Abstract
CBFβ-SMMHC is expressed in M4Eo acute myeloid leukemia (AML) as a result of inv(16), but how it contributes to leukemogenesis is unknown. p53 mutations are rare in de novo AML, but they are common in many malignancies. Expression of CBFβ-SMMHC in Ba/F3 cells reduced p53 induction in response to ionizing radiation or etoposide 3- to 4-fold. However, p53 induction was normal in Ba/F3 cells expressing a CBFβ-SMMHC variant that does not interfere with DNA binding by CBF, indicating that a CBF genetic target regulates p53 induction. The p53 gene may be regulated by CBF, because p53 mRNA levels were reduced by CBFβ-SMMHC. Reduced p53 induction was not caused by slowed cell proliferation, a consequence of CBFβ-SMMHC expression, because p53 was induced similarly in control cultures and in cultures propagated in 10-fold less interleukin-3 (IL-3). CBFβ-SMMHC did not slow apoptosis resulting from IL-3 withdrawal, where p53 induction is minimal, but slowed apoptosis in Ba/F3 cells exposed to 10 Gy of ionizing radiation or 3 to 8 μg/mL etoposide, providing 2-fold protection at 6 or 18 hours. Inhibition of apoptosis was temporary, because all the cells exposed to these doses ultimately died, and clonal survival assays performed using 0.04 μg/mL etoposide did not show protection by CBFβ-SMMHC. p21 levels were increased in cells subjected to DNA damage, regardless of CBFβ-SMMHC expression and attenuated p53 induction. Bcl-2, bcl-xL, bcl-xS, and bax levels were unaffected by CBFβ-SMMHC. Attenuated p53 induction may contribute to leukemogenesis by CBFβ-SMMHC by slowing apoptosis via a p21-independent mechanism.
CBFβ-SMMHC IS expressed in the blasts of patients with the French-American-British M4eo subtype of acute myeloid leukemia (AML) as a consequence of the inv(16)(p13q22) or the t(16;16)(p13q22) chromosomal abnormalities.1-4 In CBFβ-SMMHC the CBFβ transcription factor is fused to the tail domain of a smooth muscle myosin heavy chain (SMMHC).
Core binding factor (CBF) is a family of heterodimeric transcription factors containing a common β subunit, CBFβ, and one of three CBFα subunits, CBFα1, AML1 (CBFα2), or CBFα3.5-9CBFβ does not bind DNA directly, but increases the affinities of the CBFα subunits for DNA.7,8 The CBFα subunits contain a Runt-homology domain required for DNA binding and for heterodimerization.6,10 Mice lacking CBFβ or AML1 fail to develop definitive hematopoiesis.11-14
The tail domain of SMMHC is a coiled-coil dimerization domain that can multimerize into large filaments.15 CBFβ-SMMHC can sequester CBFα subunits into nonfunctional complexes.16,17 Mice in which the CBFβ-SMMHC cDNA has been “knocked-in” to its endogenous locus fail to develop definitive hematopoiesis or leukemia and die in utero.18This phenotype is identical to that of CBFβ- or AML1-null mice, providing additional evidence that CBFβ-SMMHC acts by inhibiting CBF functions.
AML1-ETO, expressed from the t(8;21) chromosome in AML, and TEL-AML1, expressed from the t(12;21) chromosome in B-lineage acute lymphocytic leukemias, are each capable of repressing transcriptional activation by CBFα,19-21 and AML1-ETO knock-in mice do not develop definitive hematopoiesis or leukemia.22 23 The similarity between these mice, mice expressing CBFβ-SMMHC, and mice lacking CBFβ or AML1 supports the hypothesis that all of the CBF oncoproteins act to inhibit CBF functions.
We expressed CBFβ-SMMHC inducibly from the sheep metallothionein (MT) promoter in two interleukin-3 (IL-3)–dependent hematopoietic cell lines, Ba/F3 B-lymphoid cells and 32D cl3 myeloid cells. Exposure of these lines to zinc chloride induced CBFβ-SMMHC levels more than 10-fold, to levels similar to endogenous CBFβ. As a result, cell generation time increased 1.7-fold, with a 4-fold increase in the ratio of G1 to S phase cells and evident hypophosphorylation of the Retinoblastoma (Rb) protein.24 Also, 32D cl3 granulocytic differentiation was not inhibited, even though CBF regulates genes encoding myeloid differentiation markers, such as myeloperoxidase and neutrophil elastase.25 We speculated that in M4eo AML initial genetic hits occur which allow higher-level expression of CBFβ-SMMHC and thus, inhibition of differentiation.
We now provide data that support an alternative model for how CBFβ-SMMHC contributes to leukemogenesis. p53 is widely mutated in human malignancies and yet is rarely altered in de novo AML.26 p53 is induced in response to DNA strand breaks,27-29 leading to cell cycle arrest or apoptosis.28 30-34 Thus, normal cells are afforded an opportunity to repair the damage or to die, whereas cells harboring a p53 mutation may be more susceptible to developing additional genetic alterations, leading to malignant progression. When cells expressing CBFβ-SMMHC were treated with ionizing radiation (IR) or VP-16, p53 induction was markedly reduced. Similarly, CBFβ-SMMHC did not affect the rate of apoptosis resulting from IL-3 withdrawal, but did prolong the survival of Ba/F3 cells exposed to 10 Gy of IR or 3 to 8 μg/mL of VP-16. Reduced p53 induction and slowed apoptosis did not result from slowed proliferation alone, because these effects were not observed when the growth rate of Ba/F3 cells was slowed by transfer to media containing 10-fold–reduced IL-3. Also, p53 induction was normal in Ba/F3 cells expressing a mutant version of CBFβ-SMMHC that cannot interact with CBFα subunits, indicating that a CBF genetic target regulates p53 induction. Reduced p53 induction may be caused in part by direct inhibition of p53 gene transcription, because p53 mRNA levels were reduced by CBFβ-SMMHC. Attenuated p53 induction and slowed apoptosis may contribute to leukemogenesis by CBFβ-SMMHC.
MATERIALS AND METHODS
Cell culture and transduction.
Ba/F3 cell lines were maintained in RPMI 1640 supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (HI-FBS), 1 ng/mL murine IL-3 (R & D Systems, Minneapolis, MN), and penicillin-streptomycin (P/S). Zinc chloride was added to 100 μmol/L final concentration when indicated. Ba/F3 lines expressing CBFβ-SMMHC or containing the empty MTCB6 vector were maintained in 1.2 mg/mL G418. To remove IL-3, cells were washed twice with phosphate-buffered saline (PBS) and then resuspended in RPMI 1640 with HI-FBS and P/S.
DNA-damaging agents.
Cells were exposed to gamma irradiation using a Gammacell 40 irradiator (Atomic Energy of Canada, Ottawa, Canada). VP-16 (Sigma, St Louis, MO) was kept as 10 mg/mL stock in dimethyl sulfoxide. Once added, VP-16 was not removed from the cultures. Cells cultured with zinc for 18 hours before exposure to DNA-damaging agents were also cultured with zinc chloride during, and if necessary, after, these treatments.
Cell death and apoptosis analysis, clonogenic assay, and cell cycle analysis.
At designated times after exposure to IR or during exposure to VP-16, 5 × 105 cells were washed with PBS and stained with propidium iodide (PI) and fluorescein isothiocyanate (FITC)-Annexin V using the ApoAlert Kit (Clonetech, Palo Alto, CA). The cells were resuspended in 200 μL of binding buffer, and 5 μL of FITC-Annexin V and 5 μL of 50 μg/mL PI were then added. After incubation at room temperature for 10 minutes the cells were subject to fluorescence-activated cell sorter (FACS) analysis with a FACSort apparatus (Becton Dickinson, Mountain View, CA).
For clonogenic assays, cells were diluted to 0.3 to 2 cells/mL, based on the results of preliminary experiments, so that colonies would be present in less than 50% but more than 10% of the wells. After dilution, zinc and/or VP-16 was added as indicated and each group of cells was cultured in two 96-well dishes. Colony yields were scored 8 days later.
Viable cell counting was performed by enumerating cells which exclude trypan blue dye using a hemocytometer. Cell cycle analysis was performed after a 30-minute bromo-deoxy-uridine (BrdU) pulse using FITC–anti-BrdU/PI staining and FACScan analysis as described.24
Western blotting.
Total cellular protein extracts were subjected to Western blotting as described,35 except that 5% nonfat dried milk was included with both the primary and secondary antisera. Polyclonal rabbit antiserum raised against a CBFβ-GST fusion protein was used at a ratio of 1:1,000.36 p53 (Ab-3) and p21WAF1/CIP1(Ab-5) rabbit antisera (Oncogene Research, Cambridge, MA) were used at ratios of 1:200, 1:100, and 1:200, respectively. Bcl-2 (N-19), bcl-xL/S (L-19), and bax (N-20) antisera (Santa Cruz Biotechnology, Santa Cruz, CA) were used at a ratio of 1:100. The bands were visualized with an electrochemiluminescence kit (Amersham, Arlington Heights, IL). Filters were stripped as described.24
Northern blotting.
Poly-A–containing RNA was isolated using a Mini RiboSep ULTRA kit (Collaborative Biomedical Products, Bedford, MA). Five micrograms of mRNA was subjected to Northern blotting with the murine p53 and mdm2 cDNAs (kindly provided by C. Canman and M. Kastan, St Jude Children’s Research Hospital) and the human actin cDNA as described.25
Statistical methods.
The Student’s t-test was used to compare survival and apoptosis between experimental groups.
RESULTS
CBFβ-SMMHC reduces p53 induction by DNA damage.
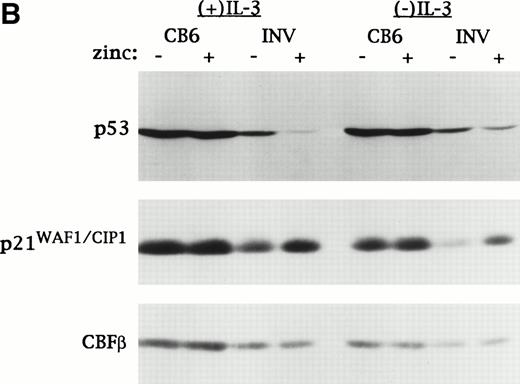
For these studies we used a pool of vector-transfected Ba/F3 cells (MTCB6) and two subclones expressing CBFβ-SMMHC from the MT promoter (MTINV-2 and MTINV-3).24 Ba/F3 lines were chosen in lieu of 32D cl3 lines for lines for these experiments because the latter were difficult to maintain, perhaps due to excess MT promoter leakiness. Parental Ba/F3 cells respond to IR by inducing p53 and undergoing either a G1 arrest in IL-3 or accelerated apoptosis in the absence of IL-3.31 To test the effect of CBFβ-SMMHC on p53 induction by IR, MTCB6, MTINV-2, or MTINV-3 cells were cultured for 18 hours in the presence or absence of zinc. These six cultures were then split into two, and one culture from each pair was irradiated to 10 Gy. Cultures initially treated with zinc were continued in zinc after irradiation. Ninety minutes later total cellular extracts were prepared and subjected to Western blotting for p53, p21, and CBFβ-SMMHC. The blot was stained with Fast Green (Sigma, St Louis, MO) as a loading control (Fig1A). In the absence of irradiation, exposure to zinc did not lead to increased p53 levels in either MTINV line, indicating that CBFβ-SMMHC alone cannot induce p53. IR induced p53 levels in the MTCB6 cells, and induction was similar in the presence or absence of zinc. p53 was induced similarly by IR in the MTINV-2 and MTINV-3 cells in the absence of zinc. However, when CBFβ-SMMHC was expressed at high levels as a result of zinc treatment, p53 induction by IR was reduced threefold to fourfold.
CBFβ-SMMHC inhibits p53 induction but induces p21WAF1/CIP1 in Ba/F3 cells exposed to IR or VP-16. (A) MTCB6 cells (vector-transfected) and MTINV-2 or MTINV-3 cells (expressing CBFβ-SMMHC from the MT promoter) were cultured ± 100 μmol/L zinc chloride for 18 hours. Half of the cells from each of these four cultures were then irradiated to 10 Gy. Cells exposed initially to zinc were continued in zinc after irradiation. Total cellular protein extracts were prepared 90 minutes later and electrophoresed on a 10% sodium dodecyl sulfate–polyacrylamide gel. The proteins were then transferred to a nitrocellulose filter, which was stained with Fast Green to control protein loaded (bottom panel) and subjected to Western blotting with antisera specific for murine p53, p21WAF1/CIP1, and CBFβ. The latter antisera detected CBFβ-SMMHC, which was induced by zinc in the MTINV-2 and MTINV-3 cells. (B) MTCB6 and MTINV-3 cells were cultured ± zinc for 18 hours. The cells were then exposed to 8 μg/mL VP-16 for 150 minutes in the presence or absence of IL-3. Total cellular extracts were then prepared and subjected to Western blotting for p53, p21, and CBFβ, with CBFβ levels serving as a control for protein loading.
CBFβ-SMMHC inhibits p53 induction but induces p21WAF1/CIP1 in Ba/F3 cells exposed to IR or VP-16. (A) MTCB6 cells (vector-transfected) and MTINV-2 or MTINV-3 cells (expressing CBFβ-SMMHC from the MT promoter) were cultured ± 100 μmol/L zinc chloride for 18 hours. Half of the cells from each of these four cultures were then irradiated to 10 Gy. Cells exposed initially to zinc were continued in zinc after irradiation. Total cellular protein extracts were prepared 90 minutes later and electrophoresed on a 10% sodium dodecyl sulfate–polyacrylamide gel. The proteins were then transferred to a nitrocellulose filter, which was stained with Fast Green to control protein loaded (bottom panel) and subjected to Western blotting with antisera specific for murine p53, p21WAF1/CIP1, and CBFβ. The latter antisera detected CBFβ-SMMHC, which was induced by zinc in the MTINV-2 and MTINV-3 cells. (B) MTCB6 and MTINV-3 cells were cultured ± zinc for 18 hours. The cells were then exposed to 8 μg/mL VP-16 for 150 minutes in the presence or absence of IL-3. Total cellular extracts were then prepared and subjected to Western blotting for p53, p21, and CBFβ, with CBFβ levels serving as a control for protein loading.
IR alone induced p21 in each of these lines. p21 levels remained high 90 minutes post-IR in the MTCB6 or MTINV cultures exposed to zinc (Fig1A), and were similarly elevated 3 hours or 4.5 hours post-IR as well (data not shown). Thus, CBFβ-SMMHC attenuated p53 induction, but not p21 induction, by IR.
To assess the effect of CBFβ-SMMHC on p53 and p21 induction by VP-16, we exposed MTCB6 or MTINV-3 cells to 8 μg/mL VP-16 for 150 minutes, in the presence or absence of IL-3. We used 150 minutes because p53 induction was delayed in response to VP-16 compared with IR (data not shown). Cells were cultured ± zinc, but all cultures were exposed to VP-16. CBFβ levels were also analyzed as a loading control (Fig 1B). Again, zinc did not affect the degree of p53 induction in MTCB6 cells. However, induction of p53 by VP-16 was reduced approximately threefold in MTINV-3 cells expressing CBFβ-SMMHC as a result of exposure to zinc, both in the presence and in the absence of IL-3. Induction of p21 by VP-16 was similar in MTCB6 cells regardless of the presence of zinc, and p21 levels were higher in MTINV-3 cells exposed to VP-16 when CBFβ-SMMHC expression was induced. As indicated by the CBFβ levels, paired samples, ± zinc, had nearly identical protein content.
CBFβ-SMMHC slows apoptosis induced by DNA damage.
CBFβ-SMMHC did not induce apoptosis in Ba/F3 cells cultured in IL-3.24 We sought to determine whether CBFβ-SMMHC alters the rate of apoptosis induced by IL-3 withdrawal. To assess survival, cells were stained with PI and subjected to FACScan analysis. On average, 94% of parental, MTCB6, or MTINV Ba/F3 cells excluded PI, and therefore were detected as viable by this assay (data not shown). The effect of zinc on the survival of MTINV-3 cells after IL-3 withdrawal was determined (Fig 2). Induction of CBFβ-SMMHC did not affect the proportion of MTINV-3 cells alive 6, 18, 24, or 42 hours after IL-3 withdrawal in duplicate experiments. At 6 hours survival was 88%, and at 18 hours survival was 39%. Also, removal of both IL-3 and serum for 6 hours reduced MTINV-3 cell survival to 78% (±5%) and induction of CBFβ-SMMHC further reduced their survival, to 57% (±6%) (data not shown). Thus, CBFβ-SMMHC did not slow the rate of cell death resulting from serum and/or IL-3 withdrawal.
CBFβ-SMMHC does not slow the rate of cell death resulting from IL-3 withdrawal in Ba/F3 cells. MTINV-3 cells were cultured ± zinc chloride for 18 hours. They were then washed three times with PBS and cultured without IL-3. Cells initially exposed to zinc were continued in zinc. At 0, 6, 18, 24, and 42 hours an aliquot of cells from each culture was analyzed for the proportion of surviving cells by PI staining and FACScan analysis. Mean results from duplicate experiments are shown. Standard errors (SEs) were smaller than the data points depicted.
CBFβ-SMMHC does not slow the rate of cell death resulting from IL-3 withdrawal in Ba/F3 cells. MTINV-3 cells were cultured ± zinc chloride for 18 hours. They were then washed three times with PBS and cultured without IL-3. Cells initially exposed to zinc were continued in zinc. At 0, 6, 18, 24, and 42 hours an aliquot of cells from each culture was analyzed for the proportion of surviving cells by PI staining and FACScan analysis. Mean results from duplicate experiments are shown. Standard errors (SEs) were smaller than the data points depicted.
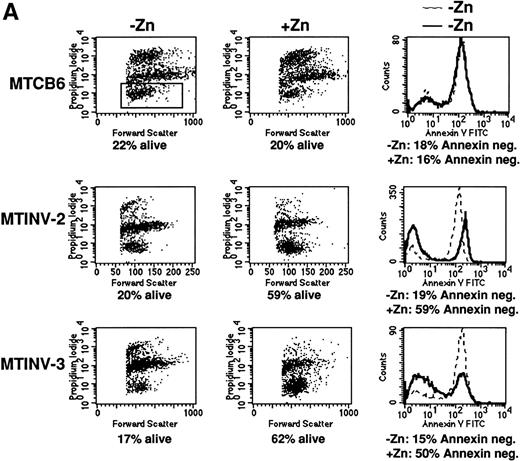
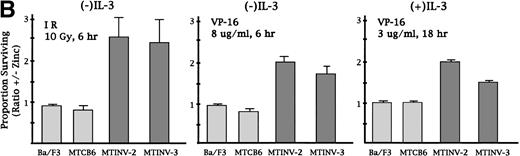
We next sought to determine whether CBFβ-SMMHC affected the rate of cell death induced by IR or VP-16. Parental Ba/F3, MTCB6, MTINV-2, and MTINV-3 cells were cultured in the presence or absence of zinc for 18 hours and then exposed to 10 Gy of IR. The cells were then continued in culture in the presence or absence of IL-3 for 6 hours. Cultures pretreated with zinc for 18 hours were continued in zinc after irradiation. The cells were stained with both PI and FITC-Annexin V before FACScan analysis, so that the proportion of viable cells and the proportion of cells undergoing apoptosis could be measured simultaneously. An example of data obtained for cells cultured without IL-3 after irradiation is shown in Fig 3A. The cluster of cells containing approximately 30 U or less of PI were considered to be alive. For all three lines, 10 Gy reduced the proportion of viable cells at 6 hours to about 20%, whereas IL-3 withdrawal alone only reduced viability to 88% at 6 hours (Fig2). The survival of MTCB6 cells was not affected by zinc, whereas the survival of both the MTINV-2 and MTINV-3 cells was increased about 3-fold when CBFβ-SMMHC was induced with zinc. These increases in survival were reflected in the proportion of cells that were negative for Annexin V binding. Because Annexin V binding is an early marker for apoptosis,37 38 this result indicates that cell death from irradiation is a result of apoptosis and that induction of CBFβ-SMMHC with zinc protected the cells from apoptosis. The results of the PI exclusion assay from three such experiments are shown in the left panel of Fig 3B. In the absence of IL-3, CBFβ-SMMHC increased survival of MTINV-2 and MTINV-3 cells irradiated to 10 Gy 2.5-fold, compared with parental Ba/F3 or MTCB6 cells (P < .05). Results with Annexin V paralleled these survival data (data not shown).
CBFβ-SMMHC slows apoptosis in Ba/F3 cells exposed to IR or VP-16. (A) MTCB6, MTINV-2, or MTINV-3 cells were cultured in the absence or presence of zinc for 18 hours. The cells were then removed from IL-3 and irradiated to 10 Gy, and 6 hours later the cells stained with PI and FITC-Annexin V and subjected to FACScan analysis. After irradiation, cells initially in zinc were continued in zinc. Data from a typical experiment are shown. The cluster of cells that excluded PI and was determined to be alive is boxed in the upper left panel. (B) The proportion of parental Ba/F3, MTCB6, MTINV-2, or MTINV-3 cells surviving 10 Gy or IR followed by culture in the absence of IL-3 for 6 hours, surviving 8 μg/mL VP-16 for 6 hours in the absence of IL-3, or surviving 3 μg/mL VP-16 for 18 hours in the presence of IL-3 are shown. Ratios of the proportion of cells surviving in the presence of zinc to the proportion of cells surviving in the absence of zinc were calculated for each cell line in each experiment. Results shown are the mean and SE from three experiments.
CBFβ-SMMHC slows apoptosis in Ba/F3 cells exposed to IR or VP-16. (A) MTCB6, MTINV-2, or MTINV-3 cells were cultured in the absence or presence of zinc for 18 hours. The cells were then removed from IL-3 and irradiated to 10 Gy, and 6 hours later the cells stained with PI and FITC-Annexin V and subjected to FACScan analysis. After irradiation, cells initially in zinc were continued in zinc. Data from a typical experiment are shown. The cluster of cells that excluded PI and was determined to be alive is boxed in the upper left panel. (B) The proportion of parental Ba/F3, MTCB6, MTINV-2, or MTINV-3 cells surviving 10 Gy or IR followed by culture in the absence of IL-3 for 6 hours, surviving 8 μg/mL VP-16 for 6 hours in the absence of IL-3, or surviving 3 μg/mL VP-16 for 18 hours in the presence of IL-3 are shown. Ratios of the proportion of cells surviving in the presence of zinc to the proportion of cells surviving in the absence of zinc were calculated for each cell line in each experiment. Results shown are the mean and SE from three experiments.
In the presence of IL-3, 10 Gy of IR did not induce significant cell death or apoptosis. Ba/F3 cells underwent a transient G1/S arrest and then resumed cycling when exposed to a similar dose of IR in IL-3.31 We also treated the cells with 50 Gy of IR in the presence of IL-3. At this dose, approximately 50% of the cells were alive 18 hours later, and induction of CBFβ-SMMHC did not confer protection from cell death (data not shown).
A similar approach was taken to determine whether CBFβ-SMMHC protected cells from VP-16. In preliminary experiments with control cells, doses and exposure times for VP-16 that allowed approximately 20% survival were identified. VP-16 was used at 3 μg/mL for 18 hours in the presence of IL-3 or at 8 μg/mL for 6 hours in the absence of IL-3. The results of three experiments are diagramed in the center and right panels of Fig 3B. PI exclusion results again correlated with FITC-Annexin V binding (data not shown). CBFβ-SMMHC protected Ba/F3 cells 1.8-fold from apoptosis and cell death occurring as consequence of VP-16 exposure, both in the presence and absence of IL-3. Protection from VP-16 at these doses was transient, because by 48 hours none of the cells were viable (data not shown).
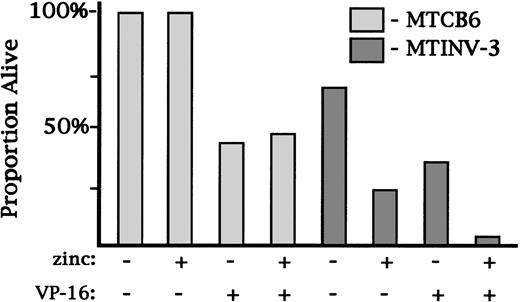
To determine whether CBFβ-SMMHC could prevent apoptosis and therefore increase clonogenic survival, we plated MTCB6 or MTINV-3 cells at low cell densities in 96-well dishes in the presence or absence of 0.04 μg/mL VP-16. The use of a dose of VP-16 in this range was necessary to allow Ba/F3 cells to survive and develop into colonies. Cells from each group were cultured ± zinc. The proportion of plated cells which gave rise to colonies 8 days later was enumerated (Fig4). The cloning efficiency of MTCB6 cells in the absence of zinc or VP-16 was 100%, and the cloning efficiency of MTINV-3 cells was 70%. Addition of zinc alone did not affect the colony yield from MTCB6 cells, but reduced the colony yield from MTINV-3 cells threefold, to 25%. This decrease was likely caused by a growth inhibitory effect of CBFβ-SMMHC. VP-16 reduced the colony yield from MTCB6 cells twofold, and zinc did not alter this effect. VP-16 alone also reduced the colony yield from MTINV-3 cells twofold, to 30% of the plated cells. Treatment of MTINV-3 cells with both zinc and VP-16 led to a colony yield of only 5%, actually lower than would have been predicted from a threefold reduction because of CBFβ-SMMHC and a twofold reduction because of VP-16. Thus, CBFβ-SMMHC did not prevent cell death resulting from VP-16 in this clonogenic assay.
CBFβ-SMMHC did not increase the clonogenic survival of Ba/F3 cells. MTCB6 or MTINV-3 cells were cultured ± zinc for 18 hours. VP-16 was then added to 0.04 μg/mL to an aliquot of each cell line. Culture was then continued in the same conditions in two 96-well dishes per group. Cells were plated at 0.3 to 2 cells/mL, based on the results of preliminary experiments, so that between 10% and 50% of the wells would develop a colony. The number of colonies were scored 8 days later, and the proportion of initially plated cells that formed colonies is shown for each cell line and culture condition.
CBFβ-SMMHC did not increase the clonogenic survival of Ba/F3 cells. MTCB6 or MTINV-3 cells were cultured ± zinc for 18 hours. VP-16 was then added to 0.04 μg/mL to an aliquot of each cell line. Culture was then continued in the same conditions in two 96-well dishes per group. Cells were plated at 0.3 to 2 cells/mL, based on the results of preliminary experiments, so that between 10% and 50% of the wells would develop a colony. The number of colonies were scored 8 days later, and the proportion of initially plated cells that formed colonies is shown for each cell line and culture condition.
Slowed proliferation does not reduce p53 induction or slow apoptosis resulting from VP-16 exposure.
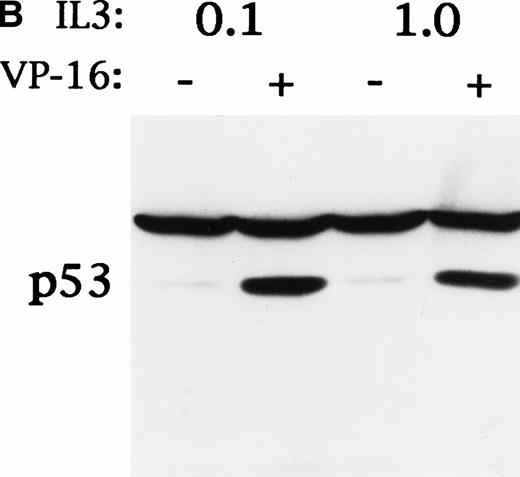
CBFβ-SMMHC increased the generation time of Ba/F3 cells from 9.6 hours to 14.4 hours and increased the ratio of G1 phase cells to S phase cells from 0.8 to 3.5.24 We sought to mimic this effect by culturing the cells in lower IL-3 concentrations (Fig5A). We found that after 48 hours of culture in 0.1 ng/mL IL-3, the generation time of Ba/F3 cells increased from 9.6 hours to 16.5 hours, and the G1/S ratio increased from 0.6 to 3.1, without evident apoptosis. We exposed parental Ba/F3 cells cultured in either 1.0 or 0.1 ng/mL IL-3 to 8 μg/mL VP-16 for 6 hours and assessed survival by PI exclusion. Slowed proliferation during G1 did not protect the cells from VP-16–mediated cell death. We also assessed p53 induction in cells cultured similarly, ± VP-16, for 150 minutes (Fig 5B). Slowed proliferation during G1 did not attenuate p53 induction by VP-16. Equal loading was evident from the levels of a cross-reactive band.
Ba/F3 cells cultured in reduced IL-3 had a slowed G1 to S transition, but did not have reduced p53 induction or increased survival in response to VP-16. (A) Parental Ba/F3 cells were cultured in 1.0 ng/mL IL-3, which allows optimum growth, or in 0.1 ng/mL IL-3. After 48 hours, cell generation time was determined by enumerating the number of viable cells present in the cultures on subsequent days. To determine the ratio of the proportion of cells in G1 phase to the proportion of cells in S phase (G1/S), the cells were exposed to 30 μmol/L BrdU for 30 minutes, fixed, stained with anti–BrdU-FITC and PI, and subjected to FACScan analysis. The proportion of cells surviving exposure to 8 μg/mL VP-16 for 6 hours was determined as in Fig 3. (B) Ba/F3 cells cultured in 1.0 ng/mL or 0.1 ng/mL IL-3 for 48 hours were continued in cultured ± 8 μg/mL VP-16 for 150 minutes. Total cellular protein extracts were then obtained and subjected to Western blot analysis for p53.
Ba/F3 cells cultured in reduced IL-3 had a slowed G1 to S transition, but did not have reduced p53 induction or increased survival in response to VP-16. (A) Parental Ba/F3 cells were cultured in 1.0 ng/mL IL-3, which allows optimum growth, or in 0.1 ng/mL IL-3. After 48 hours, cell generation time was determined by enumerating the number of viable cells present in the cultures on subsequent days. To determine the ratio of the proportion of cells in G1 phase to the proportion of cells in S phase (G1/S), the cells were exposed to 30 μmol/L BrdU for 30 minutes, fixed, stained with anti–BrdU-FITC and PI, and subjected to FACScan analysis. The proportion of cells surviving exposure to 8 μg/mL VP-16 for 6 hours was determined as in Fig 3. (B) Ba/F3 cells cultured in 1.0 ng/mL or 0.1 ng/mL IL-3 for 48 hours were continued in cultured ± 8 μg/mL VP-16 for 150 minutes. Total cellular protein extracts were then obtained and subjected to Western blot analysis for p53.
CBFβ-SMMHC does not alter bcl-2, bcl-xL, bcl-xS, or bax levels.
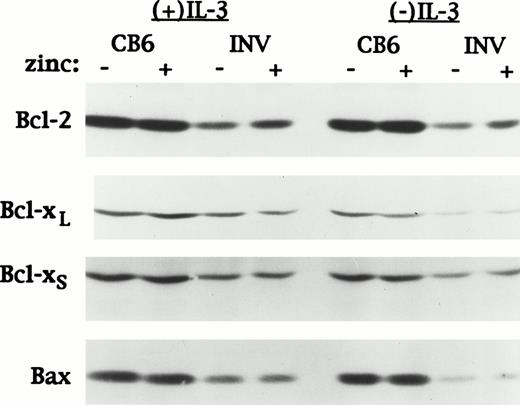
Apoptosis is regulated in part by a balance between antiapoptotic bcl-2 family members such as bcl-2 and bcl-xL and proapoptotic family members such as bax and bcl-xS.39 The bax gene is directly activated by p53.40 The same extracts used to assess the affect of VP-16 on p53 and p21 levels in Fig 1C were analyzed for expression of bcl-2, bcl-xL, bcl-xS, and bax (Fig 6). The levels of these proteins were unaffected by zinc, in MTCB6 cells, or by induction of CBFβ-SMMHC, in MTINV-3 cells. Therefore, slowed apoptosis in CBFβ-SMMHC–expressing cells exposed to VP-16 is not caused by altered expression of bcl-2, bcl-xL, bcl-xS, or bax.
CBFβ-SMMHC does not affect bcl-2, bcl-xL, bcl-xS, or bax expression in Ba/F3 cells exposed to VP-16. The same MTCB6 and MTINV-3 extracts described in Fig 1C were subjected to Western blotting for bcl-2, bcl-xL, bcl-xS, and bax.
CBFβ-SMMHC does not affect bcl-2, bcl-xL, bcl-xS, or bax expression in Ba/F3 cells exposed to VP-16. The same MTCB6 and MTINV-3 extracts described in Fig 1C were subjected to Western blotting for bcl-2, bcl-xL, bcl-xS, and bax.
A CBFβ-SMMHC variant that cannot bind CBFα does not attenuate p53 induction.
CBFβ(Δ2-11)-SMMHC, which lacks amino acids 2-11 of the CBFβ segment of CBFβ-SMMHC, does not interact with CBFα subunits, inhibit CBF DNA binding, or inhibit Ba/F3 cell proliferation.17 41 MTINV (βΔ2-11) cells, which express this CBFβ-SMMHC variant from the MT promoter, were cultured in the presence or absence of zinc for 18 hours. An aliquot from each culture was then irradiated 10 Gy. Induction of p53 90 minutes post-IR and of CBFβ(Δ2-11)-SMMHC by zinc was then assessed by Western blotting (Fig 7A). Induction of p53 was robust, even when expression of CBFβ(Δ2-11)-SMMHC was induced. This result indicates that CBFβ-SMMHC attenuates p53 induction in Ba/F3 cells by interfering with activation of a critical gene or set of genes by CBF.
CBFβ-SMMHC must interact with CBF and inhibit CBF transactivation to attenuate p53 induction, and reduces p53 mRNA expression in Ba/F3 cells. (A) MTINV(β▵2-11) Ba/F3 cells, which express a CBFβ-SMMHC variant that cannot bind CBF, were cultured ± 100 μmol/L zinc chloride for 18 hours. Half of the cells from each of these cultures were then irradiated to 10 Gy. Cells exposed initially to zinc were continued in zinc after irradiation. Total cellular protein extracts were prepared 90 minutes later and subjected to Western blotting with antisera specific for murine p53 and CBFβ-SMMHC. Fast Green staining of the blot is shown as a control for protein loading. (B) MTCB6 and MTINV-3 cells were cultured ± zinc for 18 hours. Poly-A–containing mRNA was then prepared from each culture. Five micrograms of each sample was subjected to electrophoresis on a 1% formaldehyde-agarose gel, transferred to a nylon membrane, and probed sequentially for p53, mdm2, and actin.
CBFβ-SMMHC must interact with CBF and inhibit CBF transactivation to attenuate p53 induction, and reduces p53 mRNA expression in Ba/F3 cells. (A) MTINV(β▵2-11) Ba/F3 cells, which express a CBFβ-SMMHC variant that cannot bind CBF, were cultured ± 100 μmol/L zinc chloride for 18 hours. Half of the cells from each of these cultures were then irradiated to 10 Gy. Cells exposed initially to zinc were continued in zinc after irradiation. Total cellular protein extracts were prepared 90 minutes later and subjected to Western blotting with antisera specific for murine p53 and CBFβ-SMMHC. Fast Green staining of the blot is shown as a control for protein loading. (B) MTCB6 and MTINV-3 cells were cultured ± zinc for 18 hours. Poly-A–containing mRNA was then prepared from each culture. Five micrograms of each sample was subjected to electrophoresis on a 1% formaldehyde-agarose gel, transferred to a nylon membrane, and probed sequentially for p53, mdm2, and actin.
CBFβ-SMMHC reduced p53 mRNA levels.
If CBF activates the p53 gene, then CBFβ-SMMHC would be predicted to reduce p53 mRNA levels. Alternatively, if increased mdm2 protein levels account for decreased p53 induction by DNA damage, then CBFβ-SMMHC might increase mdm2 mRNA levels. To test these possibilities, MTCB6 and MTINV-3 cells were cultured for 18 hours in the presence or absence of zinc. Poly-A–containing mRNA was then isolated from each culture, and 5 μg of each sample was subjected to Northern blotting for p53, mdm2, and actin (Fig 7B). Zinc alone did not affect p53 or mdm2 mRNA levels in CB6 cells. However, in MTINV-3 cells induction of CBFβ-SMMHC with zinc reduced the level of p53 mRNA relative to actin mRNA approximately fourfold, but did not alter the level mdm2 mRNA. Similarly, in MTINV-3 cells treated with VP-16, induction of CBFβ-SMMHC reduced p53 mRNA levels approximately twofold (data not shown).
DISCUSSION
Our results suggest that CBFβ-SMMHC contributes to leukemogenesis by attenuating the p53 response to DNA damage. Several studies indicate that apoptosis as a consequence of DNA damage in hematopoietic cells requires induction of p53. p53 is required for apoptosis of thymocytes exposed to IR or VP-16.32,33 Expression of exogenous p53 in the M1 myeloid leukemia line induces apoptosis.42 Also, inhibition of p53 induction, by expression of the viral E6 protein, inhibits apoptosis in Ba/F3 cells exposed to IR and withdrawn from IL-3.31
Because p53 mutations are rare in de novo AMLs,26 reduced induction of p53 by CBFβ-SMMHC might serve as an alternative mechanism to interfere with a p53-dependent apoptotic pathway. Indeed, reduced p53 induction resulting from CBFβ-SMMHC expression correlated with slowed apoptosis in Ba/F3 cells treated with IR or VP-16. Slowed apoptosis might enable a rare cell to express additional genetic changes, survive, and progress towards transformation. Also, in cooperation with other mutations already present in preleukemic cells, reduced levels of p53 resulting from CBFβ-SMMHC expression might directly stimulate survival. Reduced p53 levels stimulated the survival of solid tumor cells subjected to hypoxia.30 Perhaps a similar selective advantage can contribute to leukemogenesis in vivo independent of DNA damage.
The doses of IR or VP-16 we used to induce p53 represent a greater degree of DNA damage than most leukemic precursors are likely subjected to in vivo. p53 was not induced in Ba/F3 cells exposed to 0.04 μg/mL VP-16 for 1.5, 3, 4.5, or 24 hours (data not shown). Perhaps p53 is induced more effectively by physiological levels of DNA damage in preleukemic cells.
Increased expression of p21WAF1/CIP1 occurred despite reduced p53 expression in cells treated with IR or VP-16. In some experiments, induction of CBFβ-SMMHC led to even higher levels of p21 (Fig 1B), as reported previously.24 p21 protects colon carcinoma cells from apoptosis.34 However, expression of the viral E7 protein in Ba/F3 cells, which mimics p21 overexpression by inactivating Rb and releasing E2F proteins, does not sensitize them to IR in the presence or absence of IL-3.43 This finding suggests that the reduced radio- and chemo-sensitivity of CBFβ-SMMHC–expressing Ba/F3 cells results from attenuated p53 induction and not from increased p21 expression. Critically testing whether decreased p53 induction is the primary cause of slowed apoptosis after DNA damage in CBFβ-SMMHC–expressing cells would require coexpression of exogenous p53, in a manner dependent on DNA damage.
CBFβ-SMMHC decreases the proportion of Ba/F3 cells in S phase 1.7-fold,24 raising the possibility that protection from apoptosis was caused by decreased exposure to DNA damage during S phase. However, IR damages DNA in all cell cycle phases, and exposure to VP-16 was for 18 hours in the presence of IL-3, allowing sufficient time for all cells to enter S phase.
CBFβ-SMMHC increases the proportion of cells in G1 phase 1.7-fold, but does not affect the proportion of cells in G2.24 A lengthened G1 might allow increased time for DNA repair before S phase. However, DNA repair processes are rapid, and time spent in the G2 phase may be a more critical determinant of radiation and VP-16 sensitivity.44-46 Also, we found that slowing the G1 to S transition in Ba/F3 cells by reducing IL-3 in the culture media did not attenuate p53 induction or stimulate survival when the cells where exposed to VP-16.
The clinical observation that about 60% of patients with AMLs associated with inv(16) or t(8;21) are cured by conventional chemotherapy, whereas only about 30% of the remaining AML patients are cured47 suggests that CBFβ-SMMHC and AML1-ETO might sensitize hematopoietic cells to chemotherapy. Yet we observed protection from IR and VP-16, and from Ara-C and daunomycin as well (data not shown). However, the protection observed was modest and was only compared with that obtained using control Ba/F3 lines. Ba/F3 cells expressing bcr-abl(p210) were more resistant than MTINV lines to IR or VP-16 (M.B., X.W., and A.D.F., unpublished observation, August 1997). Similarly, subsets of AML that are generally refractory to chemotherapy may have stronger protective mechanisms than AMLs expressing CBFβ-SMMHC or AML1-ETO.
We have begun to investigate the mechanism of reduced p53 induction as a consequence of CBFβ-SMMHC expression. A CBFβ-SMMHC variant that cannot interact with CBFα subunits did not attenuate p53 induction (Fig 6A) and did not confer protection from IR or VP-16 (M.B. and A.D.F., unpublished observation, August 1997). These findings suggest that CBFβ-SMMHC reduces p53 induction by reducing the expression of a gene regulated by CBF. Consistent with this model, Ba/F3 cells expressing AML1-ETO also had reduced p53 induction in response to IR (M.B. and A.D.F., unpublished observation, May 1998). If direct binding of p53 by CBFβ-SMMHC is required to decrease p53 expression, then cells expressing CBFβ(Δ2-11)-SMMHC might have been expected to have attenuated p53 induction. Our Northern blot data suggest that reduction of p53 mRNA levels partly accounts for attenuated p53 induction in the presence of CBFβ-SMMHC. Additional experiments will be required to determine whether the p53 gene is regulated by CBF. Canonical CBF consensus-binding sites, 5′-ACC(A/G)CA-3′, are present at bps 656 and 1912 of the murine p53 intron 1 (GenBank #U10088) and at bp 1348 of the human intron 1 (GenBank #X54156), but not in the murine or human p53 promoter regions between bps −420 and +1.48 Even if CBF does not regulate the p53 gene, CBF oncoproteins might bind these or other sites and repress its transcription.
p53 protein levels are controlled by several mechanisms, including amino-terminal phosphorylation and mdm2-mediated degradation.49-51 Our future experiments will also elucidate the effect of CBFβ-SMMHC on these pathways and will identify the genetic targets of CBF and CBFβ-SMMHC responsible for the phenotypes of hematopoietic cells expressing CBFβ-SMMHC. Therapies designed to inhibit the effect of CBFβ-SMMHC on p53 induction and apoptosis might prove useful for leukemias expressing CBF oncoproteins.
ACKNOWLEDGMENT
We thank C. Canman and M. Kastan for Ba/F3 cells, for the p53 and mdm2 cDNA, and for helpful discussions.
Supported in part by National Institutes of Health Grant No. HL51388 (A.D.F.). A.D.F. is a Leukemia Society Scholar, and P.P.L. is a Leukemia Society Special Fellow.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Alan D. Friedman, MD, Johns Hopkins Oncology Center, Room 3-109, 600 N Wolfe St, Baltimore, MD 21287.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal