Abstract
Chronic myeloid leukemia (CML) is thought to arise from a pluripotent hematopoietic stem cell that has undergone a reciprocal translocation between the BCR gene on chromosome 22 and the ABL proto-oncogene on chromosome 9. This rearrangement results in a shortened chromosome 22, designated the Philadelphia (Ph) chromosome. The Ph chromosome has been found in cells from all hematopoietic lineages except mature T lymphocytes. To examine this issue, we combined fluorescence-activated cell sorting (FACS) and fluorescence in situ hybridization (FISH) to study lineage involvement of mature cells and stem cells in 12 patients with CML in the chronic phase. We found Ph chromosomes in myeloid cells and most B lymphocytes (CD19+) but not in mature T cells (CD3+) or natural killer (NK) cells (CD3−56+). Moreover, evidence of BCR/ABL fusion was found in pluripotent stem cells (CD34+Thy-1+), B-progenitor cells (CD34+CD19+), T/NK progenitor cells (CD34+CD7+ cells), and T progenitor cells (CD34+CD7+CD5+) with a frequency equal to that in all CD34+ cells isolated by FACS from bone marrow cells. T lymphocytes showed a marked decrease in Ph+ cells between progenitor cells and mature cells. Moreover, the ratios of Ph+ to Ph− cells in mature T cells and NK cells were below background levels, whereas Ph+ B lymphocytes also decreased during their maturation. These data suggest that Ph+ lymphocytes are eliminated during differentiation. In contrast to FISH of blood and bone marrow, which gives information principally about mature cells, the technique of “sorter FISH (FACS + FISH)” provides a powerful tool to explore the cytogenetic changes in immature cell populations of stem cell diseases based on immunophenotypes. Further clarification of genetic changes in stem cells could be achieved by using sorter FISH with monoclonal antibodies.
CHRONIC myelogenous leukemia (CML) is a malignant hematological disorder of the human hematopoietic stem cells. Studies with glucose-6-phosphate dehydrogenase (G6PD) isoenzymes1,2 have shown that these stem cells are capable of differentiation into myeloid cells, monocytes, erythrocytes, platelets, and B lymphocytes. The Philadelphia (Ph) chromosome, t(9;22)(q34;q11), is the cytogenetic hallmark of CML; this balanced translocation results in a chimeric BCR/ABL gene that expresses an 8.5-kb hybrid mRNA transcript and a 210-kD fusion protein (P210). This BCR/ABL expression is presumed to effect clonal expansion in CML by deregulation of cell proliferation.3 As a recent advance in the treatment of CML, interferon-α has been found to reduce the Ph+ clone and prolong survival of CML patients. However, most patients experience transformation from the chronic phase to an acute blastic leukemia (blast crisis) after various time periods. In a majority of patients, the blasts resemble acute myeloblastic leukemia cells, and in about one third the blasts morphologically resemble the lymphoblasts in acute lymphoblastic leukemia.4 Less commonly, the dominant phenotype is an erythroblast, a megakaryoblast, or an undifferentiated blast with no detectable lineage-associated markers.5 T-cell markers in CML blast crisis have rarely been reported.6 7 This observation supports the hypothesis that the oncogenic event in CML occurs in a pluripotent hematopoietic stem cell capable of multilineage differentiation.
Acquisition of the Ph chromosome in nonlymphoid cells has been previously observed. For lack of direct evidence, the lineage involvement of T/natural killer (NK) and B lymphocytes remains unclear. However, in most studies, direct analysis of T cells showed no evidence of the Ph chromosome.8-13 In two studies, occurrence of the Ph chromosome in phytohemagglutinin (PHA)-responding peripheral blood (PB) cells, mostly peripheral T cells, was reported.14,15In one study, direct determination of the Ph chromosome in B and T lymphocytes by means of tricolor immunophenotyping/fluorescence in situ hybridization (FISH) showed that two patients featured involvement of CD20+ lymphocytes and that CD3+ lymphocytes in all six patients were negative for the Ph chromosome.16,17In another study, T-cell clones from PB cells in the chronic phase were screened for BCR/ABL fusion transcripts by using reverse transcriptase-polymerase chain reaction (RT-PCR). The BCR/ABL transcripts could be detected in only a few T-cell clones.18 Most B-lymphoblastoid cell lines established from CML patients do not carry the Ph marker or any submicroscopic BCR/ABL rearrangement.19 Recently, Haferlach et al17reported that t(9;22) was detectable in 34% of CD3+ T lymphocytes, in 32% of CD19+ B lymphocytes, and in 82% of CD34+ progenitor cells when they used a combination of blood and bone marrow (BM) smears stained with May-Grünwald-Giemsa and FISH. The Ph+ ratio of mature T cells was very high compared with those previously reported.20 As can be seen from these findings, the involvement of lymphocytes still remains to be settled.
This study examines the Ph chromosome status in mature cells and progenitor cells of various lineages and in multipotent stem cells from CML patients. The more immature cells are more difficult to culture in vitro because they are dormant. With fluorescence-activated cell sorting (FACS), mature cells and stem cells can be sorted on the basis of immunophenotype, but the number of stem cells is not sufficient for Southern blot analysis. Moreover, because cell sorting cannot avoid a few contaminating cells, PCR or cell culture for amplification of the number of cells would hamper interpretation of the results. Although chromosome analysis is restricted to metaphase cells, FISH has the advantage of allowing for a cell-by-cell analysis of the BCR/ABL fusion signals in nondividing cells that are potentially transcriptionally inactive. For these reasons, we applied FISH to blood smear (smear FISH) for mature cells and FACS (sorter FISH) to collected cells for the stem cells of 12 patients with CML in the chronic phase who had Ph chromosome in all cells as determined by standard chromosome analysis.
MATERIALS AND METHODS
Patients and samples.
The patient characteristics are shown in Table 1. At the time of study, 4 patients had not had any prior therapy, but 6 patients had been receiving chemotherapy with hydroxyurea and 2 patients with interferon-α and hydroxyurea. Ph chromosomes were observed in all analyzed metaphases by using a standard chromosome analysis technique. PB specimens and BM were obtained from 12 patients with Ph+ CML in the chronic phase and from donors of BM transplantation after informed consent was obtained. BM mononuclear cells (BMMC) or PB mononuclear cells (PBMC) were isolated by Ficoll-Hypaque density gradient centrifugation. Phagocytic cells were removed from BMMC or PBMC with Silica Suspension (Immuno-Biological Laboratories, Gunma, Japan), which enabled us to collect more CD34+ cells effectively. These nonphagocytic cells were used for the following study with FACS.
Clinical and Laboratory Findings for 12 Patients With CML
| Patient No. . | Age/Sex . | Mos . | Therapy . | WBC . | Hb . | Plt . | Karyotype . |
|---|---|---|---|---|---|---|---|
| 1 | 71/F | 36 | IFN + HU | 5,400 | 8.3 | 66.1 | 46,XX,t(9;22)(q34;q11)[20] |
| 2 | 72/F | 80 | HU | 4,800 | 10.9 | 22.4 | 46,XX,der(9)inv(9)(p11q13)t(9;22)(q34;q11),der(22)t(9;22)(q34;q11)[20] |
| 3 | 47/M | 87 | HU | 5,000 | 11.4 | 27.5 | 46,XY,t(9;22)(q34;q11)[20] |
| 4 | 52/F | 39 | HU | 5,800 | 11.6 | 54.0 | 46,XX,t(9;22)(q34;q11)[20] |
| 5 | 82/M | 22 | HU | 8,000 | 11.1 | 27.0 | 46,XY,t(9;22)(q34;q11)[20] |
| 6 | 37/M | 86 | IFN + HU | 3,200 | 14.3 | 23.7 | 46,XY,t(9;22)(q34;q11)[20] |
| 7 | 68/F | 117 | HU | 4,600 | 9.5 | 10.7 | 46,XX,t(9;22)(q34;q11)[20] |
| 8 | 19/M | 0 | None | 138,800 | 12.4 | 54.2 | 46,XY,t(9;22)(q34;q11)[20] |
| 9 | 69/F | 65 | HU | 6,100 | 7.5 | 72.5 | 46,XX,t(8;9;22)(p23;q34;q11)[20] |
| 10 | 40/F | 0 | None | 37,800 | 8.6 | 30.9 | 46,XX,t(9;22)(q34;q11)[20] |
| 11 | 31/M | 0 | None | 67,500 | 14.6 | 32.0 | 46,XY,t(9;22)(q34;q11)[20] |
| 12 | 39/M | 0 | None | 104,800 | 13.3 | 17.1 | 46,XY,t(9;22)(q34;q11)[20] |
| Patient No. . | Age/Sex . | Mos . | Therapy . | WBC . | Hb . | Plt . | Karyotype . |
|---|---|---|---|---|---|---|---|
| 1 | 71/F | 36 | IFN + HU | 5,400 | 8.3 | 66.1 | 46,XX,t(9;22)(q34;q11)[20] |
| 2 | 72/F | 80 | HU | 4,800 | 10.9 | 22.4 | 46,XX,der(9)inv(9)(p11q13)t(9;22)(q34;q11),der(22)t(9;22)(q34;q11)[20] |
| 3 | 47/M | 87 | HU | 5,000 | 11.4 | 27.5 | 46,XY,t(9;22)(q34;q11)[20] |
| 4 | 52/F | 39 | HU | 5,800 | 11.6 | 54.0 | 46,XX,t(9;22)(q34;q11)[20] |
| 5 | 82/M | 22 | HU | 8,000 | 11.1 | 27.0 | 46,XY,t(9;22)(q34;q11)[20] |
| 6 | 37/M | 86 | IFN + HU | 3,200 | 14.3 | 23.7 | 46,XY,t(9;22)(q34;q11)[20] |
| 7 | 68/F | 117 | HU | 4,600 | 9.5 | 10.7 | 46,XX,t(9;22)(q34;q11)[20] |
| 8 | 19/M | 0 | None | 138,800 | 12.4 | 54.2 | 46,XY,t(9;22)(q34;q11)[20] |
| 9 | 69/F | 65 | HU | 6,100 | 7.5 | 72.5 | 46,XX,t(8;9;22)(p23;q34;q11)[20] |
| 10 | 40/F | 0 | None | 37,800 | 8.6 | 30.9 | 46,XX,t(9;22)(q34;q11)[20] |
| 11 | 31/M | 0 | None | 67,500 | 14.6 | 32.0 | 46,XY,t(9;22)(q34;q11)[20] |
| 12 | 39/M | 0 | None | 104,800 | 13.3 | 17.1 | 46,XY,t(9;22)(q34;q11)[20] |
We used a standard technique for chromosome analysis. Aspirated BM was cultured overnight without stimulation and chromosome analysis was performed with the trypsin-treated G-banding method as described by ISCN (1995).
Abbreviations: Mos, months since diagnosis; IFN, interferon-α; HU, hydroxyurea; WBC, white blood cell count per microliter; Hb, hemoglobin (g/dL); Plt, platelets (104/μL).
FACS.
Fluorescein-conjugated CD8 (B9.11-FITC), CD7 (3A1-FITC), CD19 (89B-FITC), CDw90 (Thy-1) (F15.42.1.5-FITC), PECy5-conjugated CD34 (581-PECy5), CD4 (13B8.2-PECy5) (Coulter Immunotech, Margency, France), and fluorescein isothiocyanate (FITC)-conjugated CD3 (Leu4-FITC), HLA-DR-FITC- and phycoerythrin (PE)-conjugated CD34 (HPCA2-PE), CD56 (Leu19-PE) and CD3 (Leu4-PE) (Becton Dickinson, Sunnyvale, CA) were used. Both FITC-conjugated and PE-conjugated nonspecific Ms IgG were obtained from DAKO (Tokyo, Japan).
Flow cytometry analysis and cell sorting were performed on an EPICS Elite (Coultronics, Margency, France). Between 1,000 and 30,000 cells per fraction were sorted and collected in fetal calf serum (FCS). The purity attained was 97%. These cells were used for cytospin preparations and stained with May-Grünwald-Giemsa.
In situ hybridization.
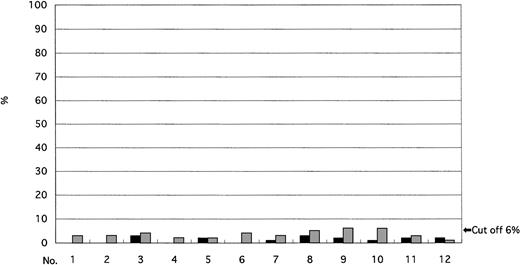
The probes used in this study were the BCR/ABL translocation probes commercially available from Oncor, Inc (Gaithersburg, MD). The probes flank the fusion site in essentially all cases of CML.21For prehybridization, slides were immersed in 0.01 N HCl/0.001% pepsin for 5 minutes, washed two times in phosphate-buffered saline (PBS) for 3 minutes, and treated with 4% paraformaldehyde/PBS for 5 minutes. After being washed with PBS, the cells were dehydrated through 70%, 80%, and 100% ethanol. The hybridization protocol followed the Oncor instructions and signals were detected with the aid of fluorescein-labeled avidin/rhodamine-labeled antidigoxigenin (Oncor). Signals were visualized under a Nikon microscope (Tokyo, Japan) with a FITC/Rhodamine dual band filter (Nikon). Evaluation of the preparations was performed by counting 100 nuclei per slide. The mean percentage of nuclei with a false-positive signal was calculated for the control PB from hematological disease-free individuals. A previous study reported false-positive cells occurring at a frequency of about 2%,21 and we found false-positive cells at a frequency of 0% to 6% (cutoff).
Purification of progenitor populations.
Stem cells were characterized by defining subpopulations of CD34+ cells. Thy-1 is a marker expressed by fetal and adult BM stem cells and the Thy-1+ subset has multilineage differentiation capacity, as shown by its ability to produce T cells, B cells, and myeloid cells.22,23 The CD34+CD10+CD19+ population represents exclusively B-lymphoid committed progenitors.24Cytoplasmic CD3+CD7+ cells were considered to represent committed T-cell progenitors based on the fact that CD2, cyCD3, CD5, and CD7 are coexpressed on all mature T cells but not on all mature B cells.25 However, these antigens are also expressed on CD34+ NK progenitors.26 In addition, CD7 is present on B-cell and myeloid progenitors.27 These data suggest that CD34+CD7+ and CD5+/CD2+populations represent T-lymphoid committed progenitors.25On the basis of these findings, we used CD34+Thy-1+ cells as the pluripotent stem cells, CD34+CD19+ as the B progenitor cells, CD34+CD7+ cells as the T/NK progenitor cells, and CD34+CD7+CD5+ as the T progenitor cells in this study.
RESULTS
The influence of treatment with interferon-α and hydroxyurea on the proliferation of cell lineage was taken into consideration, but the data for patients with and without previous treatment at the time of this study were not different.
Flow cytometry analysis of CD34+ cells obtained from BM mononuclear cells.
We isolated CD34+ cells from 8 of 12 patients. The other 4 patients were assessed only by FISH of the PB smear. Table 2 indicates that the number of CD34+ and CD34+7+ cells was quite variable and higher than that in normal donors. The other subpopulations of lymphoid progenitor cells did not differ from those in normal donors.
Flow Cytometry Analysis of Stem Cell Population Obtained From BMMC
| . | CD34+/ MNC . | CD34+7+/ MNC . | CD34+7+5+/ MNC . | CD34+19+/ MNC . | CD34+Thy+/ MNC (%) . |
|---|---|---|---|---|---|
| Patient No. | |||||
| 2 | 8.9 | 2.2 | 0.1 | 0.1 | 0.3 |
| 3 | 13.4 | 2.3 | ND | 1.6 | 0.3 |
| 4 | 10.3 | 0.9 | ND | 0.6 | 0.4 |
| 6 | 22.8 | 12.1 | 0.1 | 0.2 | 0.1 |
| 8 | 22.2 | 2.6 | ND | 0.1 | 0.0 |
| 9 | 10.7 | 1.4 | 0.3 | 0.6 | 0.5 |
| 11 | 9.8 | 1.4 | 0.1 | 2.0 | 0.0 |
| 12 | 23.3 | 4.8 | 0.4 | 0.4 | 0.1 |
| Mean | 15.2 | 3.5 | 0.2 | 0.7 | 0.2 |
| Normal donor | |||||
| 1 | 3.9 | 0.4 | ND | 0.7 | 0.3 |
| 2 | 6.8 | 0.9 | ND | 0.6 | 0.0 |
| 3 | 2.3 | 0.1 | 0.0 | 0.6 | 0.1 |
| 4 | 7.4 | 0.5 | 0.2 | ND | ND |
| Mean | 5.1 | 0.5 | 0.1 | 0.6 | 0.1 |
| . | CD34+/ MNC . | CD34+7+/ MNC . | CD34+7+5+/ MNC . | CD34+19+/ MNC . | CD34+Thy+/ MNC (%) . |
|---|---|---|---|---|---|
| Patient No. | |||||
| 2 | 8.9 | 2.2 | 0.1 | 0.1 | 0.3 |
| 3 | 13.4 | 2.3 | ND | 1.6 | 0.3 |
| 4 | 10.3 | 0.9 | ND | 0.6 | 0.4 |
| 6 | 22.8 | 12.1 | 0.1 | 0.2 | 0.1 |
| 8 | 22.2 | 2.6 | ND | 0.1 | 0.0 |
| 9 | 10.7 | 1.4 | 0.3 | 0.6 | 0.5 |
| 11 | 9.8 | 1.4 | 0.1 | 2.0 | 0.0 |
| 12 | 23.3 | 4.8 | 0.4 | 0.4 | 0.1 |
| Mean | 15.2 | 3.5 | 0.2 | 0.7 | 0.2 |
| Normal donor | |||||
| 1 | 3.9 | 0.4 | ND | 0.7 | 0.3 |
| 2 | 6.8 | 0.9 | ND | 0.6 | 0.0 |
| 3 | 2.3 | 0.1 | 0.0 | 0.6 | 0.1 |
| 4 | 7.4 | 0.5 | 0.2 | ND | ND |
| Mean | 5.1 | 0.5 | 0.1 | 0.6 | 0.1 |
Abbreviations: MNC, mononuclear cells; ND, not done.
Lineage involvement determined by FISH analysis of blood smear.
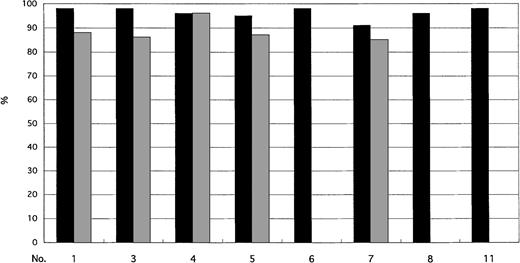
Neutrophils and monocytes were consistently highly positive for the Ph chromosome, ie, greater than 90% and 80%, respectively. Because of the marked increase in granulocytes, we did not find monocytes within the field observed by FISH in 3 patients (patients no. 6, 8, and 11; Fig 1).
Cells with the BCR/ABL fusion signal in neutrophils (▪) and monocytes (▩).
FISH analysis applied to sorted cells of B, T, and NK cells.
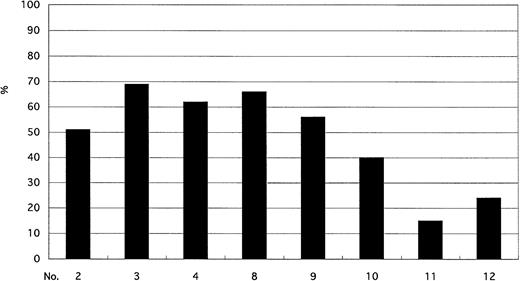
Lymphoid cells were assessed by sorter FISH, which allowed us to classify lymphocytes on the basis of May-Grünwald-Giemsa staining into B, T, or NK cells. The percentage of CD19+ B lymphocytes varied among patients, but all patients had a lower incidence of Ph+ cells among B lymphocytes than among neutrophils and monocytes (Figs 1 and 2). In contrast, the incidence of Ph+ cells among CD3+ T lymphocytes and CD3−56+ NK cells was below the cutoff value in all patients (Fig 3).
FISH analysis of stem cell CD34+ subpopulations.
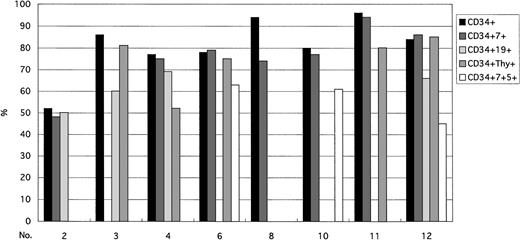
The incidence of cells bearing the Ph chromosome among CD34+ cells was as frequent as that seen in other progenitor cells (Fig 4). Not all subpopulations could always be isolated with FACS. In contrast to mature T cells, the incidence of the Ph chromosome in progenitor cells was similar to that in CD34+ cells. Compared with mature B cells, the incidence of Ph+ progenitor cells were as frequent as that in CD34 progenitor cells. The incidence of the Ph chromosome in CD34+Thy-1+ cells was already similar to the one seen in mature myeloid cells (Fig 5).
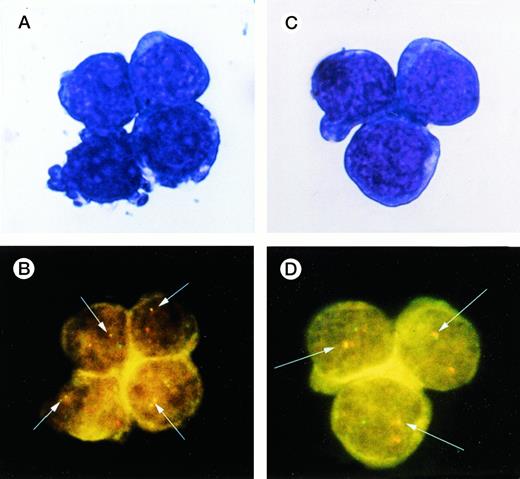
A representative image of FISH applied to CD34+Thy-1+ and CD34+7+ cells sorted from BM. (A and C) May-Giemsa stain of CD34+Thy-1+ and CD34+7+ cells. (B and D) The hybridized bcr probe was detected with rhodamine (red signal) and the hybridized abl probe with fluorescein (yellow-green signal). A yellow or red/green spot was indicative of BCR/ABL fusion (white arrows).
A representative image of FISH applied to CD34+Thy-1+ and CD34+7+ cells sorted from BM. (A and C) May-Giemsa stain of CD34+Thy-1+ and CD34+7+ cells. (B and D) The hybridized bcr probe was detected with rhodamine (red signal) and the hybridized abl probe with fluorescein (yellow-green signal). A yellow or red/green spot was indicative of BCR/ABL fusion (white arrows).
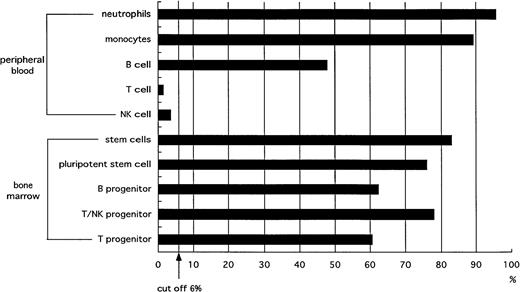
Taken all together (Fig 6), the results from the FISH analysis to detect BCR/ABL fusion show that the Ph chromosome is present in B lymphocytes (CD19+) but not in mature T cells (CD3+) or NK cells (CD3−CD56+). Moreover, BCR/ABL fusion was found in pluripotent stem cells (CD34+Thy-1+; Fig 6), B progenitor cells (CD34+CD19+), and T progenitor cells (CD34+CD7+ or CD34+CD7+CD5+ cells) at the same frequency as in all CD34+ cells sorted by FACS from BM cells. Although neutrophils and monocytes had a high percentage of Ph+ cells, the ratios of Ph+ to Ph− cells in mature T cells and NK cells were below background levels. Ph+ B lymphocytes also decreased during the maturation process.
Ratio of Ph+ cells to Ph−cells. Ph chromosomes were found in myeloid cells and B lymphocytes but not in mature T and NK cells. BCR/ABL fusion signals were found in pluripotent stem cells (CD34+Thy-1+), B progenitor cells (CD34+CD19+), and T progenitor cells (CD34+CD7+ cells or CD34+CD7+CD5+) at a frequency equal to that in all CD34+ cells. T lymphocytes showed a marked decrease of Ph+ cells between progenitor cells and mature cells. The incidence ratios of Ph+ cells in mature T cells and NK cells were below the cutoff value.
Ratio of Ph+ cells to Ph−cells. Ph chromosomes were found in myeloid cells and B lymphocytes but not in mature T and NK cells. BCR/ABL fusion signals were found in pluripotent stem cells (CD34+Thy-1+), B progenitor cells (CD34+CD19+), and T progenitor cells (CD34+CD7+ cells or CD34+CD7+CD5+) at a frequency equal to that in all CD34+ cells. T lymphocytes showed a marked decrease of Ph+ cells between progenitor cells and mature cells. The incidence ratios of Ph+ cells in mature T cells and NK cells were below the cutoff value.
DISCUSSION
CML is thought to arise from a pluripotent hematopoietic stem cell that contains the Ph chromosome. The Ph chromosome, t(9;22)(q34;q11), is the cytogenetic hallmark of CML, and this balanced translocation results in a chimeric BCR/ABL gene. This BCR/ABL expression is presumed to effect clonal expansion in CML by means of deregulation of cell proliferation.
FISH applied to blood and BM smear (smear FISH) provided various data on cell lineage involvement of the Ph chromosome, and these data provided additional support for the hypothesis that CML is a stem cell disease. Although smear FISH yielded important information, it could not provide information on changes affecting pluripotent stem cells or on progenitor cells and their mature descendants. Smear FISH also has limitations in assessing polyploid cells and eosinophils because of their autofluorescence. Cell sorting is a powerful tool to obtain stem cells of various lineages and progenitor cells of a defined maturation level. This technique is especially effective for lymphoid cells and stem cells, because it is difficult to identify subclasses of lymphocytes based on morphological observations and because immature cells are dormant and difficult to culture. However, colony assays may introduce clonal changes during culture periods. Because BCR/ABL mRNA is minimally expressed or may be absent in primitive CML progenitors, these cells may escape detection by RT-PCR.28 At the same time, cell fractions obtained by sorting based on immunophenotype may include some contaminated cells that would be amplified by culture or PCR. As a result of these considerations, we applied FISH to immunophenotype sorted cells for this study.
The Ph chromosome has been found in cells from all hematopoietic lineages except mature T lymphocytes. Our data summarized in Fig 6 show that, although mature T lymphocytes do not have the Ph chromosome, both stem cells and T progenitor cells do. There is thus an obvious discrepancy in the positivity of the Ph chromosome in T and B lymphocytes. To resolve this discrepancy between stem cells and mature lymphocytes, we examined each of the progenitor cells.
To date, several explanations have been proposed for the lack of the Ph chromosome among peripheral T lymphocytes. A long-standing hypothesis is that the majority of T cells are long-lived and born before the occurrence of clonal mutation. It is also believed that memory lymphocytes are capable of surviving for long periods of 20 years or more. But two studies in humans have reported the mean life span of lymphocytes as 530 days29 or 1,600 days,30indicating that some lymphocytes must have a half-life in excess of 3 years. We must also consider that it takes on average 6.3 years from the time that a cell acquires the Ph chromosome until a patient has clinically evident symptoms.31 The present study includes patients who have had CML for 5 to 10 years (Table 1; patients no. 2, 3, 6, 7, and 9), which is enough time for some T lymphocytes to acquire the Ph chromosome. Although we found BCR/ABL rearrangement in progenitor cells committed to the T-cell lineage, no evidence of the fusion gene was found among peripheral T lymphocytes, even in patients with a long history of CML. These data lead us to believe that the lymphocyte life span may not conclusively explain the lack of the Ph chromosome in mature T lymphocytes.
A second explanation is that the usual target of malignant transformation in CML is a more restricted stem cell committed to the myeloid and B-cell lineages but not to the T-cell lineage.32 In our study, BCR/ABL fusion was detected in CD34+Thy-1+ cells, CD34+CD19+ cells, and CD34+CD7+ or CD34+CD7+CD5+ cells at the same frequency as in all CD34 cells. These data indicate that the usual target of malignant transformation in CML is a pluripotent stem cell. Thus, a restricted stem cell target may not explain the negativity of Ph chromosome in T lymphocytes either.
Although the ontogeny of NK cells has not been fully clarified yet, CD34+ progenitor cells have been shown to differentiate into NK cells in vitro.26 33-35 Our data show that most NK cells during the chronic phase of CML are also from Ph− clones.
We must also consider the impact of age on T-cell generation and turnover. Age-related involution is characterized by a progressive reduction in thymic size and weight. After the first 20 to 30 years of life, a greater proportion of the thymus is replaced with adipose tissue. However, the exact effect of aging on thymic function remains to be settled despite this evidence of morphological changes.
Although we cannot provide a definitive explanation for this discrepancy, one possibility might be that lymphocytes with the Ph chromosome fail to differentiate. This is compatible with the finding that the B-cell population is chimeric with respect to the BCR/ABL and that Ph+ T-cell progenitors may undergo only very limited differentiation in the absence of an active thymus. Inducing the Ph+ lymphoid progenitors to differentiate in vitro could support our explanation, but the appropriate experimental procedures and equipment are not available at this time.
In conclusion, mature T cells and NK cells in most CML patients are Ph−, but most patients have a mixture of Ph+ and Ph− B cells. Moreover, in all patients with CML, BCR/ABL fusion was found in pluripotent stem cells, B progenitor cells, T/NK progenitor cells, and T progenitor cells. The application of FISH to blood and BM, smear FISH, yields important data principally about mature cells. Furthermore, the technique of sorter FISH (FACS + FISH) is a powerful tool to explore the cytogenetic changes of immature cells of stem cell diseases. Further clarification of genetic changes in stem cells could be achieved by using sorter FISH with various combinations of monoclonal antibodies.
Address reprint request to Ikuo Miura, MD, Third Department of Internal Medicine, Akita University School of Medicine, 1-1-1 Hondo, Akita, Japan; e-mail: ikuo@med.akita-u.ac.jp.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal