patients with APL (8 males and 15 females with an average age of 27.4 years; range, 4 to 76 years) were shown in Table 1. Of those, 19 patients were with de novo APL and 4 were with bone marrow relapse. All patients except 2 received ATRA therapy for remission induction. Patients with ATRA-resistance were defined as those whose APL cells failed to show maturation in vivo during remission induction with ATRA therapy or those who relapsed during ATRA therapy. Using this criteria, there were 3 patients who exhibited ATRA-resistance: 1 with de novo APL and 2 with relapse who received either a prolonged ATRA therapy or a repeated alternative treatment with ATRA administration and chemotherapy before relapse. Two major isoforms of the PML/RARα chimeric gene, a long (or B) type with fusion between PML exon 6 and RARα exon 3 and a short (or A) type with fusion between PML exon 3 and RARα exon 3 were determined as previously described.4 33
Clinical, Hematological, and Cytogenetic Features of the Patients With APL in This Study
| . | Age/Sex . | FAB . | Onset . | Karyotype . | WBC (μL) (% blasts) . | ATRA Response . | Isoforms of PML/RARα . |
|---|---|---|---|---|---|---|---|
| 1 | 14/F | M3 | De novo | t(15;17) | 2,100 (64) | Yes | Long |
| 2 | 56/F | M3 | De novo | t(15;17) | 1,400 (1) | Yes | Long |
| 3 | 10/M | M3 | Relapse | t(15;17)add(22) | 3,800 (0) | No | Short |
| 4 | 10/M | M3v | Relapse | t(15;17) | 43,600 (90) | No | Short |
| 5 | 51/M | M3 | De novo | t(15;17) | 900 (22) | Yes | Long |
| 6 | 6/M | M3 | De novo | t(15;17) | 30,400 (70) | Yes | Long |
| 7 | 5/F | M3 | De novo | t(15;17), 9p- | 4,000 (31) | Yes | Short |
| 8 | 40/F | M3/M4 | Relapse | normal | 34,100 (93) | Yes | Long |
| 9 | 64/F | M3 | De novo | t(15;17) | 900 (49) | NE | NE |
| 10 | 4/M | M3 | De novo | t(15;17) | 8,400 (52) | Yes | Long |
| 11 | 7/M | M3 | De novo | t(15;17) | 5,600 (29) | Yes | Long |
| 12 | 24/F | M3 | De novo | t(15;17) | 500 (29) | Yes | Long |
| 13 | 37/F | M3 | De novo | t(15;17), 6p- | 810 (1) | Yes | Long |
| 14 | 13/M | M3 | De novo | t(15;17) | 2,500 (62) | Yes | Long |
| 15 | 76/F | M3 | De novo | 47,XXY,t(15;17) | 1,700 (45) | No | Long |
| 16 | 46/F | M3 | Relapse | t(15;17) | 1,000 (0) | Yes | Short |
| 17 | 62/F | M3 | De novo | t(15;17) | 1,100 (13) | Yes | Long |
| 18 | 5/F | M3 | De novo | t(15;17) | 6,000 (13) | Yes | Long |
| 19 | 64/F | M3 | De novo | t(15;17) | 900 (1) | NE | Long |
| 20 | 6/F | M3 | De novo | t(15;17) | 4,100 (57) | Yes | Long |
| 21 | 10/F | M3 | De novo | t(15;17) | 4,200 (54) | Yes | Long |
| 22 | 12/F | M3 | De novo | t(15;17) | 15,000 (20) | Yes | Long |
| 23 | 10/M | M3 | De novo | t(15;17) | 2,600 (76) | Yes | NE |
| . | Age/Sex . | FAB . | Onset . | Karyotype . | WBC (μL) (% blasts) . | ATRA Response . | Isoforms of PML/RARα . |
|---|---|---|---|---|---|---|---|
| 1 | 14/F | M3 | De novo | t(15;17) | 2,100 (64) | Yes | Long |
| 2 | 56/F | M3 | De novo | t(15;17) | 1,400 (1) | Yes | Long |
| 3 | 10/M | M3 | Relapse | t(15;17)add(22) | 3,800 (0) | No | Short |
| 4 | 10/M | M3v | Relapse | t(15;17) | 43,600 (90) | No | Short |
| 5 | 51/M | M3 | De novo | t(15;17) | 900 (22) | Yes | Long |
| 6 | 6/M | M3 | De novo | t(15;17) | 30,400 (70) | Yes | Long |
| 7 | 5/F | M3 | De novo | t(15;17), 9p- | 4,000 (31) | Yes | Short |
| 8 | 40/F | M3/M4 | Relapse | normal | 34,100 (93) | Yes | Long |
| 9 | 64/F | M3 | De novo | t(15;17) | 900 (49) | NE | NE |
| 10 | 4/M | M3 | De novo | t(15;17) | 8,400 (52) | Yes | Long |
| 11 | 7/M | M3 | De novo | t(15;17) | 5,600 (29) | Yes | Long |
| 12 | 24/F | M3 | De novo | t(15;17) | 500 (29) | Yes | Long |
| 13 | 37/F | M3 | De novo | t(15;17), 6p- | 810 (1) | Yes | Long |
| 14 | 13/M | M3 | De novo | t(15;17) | 2,500 (62) | Yes | Long |
| 15 | 76/F | M3 | De novo | 47,XXY,t(15;17) | 1,700 (45) | No | Long |
| 16 | 46/F | M3 | Relapse | t(15;17) | 1,000 (0) | Yes | Short |
| 17 | 62/F | M3 | De novo | t(15;17) | 1,100 (13) | Yes | Long |
| 18 | 5/F | M3 | De novo | t(15;17) | 6,000 (13) | Yes | Long |
| 19 | 64/F | M3 | De novo | t(15;17) | 900 (1) | NE | Long |
| 20 | 6/F | M3 | De novo | t(15;17) | 4,100 (57) | Yes | Long |
| 21 | 10/F | M3 | De novo | t(15;17) | 4,200 (54) | Yes | Long |
| 22 | 12/F | M3 | De novo | t(15;17) | 15,000 (20) | Yes | Long |
| 23 | 10/M | M3 | De novo | t(15;17) | 2,600 (76) | Yes | NE |
Abbreviation: NE, not evaluated.
MATERIALS AND METHODS
Cells.
After having obtained informed consent, we obtained bone marrow or peripheral blood samples from the patients with APL before treatment. Mononuclear cells were separated by a density-centrifugation with Ficoll-Paque (density, 1.077; Pharmacia, Uppsala, Sweden) and washed with phosphate-buffered saline (GIBCO BRL, Gaithersberg, MD).
RNA extraction and cDNA synthesis.
Total RNA was isolated from mononuclear cells using an acid guanidinium thiocyanate-phenol-chloroform extraction method.34 cDNA was then synthesized by incubating 10 μg of total RNA with 20 U of AMV-reverse transcripitase (Life Sciences, St Petersburg, FL) in 20 μL of reaction mixtures containing 50 mmol/L NaCl, 20 mmol/L Tris-Cl (pH 7.4), 8 mmol/L MgCl2, 50 ng/μL of oligo-dT (Pharmacia), 2 mmol/L deoxynucleotide triphosphates, 5 mmol/L dithiothreitol, and 2 U/mL of RNAsin (Takara Syuzo, Osaka, Japan) at 42°C for 1 hour.
Oligonucleotide primers.
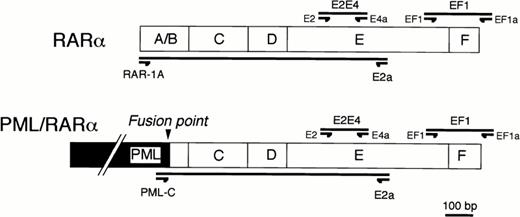
For screening the presence of mutation, two segments in the RARα/E-domain, named E2E4 and EF1 in Fig1, were amplified using two pairs of synthetic oligonucleotide primers, E2-E4a and EF1-EF1a, respectively (Sawaday Technology, Tokyo, Japan). Segments for single-strand conformation polymorphism (SSCP) analysis were designed to be no longer than 300 bp in length. The segments, E2E4 or EF1, correspond to the region at the middle part of E-domain or to the region spanning both of the carboxyl-terminal of E-domain and the F-domain of RARα gene, respectively. Because both of sense and antisense primers of these oligonucleotides correspond to the regions within the E and F-domains of RARα gene, it was expected that polymerase chain reaction (PCR) products would be a mixture of DNA fragments amplified from both of the RARα gene and PML/RARα chimeric gene. To distinguish which gene, either RARα or PML/RARα, the DNA segments were derived from, we designed the other two pairs of oligonucleotide primers with sense primers corresponding either to exon 1 of the RARα gene (RAR-1A) or to exon 3 of the PML gene (PML-C) (Fig1). Using a common antisense oligonucleotide primer (E2a) at the E-domain of the RARα gene, DNA segments could be differentially amplified from the RARα or PML/RARα genes.
Primer designs for RT-PCR/SSCP analysis (EF1 and EF1a primers for EF1 segment and E2 and E4a primers for E2E4 segment) or for differential amplification of the E-domain of RARα (RAR-1A and E2a) or of PML/RARα gene (PML-C and E2a).
Primer designs for RT-PCR/SSCP analysis (EF1 and EF1a primers for EF1 segment and E2 and E4a primers for E2E4 segment) or for differential amplification of the E-domain of RARα (RAR-1A and E2a) or of PML/RARα gene (PML-C and E2a).
PCR.
PCR reactions were performed as described, with modification.35 Reaction mixtures containing 0.2 μL of cDNA, 200 mmol/L deoxynucleotides, 1.0 mmol/L oligonucleotide primers, and 0.6 U of Taq polymerase (Perkin-Elmer Cetus, Norwalk, CT) in 20 μL of PCR buffer (10 mmol/L Tris-Cl, 50 mmol/L KCl, 1.5 mmol/L MgCl2, 5% dimethyl sulfoxide) were applied to Gene Amp PCR system 2400 (Perkin-Elmer Cetus) in 30 cycles. The conditions were denaturation for 30 seconds at 94°C, annealing for 30 seconds at 61°C for the paired primers of RAR-1A/E2a or at 57°C for other pairs of primers, and extension for 30 seconds at 72°C, respectively. The sequences of oligonucleotide primers were as follows: E2 (sense), 5′-GCATCATTAAGACTGTGGAG-3′; E4a (antisense), 5′-GCGAAGGCAAAGACCAGG-3′; EF1 (sense), 5′-AGATTACTGACCTGCGAAGC-3′; EF1a (antisense), 5′-GTAGAAAGGCAGAGAAAAGC-3′; RAR-1A (sense), 5′-ATGGCCAGCAACAGCAGCTCCTGCCCGAC-3′; PML-C (sense), 5′-CCGATGGCTTCGACGAGCTT-3′; E2a (antisense), 5′-CAGCCCCGTCTCCGCATCAT-3′.
SSCP.
For screening the presence of mutation, we used a nonradioisotopic modification of SSCP as described.36 37 Aliquots (2 μL) of PCR products were subsequently diluted with an equal volume of 95% formamide (Sigma, St Louis, MO) denatured at 90°C for 5 minutes and then electrophoresed in nondenaturing 8% polyacrylamide gel containing 10% glycerol (40 cm × 40 cm × 0.4 mm; Wako Pure Chemical, Osaka, Japan). Electrophoresis was performed at room temperature with the power constant at 20 W for 7 hours using 1× TBE. After electrophoresis, DNA bands were visualized by silver staining (Bio-Rad Laboratories, Hercules, CA).
DNA sequencing.
Reverse transcription-PCR (RT-PCR) products amplified from the patients with positive SSCP finding were subcloned into ddT-tailed pBluescript (Strategene, La Jolla, CA), and more than 5 subclones were selected randomly and sequenced using a T7 sequencing kit (U.S. Biochemical, Cleveland, OH) by a DNA sequencer (Pharmacia, Piscataway, NJ) by the dideoxy chain termination method.38
Allele-specific oligonucleotide (ASO) hybridization.
ASO hybridization was used to confirm the presence of mutation and to distinguish in which gene, either RARα or PML/RARα, the point mutations were present. DNA fragments were amplified differentially from either the RARα or PML/RARα gene, transferred onto nylon membrane after separation by gel electrophoresis with 2% agarose, and then hybridized with fluorescein-dUTP labeled 20 bp length of oligonucleotide probes identical to either wild-type or mutant sequence by using ECL kit and the manufacturer's instruction (Amersham International, Buckinghamshire, UK). After washing, chemiluminescence reaction was performed on the membrane, and then autoradiography was performed at room temperature for 30 minutes. To reduce nonspecific binding of oligonucleotide probes, an excess amount of nonlabeled wild-type or mutant oligonucleotide probes was added to the hybridization mixtures with fluorescein-dUTP labeled either mutant or wild-type oligonucleotide probes, respectively.
Transient transfection experiments for transcriptional activity.
cDNA clones of mutant PML/RARα with either G815A or A889T mutation were derived from pCMX expression vector harboring a short form of the wild-type PML/RARα cDNA using a site-directed mutagenesis kit (Stratagene, Cambridge, UK). These cDNA clones were then used in transient transfection experiments for evaluating in vitro transcriptional activity. African green monkey kidney, CV-1 cells, were grown in monolayer cultures at 37°C under 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum, 100 U/mL penicillin, and 100 μg/mL streptomycin. When CV-1 cells became 80% confluent, they were grown in DMEM with 1% calf serum treated with resin and charcoal (stripped medium).39 After 4 hours of incubation, cells were transfected with expression plasmid (2 μg), R-140 luc reporter plasmid containing a βRARE sequence40 (1.2 μg), and CMV-β-galactosidase control plasmid (pCMVβ; Clontech, Palo Alto, CA; 0.8 μg) per 3.5-cm plate using the calcium phosphate precipitation method.41 Cells then were grown for 36 hours in the absence or presence of ATRA. Luciferase and β-galactosidase activities of the cell extracts were assayed to correct for transfection efficiency.42 The corrected luciferase activities of samples were expressed as fold increases of luciferase activity to that of the samples in the absence of ligand (1-fold basal).
RESULTS
SSCP analysis for EF1 and E2E4 segments.
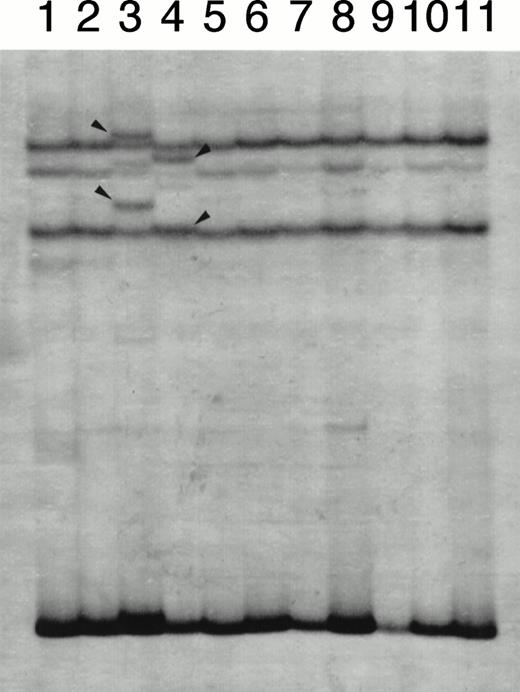
At first, we screened for mutation in the EF1 segment because this segment contains the region where mutations were detected in HL-60 or NB4 subclones with an acquired resistance to ATRA. SSCP analysis for EF1 segment showed single-strand bands with normal electrophoretic mobility in all patients (data not shown). These results indicated that mutation in the EF1 region of either the RARα or the PML/RARα chimeric gene was unlikely. By contrast, SSCP analysis for the E2E4 segment showed the bands with normal and altered mobility in 2 patients (Fig 2). Interestingly, these 2 patients were those who exhibited resistance to ATRA therapy at relapse. Because this SSCP finding was obtained at relapse in both patients, we examined whether their blood materials obtained at their initial onset would have the same SSCP findings. As shown in Fig3, SSCP findings at the initial onset did not show altered bands in both patients and were identical to that of HL-60 without mutation. This indicated that the appearance of altered bands in SSCP analysis was not a primary, but an acquired event during their clinical course.
A representative result of SSCP analysis of E2E4 segments. In addition to bands with normal mobility, bands with altered mobility were detected in lanes 3 (case no. 1) and 4 (case no. 2), which were marked with arrow heads.
A representative result of SSCP analysis of E2E4 segments. In addition to bands with normal mobility, bands with altered mobility were detected in lanes 3 (case no. 1) and 4 (case no. 2), which were marked with arrow heads.
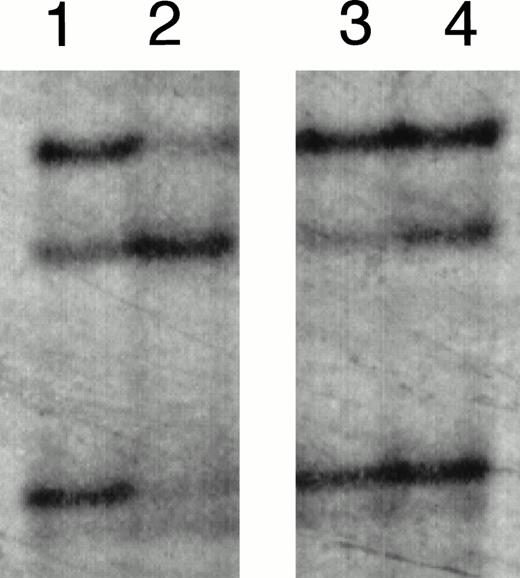
SSCP analysis of E2E4 segments amplified from the patients (cases no. 1 and 2) at the initial onset. Lanes 1 and 4, HL-60 without mutation; lanes 2 and 3, cases no. 1 and 2 at the initial onset, respectively.
SSCP analysis of E2E4 segments amplified from the patients (cases no. 1 and 2) at the initial onset. Lanes 1 and 4, HL-60 without mutation; lanes 2 and 3, cases no. 1 and 2 at the initial onset, respectively.
DNA sequence of missense mutations.
PCR products derived from the patients with positive SSCP findings were subcloned into ddT pBluescript, and DNA sequence was determined in more than five subclones randomly selected on an LB agarose plate. As shown in Fig 4, two of five subclones examined in 1 patient (case no. 1) showed a missense mutation of G815A according to the nucleotide sequence of RARα cDNA, which generated a newPst I site. However, the other three subclones had no mutation, suggesting a possibility that these subclones were amplified either from the normal RARα or wild-type PML/RARα gene. Similarly, the presence of missense mutation, A889T, was shown in the other patient (case no. 2) with a positive SSCP finding by DNA sequencing (data not shown).
DNA sequence of a subcloned E2E4 segment derived from case no. 1 who acquired ATRA resistance at relapse. The presence of G815A missense mutation was detected as compared with that of normal RARα/E-domain sequence. The position number of nucleotides was noted according to the sequence of normal RARα cDNA.
DNA sequence of a subcloned E2E4 segment derived from case no. 1 who acquired ATRA resistance at relapse. The presence of G815A missense mutation was detected as compared with that of normal RARα/E-domain sequence. The position number of nucleotides was noted according to the sequence of normal RARα cDNA.
Mutations present exclusively in PML/RARα chimeric gene.
For determining which gene, either RARα or PML/RARα chimeric gene, had the mutations, DNA segments were amplified differentially either from the RARα or the PML/RARα gene and then examined by digesting with Pst I restriction enzyme or by hybridizing with wild-type or mutant oligonucleotide probes. In case no. 1 with G815A mutation, an additional Pst I site was detected in DNA segment amplified from the PML/RARα gene, but not in that derived from the RARα gene (Fig 5). These results indicated that G815A mutation was present exclusively in the RARα/E-domain of PML/RARα chimeric gene. Similarly, in case no. 2 with A889T mutation, the oligonucleotide probe containing A889T mutation was exclusively hybridized to DNA segments amplified from the PML/RARα chimeric gene, but not to that derived from RARα gene (Fig6). However, in contrast to case no. 1, case no. 2 had the PML/RARα chimeric gene that could hybridize to both wild-type and mutant probes, suggesting that two distinct subclones of APL cells with or without A889T mutation in the RARα/E-domain of PML/RARα gene were present simultaneously in this patient at relapse.
Detection of G815A mutation by Pst I digestion in case no. 1. DNA segments amplified differentially from RARα in lanes 1 through 4 or from PML/RARα chimeric gene in lanes 5 through 8. Lanes 1 and 3, HL-60; lanes 5 and 7, an ATRA-sensitive control patient harboring a short PML/RARα isoform; lanes 2, 4, 6, and 8, case no. 1. PCR products in lanes 2, 4, 6, and 8 were digested with Pst I before electrophoresis. The Pst I digestion pattern of RARα-derived segments in case no. 1 (lanes 3 and 4) was similar to that of HL-60 (lanes 1 and 2). By contrast, the 873-bp segment derived from PML/RARα in case no. 1 was digested with Pst I into three fragments due to two Pst I sites including one additional site (lanes 7 and 8), which was not detected in a control patient without mutation (lanes 5 and 6).
Detection of G815A mutation by Pst I digestion in case no. 1. DNA segments amplified differentially from RARα in lanes 1 through 4 or from PML/RARα chimeric gene in lanes 5 through 8. Lanes 1 and 3, HL-60; lanes 5 and 7, an ATRA-sensitive control patient harboring a short PML/RARα isoform; lanes 2, 4, 6, and 8, case no. 1. PCR products in lanes 2, 4, 6, and 8 were digested with Pst I before electrophoresis. The Pst I digestion pattern of RARα-derived segments in case no. 1 (lanes 3 and 4) was similar to that of HL-60 (lanes 1 and 2). By contrast, the 873-bp segment derived from PML/RARα in case no. 1 was digested with Pst I into three fragments due to two Pst I sites including one additional site (lanes 7 and 8), which was not detected in a control patient without mutation (lanes 5 and 6).
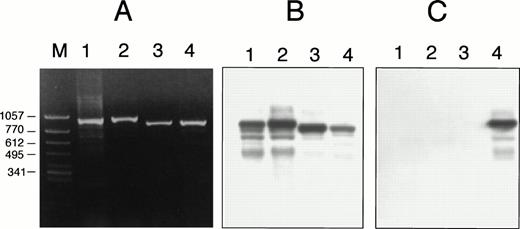
Detection of A889T by ASO hybridization in case no. 2. Using a control sample (ATRA-sensitive APL cells with a short PML/RARα isoform) (lanes 1 and 3) and case no. 2 (lanes 2 and 4), DNA segments amplified differentially from RARα (lanes 1 and 2) or PML/RARα chimeric gene (lanes 3 and 4). (A) Ethidium bromide staining of agarose gel electrophoresis. (B) The membrane blotted from (A) was hybridized with oligonucleotide probe specific for the wild-type sequence. (C) The membrane same as (B) was hybridized with oligonucleotide probe for A889T. The sequence of wild-type probe was 5′-CGGACCCAGATGCACAACGC-3′, the center adenine of which was replaced with thymidine in the mutant probe. The mutant probe positively hybridized only to the DNA segment that was amplified from PML/RARα chimeric gene of case no. 2 (lane 4).
Detection of A889T by ASO hybridization in case no. 2. Using a control sample (ATRA-sensitive APL cells with a short PML/RARα isoform) (lanes 1 and 3) and case no. 2 (lanes 2 and 4), DNA segments amplified differentially from RARα (lanes 1 and 2) or PML/RARα chimeric gene (lanes 3 and 4). (A) Ethidium bromide staining of agarose gel electrophoresis. (B) The membrane blotted from (A) was hybridized with oligonucleotide probe specific for the wild-type sequence. (C) The membrane same as (B) was hybridized with oligonucleotide probe for A889T. The sequence of wild-type probe was 5′-CGGACCCAGATGCACAACGC-3′, the center adenine of which was replaced with thymidine in the mutant probe. The mutant probe positively hybridized only to the DNA segment that was amplified from PML/RARα chimeric gene of case no. 2 (lane 4).
Ligand-dependent transcriptional activity.
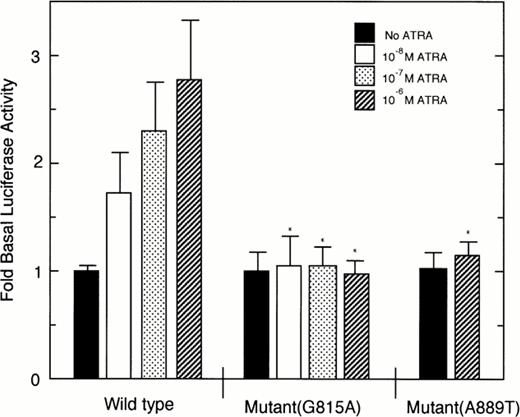
As shown in Fig 7, ligand-dependent transcriptional activity of the wild-type PML/RARα chimeric protein was increased in a manner dependent on ATRA concentrations. By contrast, the mutant PML/RARα chimeric proteins harboring either G815A or A889T mutation showed no increase of luciferase activity in the presence of ATRA as compared with that of wild-type PML/RARα with significant difference.
Ligand-dependent transcriptional activity of wild-type or mutant PML/RARα chimeric protein on βRARE. Luciferase activity of samples was normalized to β-galactosidase activity and then calculated as fold increases of luciferase activity to that of samples in the absence of ATRA. Each point represents the mean of triplicated results, and bars denote SD. *Significant difference as compared with the luciferase activities of wild-type PML/RARα in the presence of the corresponding concentrations of ATRA.
Ligand-dependent transcriptional activity of wild-type or mutant PML/RARα chimeric protein on βRARE. Luciferase activity of samples was normalized to β-galactosidase activity and then calculated as fold increases of luciferase activity to that of samples in the absence of ATRA. Each point represents the mean of triplicated results, and bars denote SD. *Significant difference as compared with the luciferase activities of wild-type PML/RARα in the presence of the corresponding concentrations of ATRA.
Summary of the patients with missense mutations.
Similarly and interestingly, as shown in Table2, both patients with missense mutations expressed a short PML/RARα isoform and had received a prolonged ATRA therapy before relapse with ATRA-resistance. Case no. 1, a 10-year-old boy, could not continue to receive chemotherapy because of liver abscess after chemotherapy for remission induction, and, thereafter, he had been undergoing ATRA therapy alone for 7 months until he relapsed with ATRA-resistance. Case no. 2, a 10-year-old boy, was successfully induced into complete remission by differentiation-induction therapy with ATRA and was then treated with an alternative regimen consisting of an intensive chemotherapy and an intermittent administration of ATRA (45 mg/m2 of ATRA for 2 weeks in every 2 months) for 11 months before relapse with ATRA-resistance. These 2 patients, cases no. 1 and 2, showed missense mutation, G815A or A889T, in the RARα/E-domain of the chimeric gene, which lead to amino acid replacement, Arg272Gln or Met297Leu, according to the amino acid sequence of RARα protein, respectively.
Profile of the Patients With an Acquired ATRA-Resistance Who Had Missense Mutations in the RARα/E-Domain of the PML/RARα Chimeric Gene
| Case No. . | Age/Sex . | PML/RARα Isoform . | ATRA Therapy Before Relapse . | ATRA Sensitivity . | Mutations* . | ||
|---|---|---|---|---|---|---|---|
| Initial . | Relapse . | DNA . | Amino Acid . | ||||
| 1 | 10 yr/M | Short form | Yes† | NE | No | G815A | Arg272Gln |
| 2 | 10 yr/M | Short form | Yes‡ | Yes | No | A889T | Met297Leu |
| Case No. . | Age/Sex . | PML/RARα Isoform . | ATRA Therapy Before Relapse . | ATRA Sensitivity . | Mutations* . | ||
|---|---|---|---|---|---|---|---|
| Initial . | Relapse . | DNA . | Amino Acid . | ||||
| 1 | 10 yr/M | Short form | Yes† | NE | No | G815A | Arg272Gln |
| 2 | 10 yr/M | Short form | Yes‡ | Yes | No | A889T | Met297Leu |
Abbreviation: NE, not evaluated.
The position of DNA or amino acids was numbered according to the sequence of RARα cDNA or protein, respectively.
Case no.1 received a prolonged ATRA therapy (for 7 months) after chemotherapy-induced hematological remission.
Case no. 2 underwent alternative treatment with ATRA and chemotherapy for 11 months after complete remission induced by ATRA therapy.
DISCUSSION
In this study, we demonstrated the presence of point mutations in the E-domain of the RARα portion of PML/RARα chimeric gene in APL with ATRA-resistance, the characteristics of which were the following. First, the mutations were detected exclusively in patients who exhibited an acquired ATRA-resistance, but not in patients who responded to ATRA therapy or exhibited poor response to the initial ATRA therapy. Second, the mutations were detected at the relapse with ATRA-resistance, but not at the initial onset. Third, the mutations were present in the E-domain of the RARα portion of PML/RARα chimeric gene, but not in the normal RARα gene. These findings clearly indicated that the appearance of APL subclone with these mutations was not a primary, but an acquired event, and is closely related to the relapse of APL patients exhibiting an acquired resistance to ATRA therapy.
A continuous intake of pharmacological doses of ATRA may induce alteration(s) in the systemic response of ATRA metabolism, which may result in the lowering plasma ATRA levels by a rapid clearance of ATRA from the plasma.23,43 With a prolonged 7-month period of ATRA administration in case no. 1, it was likely that the patient's plasma concentrations of ATRA at relapse might be lower than that required for in vitro cytodifferentiation of APL cells. However, the treatment protocol applied for case no. 2 has been demonstrated to prevent a progressive reduction of the plasma concentration of ATRA by the combination of an intermittent administration of ATRA with multidrug-combined chemotherapy alternatively.43 More importantly, APL cells of most patients with a clinical resistance to ATRA are either resistant or less sensitive to cytodifferentiation induced by ATRA in vitro.19,27 Thus, although we did not measure the plasma levels of ATRA in our patients, the intracellular alteration(s) of APL cells should be thought more significant as the mechanism(s) that causes an acquired ATRA-resistance in patients with APL.20
In APL cells, the intracellular ATRA can be oxidized to inactive metabolites by cytochrome P-450 isoform(s), and this process may be facilitated by CRABPs, which can be induced with an intake of pharmacological doses of ATRA.20,24 Alternatively, ATRA can be inactivated by the oxidative interaction with lipid hydroperoxides.20,26 Delva et al27 have reported an increased expression of CRABP-II at the time of relapse of APL as compared with the levels before ATRA therapy, suggesting an involvement of CRABP-mediated metabolism in the mechanism(s) of an acquired resistance to ATRA. By contrast, Kizaki et al28reported more recently that the levels of cytochrome P-450 activities were not significantly different between wild-type and ATRA-resistant HL-60 cells and, more interestingly, that multidrug-resistance-1 gene was expressed in ATRA-resistant APL cells, as compared with ATRA-sensitive cells. Although these findings obviously indicate that an altered intracellular metabolism(s) could underlie an acquired ATRA-resistance by APL cells, it remains to be answered whether alteration(s) in intracellular metabolism of ATRA would be quantitatively sufficient to prevent pharmacological doses of ATRA from reaching into the nucleus as an unbound form.20
Another possible mechanism is an acquired molecular alteration of the nuclear retinoid receptors of APL cells, which may impair an interaction of ligand to retinoid receptors. This possibility has been suggested by previous studies using ATRA-resistant APL cell lines that were developed by growing cells in the continuous presence of ATRA in vitro. HL-60 subclones with an acquired ATRA-resistance showed either missense or nonsense point mutations in the middle region or in the carboxyl-terminal of the RARα/E-domain, respectively.29-31 A truncated RARα protein caused by the latter nonsense mutation lacks 52 amino acids of the carboxyl end of the RARα protein, leading to a loss of transcription activity.29 More recently, Shao et al32 have reported an ATRA-resistant subclone of NB4 with a missense point mutation in the carboxyl-terminal of the RARα/E-domain in the PML/RARα chimeric gene, but not of the normal RARα gene. Interestingly, this mutation is located close to the nonsense mutation of the HL-60 subclone with truncated RARα and impairs the ATRA-binding activity of the mutated PML/RARα fusion protein.32
The mutations detected in our study were missense point mutations of G815A or A889T according to the sequence of RARα cDNA, respectively, which lead to amino acid replacement of Arg272Gln or Met297Leu using the amino acid sequence of RARα protein, respectively. These mutations, which are close to each other (74 bp apart) in the middle region of the E-domain, make a marked contrast to the location of mutations detected in ATRA-resistant HL-60 or NB4 subclones, the carboxyl-terminal of the E-domain. Conversely, the mutations of our patients are localized close to the other missense mutation detected in another subclone of ATRA-resistant HL-60.31 Therefore, the collection of these mutations of our patients and ATRA-resistant APL cell lines may imply the presence of two particular regions, either the middle or the carboxyl-terminal of the E-domain, where mutations may cluster in ATRA-resistant APL cells (Fig8). Because the amino acid sequence of the RARα/E-domain is evolutionary conserved,44 any missense mutations may alter the essential function of the E-domain, which could be the case demonstrated in our patients exhibiting an acquired resistance to ATRA.
A comparison between TR-β gene with mutations in RTH and RARα or PML/RARα genes on the alignment of sequence homology. The mutations in TR-β gene of RTH were clustering almost exclusively in the two regions, I and II, of the hormone binding domain.46 The mutations in the RARα/E-domain of ATRA-resistant APL were indicated as follows: 1, case no. 1; 2, case no. 2; 3 and 5, ATRA-resistant HL-60 subclones29-31; and 4, a ATRA-resistant NB4 subclone.32
A comparison between TR-β gene with mutations in RTH and RARα or PML/RARα genes on the alignment of sequence homology. The mutations in TR-β gene of RTH were clustering almost exclusively in the two regions, I and II, of the hormone binding domain.46 The mutations in the RARα/E-domain of ATRA-resistant APL were indicated as follows: 1, case no. 1; 2, case no. 2; 3 and 5, ATRA-resistant HL-60 subclones29-31; and 4, a ATRA-resistant NB4 subclone.32
The ligand-dependent transcriptional activity of the PML/RARα chimeric protein is shown in vitro to be increased in a manner similar to that of normal RARα, the E-domain of which functions for ligand-binding and receptor-dimerization.45 With transient transfection analysis in vitro, we demonstrated that the mutant PML/RARα lacked the activity of ligand-dependent transcription in vitro, suggesting that the change of the amino acid, Arg272Gln or Met297Leu, of RARα portion in the chimeric protein may impair these functions of PML/RARα.
An analogy between APL with an acquired ATRA-resistance and an inherited disorder with resistance to thyroid hormone (RTH) may give us important clues for understanding the function of the mutations shown by this study. RTH is an inherited disorder characterized with an impaired TH binding to thyroid hormone receptor-β (TR-β), a member of steroid/thyroid hormone receptor family with a homology to RARα gene.46 RTH is primarily caused by inherited mutations of TR-β gene, which cluster almost exclusively in the two regions of the ligand-binding domain of TR-β (Fig 8).46,47 These mutations biochemically inactivate the ligand-binding of the receptor, while preserving other functions, such as receptor-dimerization or DNA-binding, that are indispensable to the dominant negative action of the mutant receptor.46 Interestingly, in the alignment of TR-β ligand-binding domain and the RARα/E-domain on sequence homology, the mutations in ATRA-resistant patients and APL cell lines may cluster in accordance with the regions in RTH, denoted as I or II in Fig 8, respectively. Therefore, it is suggested that these two regions may play roles indispensable to the function of ATRA-binding, but not receptor-dimerization, of the PML/RARα chimeric protein.
The comparative analysis of the crystal structure of RARγ with and without bound ligand may be a further help to the understanding of how these cluster regions with mutations would play an essential role in the process of ligand-binding, despite their dispositions at a distance apart each other. After the electrostatic attraction of ATRA to the ligand-binding pocket of α-helical structures formed by the middle part of RARα/E-domain, ATRA is held into the cavity by the reposition of α-helical structures of the carboxyl-terminal of RARα protein, which is caused by the conformation change associated with the ligand proceeding into the binding cavity.48 Importantly, in this mouse trap mechanism of RARγ for ligand-binding, these two α-helical structures are consistent to the two mutation cluster regions in the E-domain of RAR, which we have found in APL cells with an acquired ATRA-resistance. More specifically, arginine at position 272 of RARα (Arg274 of RARγ), which was replaced with glutamine in case no. 1, is important in making van der Waals contact with the carbon molecule of the acyl chain of ATRA.48 Furthermore, the site-directed mutagenesis at Arg272 of RARα has been shown to impair the binding of ATRA49; therefore, Arg272Gln mutation may inhibit the ligand-binding function of PML/RARα chimeric protein. However, the role of Met297 of RARα/E-domain of the chimeric protein remains to be investigated.
For understanding how APL subclones with these mutations could expand during clinical course, it may be of note that, in case no. 2, APL subclones with either mutant or wild-type RARα/E-domain of the chimeric gene coexisted at relapse, while the chimeric gene of APL cells in case no. 1 was totally mutated at relapse. Thus, a biological pressure of ATRA therapy for selecting ATRA-resistant APL subclones may be increased as the period of ATRA administration is prolonged, because APL subclones with the mutations could have an advantage in surviving or proliferating under an environment with pharmacological concentrations of ATRA, as compared with that of ATRA-sensitive APL clone.6 However, the mechanism(s) inducing mutations in the RARα/E-domain of PML/RARα chimeric gene remains unknown.
Further study is needed to clarify the clinical significance of these mutations in larger populations of patients with APL. Moreover, it remains to be answered how these mutations could impair the functions of the RARα/E-domain in molecular levels. The answers to these questions would be of aid in establishing therapeutic approach to diminish the acquired resistance to ATRA, but also for understanding ATRA-induced cytodifferentiation of APL at the molecular level.
ACKNOWLEDGMENT
The authors are grateful to Dr T.R. Breitman for his thoughtful suggestion and Dr S. Kure for his insightful advice. We also thank the following doctors for kindly providing us with the blood materials: Drs K. Endo, K. Meguro, and Y. Saito (Internal Medicine, Tohoku University School of Medicine); Drs N. Takano and M. Endo (Department of Pediatrics, Iwate Medical University); Drs S. Yokoyama and S. Sato (Department of Pediatrics and Third Internal Medicine, Yamagata University); Drs E. Tamate and T. Sugawara (Department of Internal Medicine, Furukawa City Hospital); Dr T. Kawakami (Department of Pediatrics, Tottori University); Dr K. Asami (Department of Pediatrics, Niigata Cancer Center); and Dr S. Sibuya (Department of Pediatrics, Kanazawa Medical University).
Supported by grants from the Ministry of Education, Science and Culture, and the Ministry of Health and Public Welfare, Japan.
Address reprint requests to Masue Imaizumi, MD, Department of Pediatrics, Tohoku University School of Medicine, 1-1 Seiryo-machi Aoba-ku, Sendai 980-8574, Japan; e-mail:mimaizumi@ped.med.tohoku.ac.jp.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.
© 1998 by the American Society of Hematology.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal