Abstract
The PAX-5 gene codes for the transcription factor BSAP, which is expressed throughout B-cell development. Although loss-of-function mutation in the mouse showed an essential role forPax-5 in early B lymphopoiesis, gain-of-function mutations have implicated the human PAX-5 gene in the control of late B-cell differentiation. PAX-5 (on 9p13) has been involved together with the immunoglobulin heavy-chain (IgH) gene (on 14q32) in the recurring t(9;14)(p13;q32) translocation that is characteristic of small lymphocytic lymphoma with plasmacytoid differentiation. Here we have characterized a complex t(2;9;14)(p12;p13;q32) translocation present in a closely related non-Hodgkin’s lymphoma referred to as splenic marginal zone lymphoma (MZL). In this MZL-1 translocation, the two promoters of PAX-5 were replaced on the derivative chromosome 14 by an immunoglobulin switch Sμ promoter that was linked to the structural PAX-5 gene upstream of its translation initiation codon in exon 1B. Expression analyses confirmed thatPAX-5 transcription was upregulated due to efficient initiation at the Sμ promoter in the malignant B lymphocytes of patient MZL-1. For comparison we have analyzed PAX-5 expression in another B-cell lymphoma, KIS-1, indicating that transcription from the distalPAX-5 promoter was increased in this tumor in agreement with the previously characterized translocation of the immunoglobulin Eμ enhancer adjacent to PAX-5 exon 1A. In both lymphomas, the J-chain gene, which is thought to be under negative control by BSAP, was not expressed, whereas transcription of the putative target genep53 was unaffected by PAX-5 overexpression. Together these data indicate that the t(9;14)(p13;q32) translocation contributes to lymphoma formation as a regulatory mutation that leads to increasedPAX-5 expression in late B-cell differentiation due to promoter replacement or enhancer insertion.
TUMORS OF HEMATOPOIETIC origin are frequently associated with specific chromosomal translocations that result in the activation of proto-oncogenes controlling differentiation, proliferation, or cell survival. In B-cell non-Hodgkin’s lymphoma (NHL), these proto-oncogenes are often deregulated by translocation adjacent to regulatory elements of immunoglobulin genes. Most frequently, the immunoglobulin heavy-chain (IgH) gene on chromosome 14q32 is involved in NHL that encompasses a large spectrum of diseases with diverse morphological and clinical manifestations. Different subtypes of NHL have been correlated with characteristic chromosomal translocations and thus with the activation of specific oncogenes. For instance, the c-MYC gene on 8q24 is translocated in Burkitt’s lymphoma, the BCL-1(PRAD1) gene coding for cyclin D1 (on 11q13) is involved in mantle cell lymphoma, and the antiapoptotic BCL-2 gene on 18q21 is activated in follicular lymphoma. In addition, the BCL-6gene coding for a zinc finger transcription factor is frequently altered by rearrangement in diffuse large-cell lymphoma.1-3
The t(9;14)(p13;q32) translocation has been closely associated with a relatively rare subtype of NHL that was initially referred to as small lymphocytic lymphoma (SLL) with plasmacytoid differentiation.4 To date, this entity is listed under B-SLL in the revised REAL classification of lymphomas.5,6 This subtype of NHL is thought to originate from peripheral B lymphocytes that have been stimulated to undergo plasma cell differentiation.4,5 Consequently, this form of B-SLL is characterized by the expression of cytoplasmic and cell-surface immunoglobulin, by immunoglobulin secretion, and by an indolent clinical course that may progress with time towards a more aggressive large-cell lymphoma.4,5 The t(9;14)(p13;q32) translocation was first reported in the diffuse large-cell lymphoma KIS-1.7 The molecular characterization of this translocation showed that the IgH locus was translocated in a head-to-head configuration to the PAX-5 gene on chromosome 9p13. As a result, the potent Eμ enhancer of the IgH gene was brought into close proximity to the PAX-5promoters.8 In addition, this analysis showed that transcription of the PAX-5 gene is initiated from two promoters and thus results in splicing of two alternative 5′ exons (1A or 1B) to common coding sequences (exons 2-10).8 Molecular analysis of a second t(9;14) translocation identified breakpoints in the switch Sμ region of the IgH locus and in the downstream exon 1B of PAX-5.9 Moreover, a consistent involvement of the PAX-5 locus in t(9;14) translocations of B-SLL was confirmed by fluorescence in situ hybridization (FISH) analyses using a 1 Mb-long yeast artificial chromosome probe spanning the entire PAX-5 locus.9
PAX-5 codes for the transcription factor BSAP, which recognizes its target genes via the highly conserved paired domain.10Targeted inactivation of Pax-5 in the mouse germline showed an important role for this gene in B-cell and midbrain development.11 During B lymphopoiesis, the Pax-5gene is expressed from the earliest B-lineage–committed precursor cell up to the mature B-cell stage and is subsequently downregulated during terminal plasma cell differentiation.12,13 Consistent with this expression pattern, Pax-5 is required for progression of B-cell development beyond an early progenitor cell stage.14Although several genuine BSAP (Pax-5) target genes have been identified at the pro-B–cell stage by genetic means,15 little is known about the function of Pax-5 in late B-cell differentiation. At the mature B-cell stage, BSAP (Pax-5) has been implicated in the control of cell proliferation and in the regulation of the J-chain gene, the IgH germline ε promoter, and the 3′ enhancers of the IgH and Igκ loci.10 Because gain-of-function mutations are likely to provide new insight into the role of Pax-5 in late B-cell differentiation, we were interested in studying the effect of t(9;14)(p13;q32) translocations on the expression of PAX-5 and its target genes in human NHL.
Here, we describe the molecular characterization of a complex t(2;9;14)(p12;p13;q32) translocation that was identified in a patient diagnosed with marginal-zone lymphoma (MZL), a disease closely related to B-SLL. This translocation generated a novel transcription unit by linking a switch-Sμ promoter of the IgH locus (14q32) to the structural PAX-5 gene in exon 1B (9p13). Efficient transcription initiation from the Sμ promoter was directly shown in the malignant B lymphocytes that constituted the majority of mononuclear blood cells (MNC) in patient MZL-1. In contrast,PAX-5 transcription was increased from the upstream exon 1A promoter in the KIS-1 lymphoma, thus reflecting the close proximity of this promoter to the translocated Eμ enhancer in these cells. Hence, deregulation of PAX-5 expression by t(9;14) translocations can be brought about by either promoter replacement or enhancer insertion. The possibility is discussed that the expression of the PAX-5gene under the control of IgH regulatory elements contributes to lymphomagenesis by interfering with the inactivation ofPAX-5 transcription and thus with terminal plasma cell differentiation.
MATERIALS AND METHODS
Clinical data.
An 83-year-old woman presented with anemia, marked leukocytosis, and splenomegaly at the Hematology Department of the Vienna Medical School (Vienna, Austria) in December 1995. The white blood cell count was 31 × 109/L with 63% atypical lymphocytes that were CD19+, IgM+, κ+, and displayed low CD5 expression (Fig 1A). The serum did not contain clonal IgM paraprotein. Bone marrow histology showed multifocal lymphoid infiltrates within an otherwise normal parenchyma. These infiltrates accounted for ∼40% of the total bone marrow cells and were predominantly concentrated in the central part of the marrow spaces. The infiltrating cells were frequently located in clusters within the sinusoids of the bone marrow (Fig 2A,B), which were recently described as a characteristic hallmark of splenic marginal zone lymphoma (MZL).16 The malignant cells in the blood were characterized as medium-sized or moderately enlarged B lymphocytes with round or slightly indented nuclei, clear cytoplasm, and frequent cytoplasmic projections or thin villi (Fig 2D). Based on all these data, the B-CLL tumor of this patient was diagnosed as splenic MZL in agreement with the REAL classification.5 6 This patient, referred to as MZL-1, was in excellent clinical condition and received only supportive treatment with erythropoietin until September 1996, when thrombocytopenia and B-cell symptoms required chemotherapeutic treatment. The patient received three courses of oral therapy with chlorambucil (20 mg on day 1) and prednisone (50 mg on days 1 to 5). After this treatment, the blood-cell count improved dramatically, because the patient had a normal leukocyte count with 0% to 10% atypical lymphocytes in monthly controls until July 1997. All analyses described herein were performed with MNC of patient MZL-1 before chemotherapy. Informed consent for the scientific use of these cells was obtained from the patient.
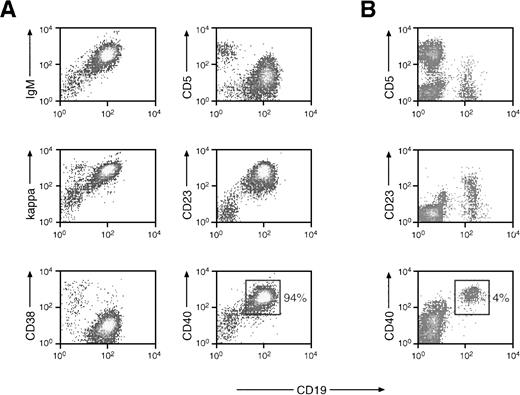
The mononuclear blood cells of patient MZL-1 consist predominantly of malignant B lymphocytes. (A) Flow cytometric analysis of peripheral blood cells of patient MZL-1. Mononuclear cells from the blood of patient MZL-1 were analyzed by flow cytometry using a FITC-conjugated anti-CD19 (HD37) antibody in combination with a PE-labeled anti-CD5 (DK23) antibody or with unlabeled anti-IgM (AF6), anti-κ (6E1), anti-CD23 (9P25), anti-CD38 (HB-7), and anti-CD40 (MoAb89) antibodies. The unlabeled MoAbs were visualized by indirect staining with a secondary PE-conjugated goat antimouse antibody. (B) Flow cytometric analysis of peripheral mononuclear cells of a normal individual. The percentage of CD40+ B lymphocytes is indicated.
The mononuclear blood cells of patient MZL-1 consist predominantly of malignant B lymphocytes. (A) Flow cytometric analysis of peripheral blood cells of patient MZL-1. Mononuclear cells from the blood of patient MZL-1 were analyzed by flow cytometry using a FITC-conjugated anti-CD19 (HD37) antibody in combination with a PE-labeled anti-CD5 (DK23) antibody or with unlabeled anti-IgM (AF6), anti-κ (6E1), anti-CD23 (9P25), anti-CD38 (HB-7), and anti-CD40 (MoAb89) antibodies. The unlabeled MoAbs were visualized by indirect staining with a secondary PE-conjugated goat antimouse antibody. (B) Flow cytometric analysis of peripheral mononuclear cells of a normal individual. The percentage of CD40+ B lymphocytes is indicated.
Morphological characterization of the malignant B lymphocytes in the bone marrow and blood of patient MZL-1. (A,B) Immunohistochemical analysis of bone marrow biopsies. Sections were stained with anti-CD79a [Ig-] (A) and anti-DBA.44 (B) antibodies. Both antibodies stained lymphoid cells that were predominantly located within the sinusoids of the bone marrow. The sinusoids are indicated by arrowheads (cross-section) or arrows (oblique [A] and longitudinal [B] sections), respectively. The clear spheroidal areas are occupied by fat cells. (C) Staining of CD5+CD19+peripheral blood cells. CD5+CD19+ cells were sorted from mononuclear cells of the peripheral blood and stained with the anti-DBA.44 antibody.25 About 70% of all sorted cells were strongly positive for the DBA.44 antigen. (D) Morphology of the malignant B lymphocytes. Blood smears of patient MZL-1 were analyzed by the modified Wright staining. Four representative B cells are shown at high magnification together with adjacent erythrocytes. These B cells contain a round or ovoid nucleus that is surrounded by a moderately increased amount of basophilic cytoplasm. Most malignant B cells have the appearance of villous lymphocytes as evidenced by the presence of cytoplasmic projections or short thin villi, which are often asymmetrically located on one side of the cell.
Morphological characterization of the malignant B lymphocytes in the bone marrow and blood of patient MZL-1. (A,B) Immunohistochemical analysis of bone marrow biopsies. Sections were stained with anti-CD79a [Ig-] (A) and anti-DBA.44 (B) antibodies. Both antibodies stained lymphoid cells that were predominantly located within the sinusoids of the bone marrow. The sinusoids are indicated by arrowheads (cross-section) or arrows (oblique [A] and longitudinal [B] sections), respectively. The clear spheroidal areas are occupied by fat cells. (C) Staining of CD5+CD19+peripheral blood cells. CD5+CD19+ cells were sorted from mononuclear cells of the peripheral blood and stained with the anti-DBA.44 antibody.25 About 70% of all sorted cells were strongly positive for the DBA.44 antigen. (D) Morphology of the malignant B lymphocytes. Blood smears of patient MZL-1 were analyzed by the modified Wright staining. Four representative B cells are shown at high magnification together with adjacent erythrocytes. These B cells contain a round or ovoid nucleus that is surrounded by a moderately increased amount of basophilic cytoplasm. Most malignant B cells have the appearance of villous lymphocytes as evidenced by the presence of cytoplasmic projections or short thin villi, which are often asymmetrically located on one side of the cell.
Immunohistochemistry.
The formalin-fixed, decalcified bone marrow trephine biopsy was paraffin-embedded, sectioned, and stained with hematoxylin and eosin. For immunohistochemistry, the sections were pretreated by microwaving in citrate buffer (10 mmol/L, pH 6.0) twice for 5 minutes at 600 W each. The sections were incubated with the monoclonal antibodies (MoAbs) DBA.44 (DAKO, Glostrup, Denmark; 1:10) and CD79a (DAKO; 1:25) for 1 hour followed by biotinylated horse antimouse immunoglobulin as the secondary antibody and by Vectorstain Elite reagent (Vector Labs, Burlingame, CA) and 3-amino-9-ethyl-carbazole (for DBA.44) or 3,3’-diamino-benzidine (for CD79a) as chromogens in the presence of H2O2.
Frozen mononuclear cells of patient MZL-1 were thawed and sorted for CD5+ CD19+ cells on a FACS Vantage TSO flow cytometer (Becton Dickinson). Cytospin preparations of the sorted CD5+ CD19+ cells were fixed with acetone on glass slides for 10 minutes. The cells were incubated with the anti-DBA.44 antibody for 1 hour, followed by incubation with biotinylated horse antimouse immunoglobulin as a secondary antibody, and then by alkaline phosphatase-conjugated streptavidin (Super Sensitive HRP Label, Biogenex, San Ramon, CA). The reaction product was visualized by new-fuchsin (DAKO) as a chromogen.
Cytogenetic analysis.
Bone marrow cells of patient MZL-1 were cultured and stimulated as previously described.17 Metaphase chromosomes were prepared and G-banded with Trypsin/Giemsa according to standard techniques and karyotyped with a PSI image analysis system (PSI, Halladale, UK). All 20 metaphase nuclei analyzed showed the same pseudodiploid karyotype: 46, XX, dup(1)(q12q32) add(1)(?p36), t(2;9;14)(p12;p13;q32), del(7)(q22q33), der(17)t(12;17)(q21;p12).
Cell lines.
The origin and culture conditions for the human B-cell lines BJA-B, Raji, Namalwa, Ramos, and HS-Sultan, the myeloma cell line RPMI 8226, and the cervical carcinoma cell line HeLa have previously been described.12,18 The human diffuse large-cell lymphoma line KIS-119 and the myeloma cell line U226 (obtained from the German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany) were grown in RPMI 1640 medium supplemented with 10% fetal calf serum and 1 mmol/L glutamine (GIBCO BRL).
Antibodies and flow cytometric analysis.
MoAb directed against the following human cell surface markers were used for flow cytometric analysis. The FITC-conjugated anti-CD19 (HD37) and PE-conjugated anti-CD5 (DK23) antibodies were purchased from DAKO, the anti-CD38 (HB-7) MoAbs from Becton Dickinson (San Jose, CA), and the anti-CD23 (9P25), anti-CD40 (MoAb89), anti-IgM (AF6) and anti-Igκ (6E1) from Immunotech (Marseille, France). A PE-conjugated F(ab′)2 goat antimouse IgG antibody (Immunotech) was used for indirect staining of the unlabeled MoAbs.
Mononuclear cells were prepared from the peripheral blood of patient MZL-1 and of a normal control individual by purification on Ficoll gradients. Single-cell suspensions were incubated with the unlabeled MoAb, washed, stained with the PE-conjugated F(ab’)2 goat antimouse IgG antibody, washed, and incubated with the FITC-conjugated anti-CD19 (HD37) antibody followed by analysis in a FACScan flow cytometer (Becton Dickinson), as previously described.11
Preparation of genomic DNA.
DNA was isolated from ∼107 cells of cultured human cell lines, from mononuclear cells of peripheral blood, or from frozen lymph node biopsies that were ground into a powder in a mortar under liquid nitrogen. These cell pellets were lysed and digested overnight at 37°C in 500 μL of a buffer containing 150 mmol/L NaCl, 50 mmol/L Tris pH 8.0, 1 mmol/L EDTA, 1% sodium dodecyl sulfate (SDS), and 2 mg/mL of proteinase K. Genomic DNA was subsequently isolated by phenol-chloroform extraction and ethanol precipitation.
Southern blot analysis.
Restriction digestions were performed overnight with 10 μg of genomic DNA at 37°C in a volume of 400 μL followed by ethanol precipitation and agarose gel electrophoresis. The separated DNA was blotted onto Gene-Screen Plus membranes (DuPont, Boston, MA) by the alkaline transfer method. DNA probes were radiolabeled by random priming and hybridized overnight to the immobilized DNA at 65°C in Church buffer (0.5 mol/L sodium phosphate pH 7.2, 7% SDS, 1 mmol/L EDTA pH 8.0). Posthybridization washes were performed at 65°C for 3 × 20 minutes in 40 mmol/L sodium phosphate pH 7.2, 1% SDS. The same membranes were rehybridized with different probes after stripping in 0.1 mol/L NaOH/0.1% SDS for 20 minutes and neutralization in 0.5 mol/L Tris pH 8, 0.1% SDS for 15 minutes.
The 1.6-kb SacI-BstEII DNA fragment of clone p8.5-17 was used as the exon 1A-specific PAX-5probe #1 (pKIS). The exon 1B-specific probe #2 consisted of the 1.8-kbPstI DNA insert of clone pD10-P1.8,8 whereas the intron 2-specific probe #3 corresponded to a 1.23-kbSacI-HindIII DNA fragment of clone pD10-N2.3.8
Cloning of translocation breakpoints.
HindIII-digested DNA prepared from MNC of patient MZL-1 was fractioned on a 0.9% low-melting agarose gel, and the DNA fragments of 8 to 9 kb size were excised and purified. The size-fractionatedHindIII fragments were ligated into the HindIII site of the λZAP Express vector (Stratagene) and packaged into phage by the use of the Gigapack III Gold packaging extracts (Stratagene). A library of 2−4 × 10 6recombinant phages was separately screened with the32P-labeled exon 1B-specific probe #2 and the intron 2-specific probe #3 for phages carrying the two reciprocal translocation events. After plaque purification, the 8.6-kb DNA insert corresponding with the translocation breakpoint on chromosome der(9) was excised in vivo with the f1 helper phage R408 (Stratagene, La Jolla, CA) from the λZAP Express vector into plasmid pBK-CMV to obtain clone pTBP-der(9). The 8.9 kb HindIII fragment carrying the reciprocal translocation breakpoint was recloned from phage DNA into the pBluescript II KS(+) vector to generate clone pTBP-der(14). The DNA sequence of the cloned breakpoints was determined on an automated sequencer (PE Applied Biosystems, Foster City, CA) by primer walking.
Riboprobes and RNase protection assay.
The following oligonucleotide pairs were used for PCR amplification of the indicated human riboprobes:
The indicated oligonucleotides and total RNA of the human B-cell line BJA-B were used to generate the following probes by RT-PCR amplification and cDNA insertion into the polylinker of pSP64 in antisense orientation relative to SP6 transcription: p53 (321 bp), Ex1A (348 bp), J-chain (220 bp), and S16 (109 bp). The probe Ex2-4 ofPAX-5 was previously described.20 The probe Ex1B was obtained by inserting a 915-bp Eag I-Pst I fragment of clone pD10-P1.8 into the polylinker of pSP64.8A 440-bp Sau3A-Pst I fragment spanning the IgH Sμ-PAX5 exon 1B breakpoint was subcloned from clone pTBP-der(14) into pSP64 to obtain the Sμ-Ex1B riboprobe.
Total RNA was prepared from human mononuclear cells and B-cell lines using the TRIzol reagent (GIBCO BRL), and 10 μg of each RNA preparation was used for RNase protection assay according to Vitelli et al.21 The hybridization temperature was 60°C for all riboprobes except for the Sμ/Ex1B and Ex1B probes that were hybridized at 70°C.
S1 nuclease analysis.
An S1 DNA probe containing the 5′ end of PAX-5 exon 1B was obtained by 5′ end labeling the EagI end of a 563-bpApaI-EagI DNA fragment isolated from clone pD10-P1.8.8 The S1 nuclease protection analysis was performed as previously described22 except that the RNA-DNA hybridization was performed at 60°C. A DNA sequencing ladder was generated by depurination of the labeled DNA probe at G- and A-residues as described.23
Electrophoretic mobility shift assay (EMSA) analysis.
Nucleotide sequence accession numbers.
RESULTS
Characterization of the lymphoma cells of patient MZL-1.
Because the specific t(9;14)(p13;q32) translocation involving thePAX-5 locus was highly correlated with SLL of the plasmacytoid subtype,4,8 9 we isolated DNA from affected lymph nodes of 18 B-SLL patients for Southern blot analysis with DNA probes specific for the 5′ region of the PAX-5 gene (see below). In addition, DNA prepared from the peripheral blood of two patients diagnosed with MZL and of one patient with prolymphocytic leukemia (PLL) were included in the Southern blot analysis. Of all these patients, an aberrantly migrating DNA fragment indicative of a possible translocation was detected in only one patient, here referred to as MZL-1 (see below). In agreement with this observation, peripheral B lymphocytes of this patient were shown, by cytogenetic analysis, to contain a complex translocation involving 9p13 (PAX-5; see below). Patient MZL-1 initially presented with anemia, leukocytosis, and splenomegaly. In studying the malignant B lymphocytes of patient MZL-1, we characterized the mononuclear cells from the peripheral blood by flow cytometry (Fig 1) and analyzed bone marrow biopsies by immunohistochemistry (Fig 2). Although a normal control individual contains only ∼4% of CD40+ B cells in the MNC fraction (Fig 1B), the blood of patient MZL-1 was almost exclusively composed of malignant B cells (up to 94%; Fig 1A). These activated B lymphocytes were weakly positive for CD5 expression (CD5low) and were monoclonal with regard to the expression of a B-cell receptor consisting of IgM and Igκ. In addition, these cells were strongly positive for CD19, CD23, CD40, and CD72, whereas they were negative for CD3, CD10, CD21, CD28, CD38, CD43, CD56, Igλ, IgA, and IgG expression (Fig 1A; data not shown).
Analysis of bone marrow biopsies of patient MZL-1 showed massive infiltrates of B lymphocytes that accounted for ∼40% of all bone marrow cells (Fig 2A and 2B), whereas only a few scattered mature B cells are usually detected in normal bone marrow (data not shown). The malignant B lymphocytes of patient MZL-1 were predominantly clustered and aligned within the sinusoids of the bone marrow (Fig 2A and 2B), which is considered to be a characteristic hallmark of lymphoma of splenic origin.16 The infiltrating B lymphocytes were reactive with antibodies directed against the B-cell marker CD79a (Ig-α; Fig 2A) and the antigen DBA.44 (Fig 2B). The DBA.44 antigen was also detected on sorted CD19+ CD5+ B lymphocytes from the peripheral blood of patient MZL-1, suggesting that these peripheral B lymphocytes and the infiltrating B cells in the bone marrow are of the same orgin (Fig 2C). The DBA.44 antigen is normally found only on a subset of B lymphocytes in the mantle and marginal zones of lymphoid follicles.25 Interestingly, the leukemic B cells of patient MZL-1 resembled villous lymphocytes as evidenced by the presence of cytoplasmic projections and fine villi (Fig 2D). The characteristic morphology, bone marrow infiltration pattern, and cell-surface phenotype of the malignant B lymphocytes therefore support the diagnosis of splenic marginal-zone lymphoma according to the REAL classification.5
Identification of a complex t(2;9;14)(p12;p13;q32) translocation in the MZL-1 lymphoma.
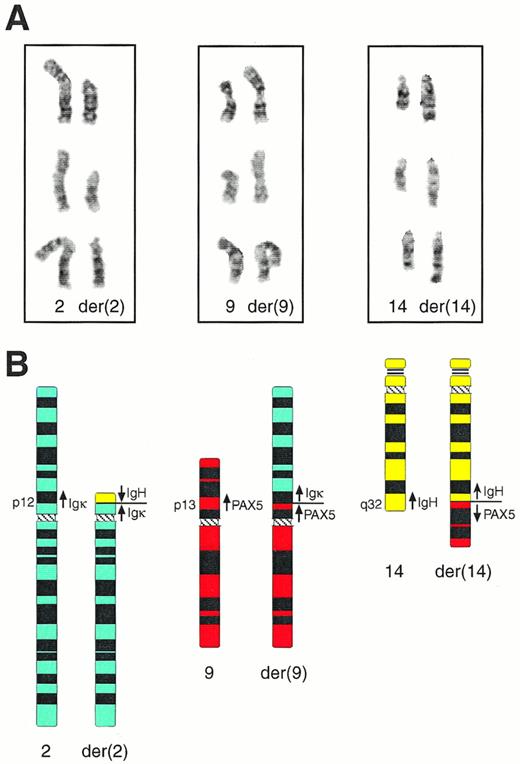
Cytogenetic analysis of bone marrow cells of patient MZL-1 showed four visible chromosomal abnormalities (for complete karyotype see Materials and Methods). A partial duplication was observed on chromosome 1 [dupl(1)(q12q32)], a deletion on chromosome 7 [del(7)(q22q33)], a nonreciprocal translocation on chromosome 17 [der(17)t(12;17)(q21;p12)], and a complex reciprocal translocation involving chromosomes 2, 9, and 14 [t(2;9;14)(p12;p13;q32); Fig 3]. Interestingly, the Igκ, PAX-5, and IgH genes are located on 2p12,26 9p13,27 and 14q32,7 respectively, suggesting that these genes may have participated in a three-way translocation during the genesis of the MZL-1 lymphoma. Because the transcriptional polarity of the three genes on the respective chromosomes is known,8 26 their involvement in the complex t(2;9;14) translocation makes the following prediction, which was subsequently verified by molecular analysis of the breakpoint sequences (see below). The IgH gene has been translocated in opposite orientation next to the Igκ andPAX-5 genes on the derivative chromosomes der(2) and der(14), respectively, whereas the Igκ and PAX-5 loci have been juxtaposed in the same transcriptional direction on chromosome der(9).
Cytogenetic anallysis of the complex t(2;9;14) translocation of patient MZL-1. (A) Partial karyotype of three representative metaphase nuclei. The normal homologues (left) and the derivative (der) chromosomes (right) of the t(2;9;14)(p12;p13;q32) translocation are shown in their G-banded form. (B) Schematic representation of the t(2;9;14) translocation. Ideograms of the normal and derivative chromosomes are shown together with the location and transcriptional direction of the Igκ, IgH, andPAX-5 genes.
Cytogenetic anallysis of the complex t(2;9;14) translocation of patient MZL-1. (A) Partial karyotype of three representative metaphase nuclei. The normal homologues (left) and the derivative (der) chromosomes (right) of the t(2;9;14)(p12;p13;q32) translocation are shown in their G-banded form. (B) Schematic representation of the t(2;9;14) translocation. Ideograms of the normal and derivative chromosomes are shown together with the location and transcriptional direction of the Igκ, IgH, andPAX-5 genes.
Replacement of the endogenous PAX-5 promoters by a switch Sμ promoter of the IgH locus in the MZL-1 translocation.
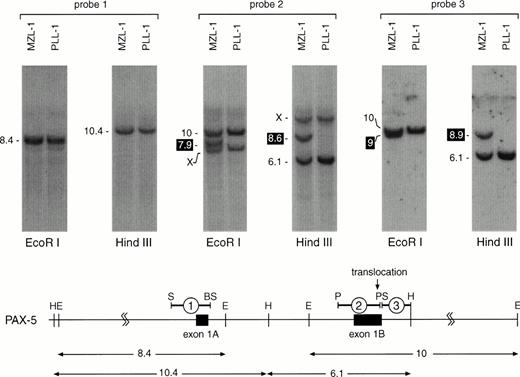
For Southern blot screening we chose a strategy that allowed us to analyze lymphoma DNA for translocation breakpoints within 22 kb of thePAX-5 5′ region (from −6 to +16 kb relative to the transcription start of exon 1A; Fig 4). The result of such an analysis is shown in Fig 4 for patients MZL-1 and PLL-1. DNA isolated from MNC of these two patients was digested withHindIII or EcoRI followed by Southern blot hybridization with three different DNA probes. Whereas the exon 1A-specific probe #1 detected only the predicted DNA fragments of thePAX-5 locus, a novel DNA fragment was observed with the exon 1B-specific probe #2 in each restriction digest of the MZL-1 DNA compared with the ‘control’ PLL-1 DNA. Interestingly, the adjacent intron probe #3 detected extra HindIII and EcoRI fragments of different lengths in the MZL-1 DNA. These data therefore indicate that the translocation breakpoint in patient MZL-1 must reside within the 3′ end of PAX-5 exon 1B, most likely between the DNA probes #2 and #3. Hence, each of the two DNA probes is specific for one of the reciprocal PAX-5 translocation events. Moreover, quantitation of the hybridization signals showed that the translocated DNA fragment corresponded to ∼80% of the wild-type PAX-5DNA, thus confirming that the majority of the MNC of patient MZL-1 consisted of malignant B lymphocytes, in agreement with the flow cytometric data (Fig 1A).
Localization of the MZL-1 translocation breakpoint in exon 1B of PAX-5. DNA from mononuclear cells of patients MZL-1 and PLL-1 was digested with HindIII or EcoRI followed by Southern blot analysis with the indicated DNA probes. The sizes of the hybridizing DNA fragments are indicated in kb to the left. The DNA fragments containing the reciprocal translocation breakpoints are highlighted in black. X denotes an unrelated DNA fragment that cross-hybridizes with the GC-rich DNA probe #2. A restriction map of the 5′ region of the PAX-5 locus8 is shown below together with the origin of the DNA probes and the hybridizingHindIII and EcoRI DNA fragments (sizes given in kb). Abbreviations: H, HindIII; E, EcoRI; S, SacI; BS, BstEII; P,PstI.
Localization of the MZL-1 translocation breakpoint in exon 1B of PAX-5. DNA from mononuclear cells of patients MZL-1 and PLL-1 was digested with HindIII or EcoRI followed by Southern blot analysis with the indicated DNA probes. The sizes of the hybridizing DNA fragments are indicated in kb to the left. The DNA fragments containing the reciprocal translocation breakpoints are highlighted in black. X denotes an unrelated DNA fragment that cross-hybridizes with the GC-rich DNA probe #2. A restriction map of the 5′ region of the PAX-5 locus8 is shown below together with the origin of the DNA probes and the hybridizingHindIII and EcoRI DNA fragments (sizes given in kb). Abbreviations: H, HindIII; E, EcoRI; S, SacI; BS, BstEII; P,PstI.
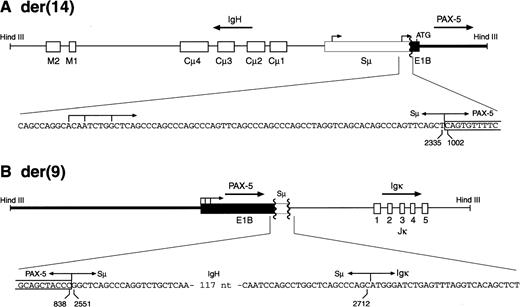
To identify the partner genes involved in the MZL-1 translocation, we took advantage of the fact that the two HindIII fragments containing the reciprocal PAX-5 translocation events were of similar size (8.6 and 8.9 kb; Fig 4). We therefore used 8 to 9 kb size-selected HindIII fragments of MZL-1 DNA to generate a genomic phage library that was subsequently screened with the DNA probes #2 and #3. Sequence analysis of the cloned HindIII fragments showed that the reciprocal MZL-1 translocations are composed of the gene arrangements shown in Fig 5. As predicted by the cytogenetic analysis (Fig 3), the PAX-5 andIgH genes are arranged in a head-to-head configuration on the 8.9-kb HindIII fragment isolated from the derivative chromosome 14 (Fig 5A). In agreement with the Southern blot data (Fig4), the breakpoint was located in the 3′ region of exon 1B of the PAX-5 gene and in the 5′ part of the switch Sμ region of the IgH locus (Figs5A and 6B). Interestingly, the Sμ region of the humanIgH gene has been shown to harbor two antisense promoters, which have been occasionally juxtaposed in a similar head-to-head translocation next to the c-MYC gene in Burkitt’s lymphoma, thus resulting in the synthesis of a hybrid Sμ/c-MYCtranscript.28 The breakpoint of the MZL-1 translocation was mapped ∼70 nucleotides downstream of the heterogeneous start sites of the second antisense Sμ promoter. Hence, in the MZL-1 translocation, the PAX-5 gene on the derivative chromosome 14 was severed from its own promoters and brought under the control of immunoglobulin Sμ promoters.
The breakpoints of the reciprocal PAX-5translocations in patient MZL-1. The two translocation breakpoints on the derivative chromosomes 14 (A) and 9 (B) were cloned asHindIII DNA fragments and sequenced. Only the relevant sequences across the breakpoints are shown below a schematic diagram of each translocation event. Numbers refer to the corresponding nucleotide positions of the PAX-5 exon 1B (Fig 6B) or immunoglobulin Sμ sequence,28 respectively. Arrows denote the transcription start sites of the antisense promoter in the immunoglobulin Sμ region as determined by Apel et al.28
The breakpoints of the reciprocal PAX-5translocations in patient MZL-1. The two translocation breakpoints on the derivative chromosomes 14 (A) and 9 (B) were cloned asHindIII DNA fragments and sequenced. Only the relevant sequences across the breakpoints are shown below a schematic diagram of each translocation event. Numbers refer to the corresponding nucleotide positions of the PAX-5 exon 1B (Fig 6B) or immunoglobulin Sμ sequence,28 respectively. Arrows denote the transcription start sites of the antisense promoter in the immunoglobulin Sμ region as determined by Apel et al.28
Characterization of exon 1B of PAX-5. (A) Heterogeneous transcription initiation. Total RNA (20 μg) isolated from the B-lymphoid BJA-B and KIS-1 cell lines as well as from control HeLa cells was analyzed by S1 nuclease protection assay with a DNA probe labeled at the EagI site in exon 1B. The S1-resistant DNA fragments were separated by electrophoresis together with a G+A sequence ladder obtained by partial depurination of the same end-labeled DNA probe. Arrowheads point to the most abundant transcription start sites. (B) DNA sequence of exon 1B. The most frequently used transcription start sites are denoted by arrows. Exon 1B sequences are shown in capital letters, whereas the flanking promoter and intron sequences are shown in lowercase letters. The numbers to the left refer to the nucleotide position relative to the first prominent transcription start site. The deduced amino acids of exon 1B, the reciprocal breakpoints [der(9) and der(14)] of the MLZ-1 translocation and the restriction sites relevant for probe design are indicated together with a consensus recognition sequence for the transcription factor Sp155 that was shown, by EMSA assay, to bind to this −40 promoter region in B-cell nuclear extract (data not shown). The invariant GT dinucleotide of the 5′ splice junction is underlined.
Characterization of exon 1B of PAX-5. (A) Heterogeneous transcription initiation. Total RNA (20 μg) isolated from the B-lymphoid BJA-B and KIS-1 cell lines as well as from control HeLa cells was analyzed by S1 nuclease protection assay with a DNA probe labeled at the EagI site in exon 1B. The S1-resistant DNA fragments were separated by electrophoresis together with a G+A sequence ladder obtained by partial depurination of the same end-labeled DNA probe. Arrowheads point to the most abundant transcription start sites. (B) DNA sequence of exon 1B. The most frequently used transcription start sites are denoted by arrows. Exon 1B sequences are shown in capital letters, whereas the flanking promoter and intron sequences are shown in lowercase letters. The numbers to the left refer to the nucleotide position relative to the first prominent transcription start site. The deduced amino acids of exon 1B, the reciprocal breakpoints [der(9) and der(14)] of the MLZ-1 translocation and the restriction sites relevant for probe design are indicated together with a consensus recognition sequence for the transcription factor Sp155 that was shown, by EMSA assay, to bind to this −40 promoter region in B-cell nuclear extract (data not shown). The invariant GT dinucleotide of the 5′ splice junction is underlined.
The gene configuration on the derivative chromosome 9 was also consistent with the result of the cytogenetic analysis (Fig 3), because the two PAX-5 promoters were positioned in the same transcriptional orientation next to the Jκ segments of theIgκ gene (Fig 5B). This gene arrangement is, however, unlikely to give rise to a functional transcription unit because the translation start codon and the 3′ splice site have been eliminated from PAX-5 exon 1B during the translocation process. Surprisingly, the PAX-5 and Igκ genes were separated by a 162 bp long sequence originating from the switch Sμ region of the IgH locus (Fig 5B). This finding suggests that the complex t(2;9;14)(p12;p13;q32) translocation has been brought about by two consecutive rearrangements, each involving the Sμ sequences of theIgH gene. The first reciprocal translocation must have occurred between the PAX-5 and IgH genes, because the alternative possibility, translocation between the IgH andIgκ loci, would have created a nonviable, dicentric chromosome (Fig 3B). Subsequently, the translocated Sμ sequences on the der(9) chromosome must have participated in a second rearrangement involving the Igκ gene. The involvement of the Sμ region in both translocations strongly argues that the complex t(2;9;14) translocation has been generated by misguided class-switch recombination. In agreement with this idea, MZL has been postulated to arise from activated B cells that normally undergo immunoglobulin class switching.5 Moreover, errors of switch recombination have previously been implicated as a major cause for translocations in mature B-cell malignancies.29,30 Furthermore, it is worthwhile to note that the reciprocal translocation between thePAX-5 and IgH genes was accompanied by the loss of 163 and 215 bp from exon 1B and the Sμ region, respectively (Figs 5 and6B). Hence, the MZL-1 translocation is not balanced at the fine structural level in agreement with similar analyses of other leukemias.31
Deregulation of PAX-5 expression by IgH control elements in B lymphocytes carrying t(9;14) translocations.
In a next step, we investigated the transcription efficiency of the wild-type and translocated PAX-5 alleles in B lymphocytes carrying t(9;14) translocations. The PAX-5 gene has been shown to be transcribed from two distinct promoters.8Transcription from the upstream exon 1A promoter is initiated from a single site 3′ of a conserved TATA-box.8 In contrast, transcription from the downstream exon 1B promoter is heterogeneously initiated in the B-lymphoid cell lines BJA-B and KIS-1, as shown by S1 nuclease protection analysis (Fig 6A). In agreement with this finding, the downstream promoter of PAX-5 lacks any TATA-like sequence, but instead contains a functional binding site for the transcription factor Sp1 in the −40 region (Fig 6B). Sequence analysis of exon 1B, furthermore, showed an extraordinarily long leader region of 1.1-kb size (Fig 6B).
Two different types of B cells were available for studying the effect of the t(9;14)(p13;q32) translocation on PAX-5 transcription, namely the MNC of patient MZL-1 and the KIS-1 cell line that was established from a diffuse large-cell lymphoma.19 In KIS-1 cells, the Eμ enhancer of the IgH locus was inserted upstream of the exon 1A promoter of PAX-5,8 whereas the immunoglobulin Sμ promoters replaced the downstream PAX-5promoter in the B lymphocytes of patient MZL-1 (Fig 5A). To quantitate the transcripts originating from the different promoters, we generated riboprobes that were specific for the upstream PAX-5 promoter (Ex1A), the downstream PAX-5 promoter (Ex1B), and the immunoglobulin Sμ promoter (Sμ/Ex1B; Fig 7). In addition, we used a riboprobe (Ex2-4) derived from the common coding sequences of exons 2-4 to map the total amount of PAX-5 mRNA. The results of the various RNase protection analyses are shown in Fig 7. The PAX-5transcript levels in the different B cells were quantitated and normalized to the expression of the small ribosomal protein S16 gene.32 In parallel, the B-cell lines BJA-B and Raji as well as primary B lymphocytes from two B-CLL patients were analyzed as control cells, indicating that PAX-5 is expressed at a constant level in these B lymphocytes in agreement with previous analyses.12 13 Compared with these control cells, the transcription from the upstream PAX-5 promoter was 8.3-fold increased in the KIS-1 cells, which reflects the close proximity of the Eμ enhancer to the exon 1A promoter in this lymphoma (Fig 7A). In contrast, the downstream PAX-5 promoter was upregulated to a lower degree (Fig 7B). Taking the transcription from both promoters and mRNA splicing into account, we observed a 5.5-fold overexpression of total PAX-5 mRNA in KIS-1 cells (Fig 7C). In comparison, thePAX-5 mRNA level was 8.4-fold increased in the B lymphocytes of patient MZL-1 (Fig 7C) as a result of efficient heterogeneous initation of transcription at the Sμ promoter (Fig 7D). Hence, the two different t(9;14) translocations analyzed did indeed lead to deregulation of PAX-5 transcription by bringing thePAX-5 gene under the control of immunoglobulin regulatory elements.
Deregulation of PAX-5 transcription by t(9;14) translocations. PAX-5 transcript levels were quantitated by RNase protection assay in total RNA isolated from mononuclear blood cells of patient MZL-1, from the large-cell lymphoma cell line KIS-1, from the B-cell lines Raji and BJA-B, from mononuclear blood cells of two patients diagnosed with B-CLL and from control HeLa cells. The 5′ ends of the PAX-5A (A) and PAX-5B (B) transcripts, the spliced mRNA sequences of the downstream paired domain (C) and the hybrid Sμ-PAX5B transcripts (D) were mapped with the riboprobes depicted below. The size of the expected RNase-protected fragments is given in numbers of nucleotides. pUC19 DNA digested withMspI was used as end-labeled DNA size marker (lane M; sizes given in nucleotides). Transcripts coding for the small ribosomal protein S1632 were comapped and used as an internal reference for quantitation of the PAX-5 RNA signals by PhosphorImager analysis. The induction of the different PAX-5transcripts in the KIS-1 and MZL-1 cells was calculated relative to the average PAX-5 expression level of the two B cell lines Raji and BJA-B and is shown below the relevant part of the S16 autoradiograph. The Sμ promoter lacks a TATA-box28 and thus results in heterogeneous transcription initiation in MZL-1 cells (D). Abbreviations: E, EcoRI; B, BamHI; EA,EagI; S, Sau3A; P, PstI.
Deregulation of PAX-5 transcription by t(9;14) translocations. PAX-5 transcript levels were quantitated by RNase protection assay in total RNA isolated from mononuclear blood cells of patient MZL-1, from the large-cell lymphoma cell line KIS-1, from the B-cell lines Raji and BJA-B, from mononuclear blood cells of two patients diagnosed with B-CLL and from control HeLa cells. The 5′ ends of the PAX-5A (A) and PAX-5B (B) transcripts, the spliced mRNA sequences of the downstream paired domain (C) and the hybrid Sμ-PAX5B transcripts (D) were mapped with the riboprobes depicted below. The size of the expected RNase-protected fragments is given in numbers of nucleotides. pUC19 DNA digested withMspI was used as end-labeled DNA size marker (lane M; sizes given in nucleotides). Transcripts coding for the small ribosomal protein S1632 were comapped and used as an internal reference for quantitation of the PAX-5 RNA signals by PhosphorImager analysis. The induction of the different PAX-5transcripts in the KIS-1 and MZL-1 cells was calculated relative to the average PAX-5 expression level of the two B cell lines Raji and BJA-B and is shown below the relevant part of the S16 autoradiograph. The Sμ promoter lacks a TATA-box28 and thus results in heterogeneous transcription initiation in MZL-1 cells (D). Abbreviations: E, EcoRI; B, BamHI; EA,EagI; S, Sau3A; P, PstI.
We next investigated whether the increased PAX-5 mRNA production was also reflected at the protein level. For this purpose, the amount of BSAP (Pax-5) protein was quantitated by the sensitive EMSA method in nuclear extracts prepared from the different types of B cells (Fig 8). A moderate, but consistent 2-2.5-fold increase in BSAP protein synthesis was observed in the KIS-1 and MZL-1 B lymphocytes as compared with the control BJA-B and Raji cells. The discrepancy between the level of mRNA and DNA-binding activity may indicate that PAX-5 expression is subject to translational or posttranslational control in B cells.
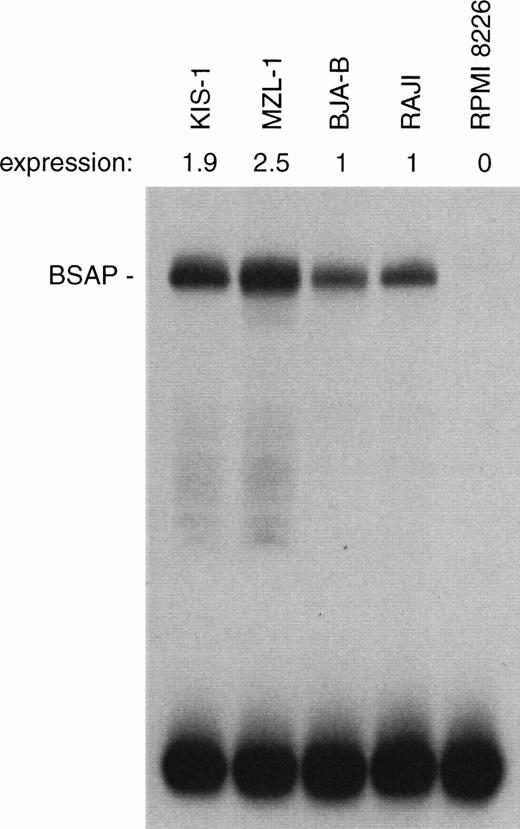
Moderate increase of BSAP protein synthesis in B lymphocytes carrying the t(9;14) translocation. The same amount (1 μg) of nuclear protein isolated from mononuclear cells of patient MLZ-1 or the indicated B-lymphoid cell lines was analyzed by EMSA assay for binding of BSAP to the paired domain recognition sequence of theH2A-2.2 gene. The radioactivity in the BSAP-DNA complex was quantitated by PhosphorImager analysis, and the BSAP expression level of the KIS-1 and MZL-1 cells is given relative to that of the control B cell lines BJA-B and Raji. PAX-5 is not expressed in the myeloma cell line RPMI 8226.
Moderate increase of BSAP protein synthesis in B lymphocytes carrying the t(9;14) translocation. The same amount (1 μg) of nuclear protein isolated from mononuclear cells of patient MLZ-1 or the indicated B-lymphoid cell lines was analyzed by EMSA assay for binding of BSAP to the paired domain recognition sequence of theH2A-2.2 gene. The radioactivity in the BSAP-DNA complex was quantitated by PhosphorImager analysis, and the BSAP expression level of the KIS-1 and MZL-1 cells is given relative to that of the control B cell lines BJA-B and Raji. PAX-5 is not expressed in the myeloma cell line RPMI 8226.
Influence of the t(9;14) translocation on the expression of putative BSAP target genes.
Targeted inactivation of the mouse Pax-5 gene has provided an elegant genetic tool for dissecting the function of BSAP (Pax-5) in gene regulation during early pro–B-cell development.14,15In contrast, little is known about the role of BSAP during the late phase of B-cell differentiation that has so far been inaccessible to loss-of-function analysis. Protein-DNA binding and transient-transfection experiments have, however, implicated BSAP in the repression of the murine J-chain gene33 and in the downmodulation of the IgH and Igκ 3′ enhancers at the mature B-cell stage.10 The gain-of-function mutations of the human t(9;14)(p13;q32) translocations have now offered an alternative approach for investigating the role of BSAP in gene regulation at late developmental stages. The J-chain gene is known to code for an immunoglobulin joining (J) protein that is essential for the assembly and secretion of pentamer IgM antibodies.34 In agreement with previous expression analyses,35 RNase protection experiments showed that the human J-chain gene is broadly expressed in a range of B-cell and myeloma cell lines (Fig 9A). Moreover, the J-chain gene was also highly expressed in MNC of a healthy donor. In contrast, transcripts of the J-chain gene could neither be detected in KIS-1 cells nor in MNC of the patient MZL-1 (Fig 9A). Hence, these data indicate that overexpression of BSAP (Pax-5) is associated with repression of J-chain gene transcription in B lymphocytes carrying the t(9;14) translocation.
J-chain and p53 gene expression in KIS-1 and MZL-1 lymphomas. (A) Absence of J-chain gene expression in B lymphocytes carrying the t(9;14) translocation. Human B-cell lines (BJA-B, Raji, Ramos, HS-Sultan), myeloma cell lines (U226, RPMI 8226), KIS-1 cells and mononuclear blood cells of patient MZL-1 as well as of a control individual (MNC) were analyzed by RNase protection assay for J-chain and S16 mRNA expression. (B) Normal p53expression in tumors overexpressing PAX-5. The p53 mRNA levels were quantitated by RNase protection analysis in a subset of the cells studied in panel A. MB-238 refers to the tumor that exhibits the highest PAX-5 expression level within a large medulloblastoma collection.20 The KIS-1 cells carry a mutation resulting in a truncated p53 mRNA (p53▵) that is detected on the longer autoradiographic exposure shown to the left.
J-chain and p53 gene expression in KIS-1 and MZL-1 lymphomas. (A) Absence of J-chain gene expression in B lymphocytes carrying the t(9;14) translocation. Human B-cell lines (BJA-B, Raji, Ramos, HS-Sultan), myeloma cell lines (U226, RPMI 8226), KIS-1 cells and mononuclear blood cells of patient MZL-1 as well as of a control individual (MNC) were analyzed by RNase protection assay for J-chain and S16 mRNA expression. (B) Normal p53expression in tumors overexpressing PAX-5. The p53 mRNA levels were quantitated by RNase protection analysis in a subset of the cells studied in panel A. MB-238 refers to the tumor that exhibits the highest PAX-5 expression level within a large medulloblastoma collection.20 The KIS-1 cells carry a mutation resulting in a truncated p53 mRNA (p53▵) that is detected on the longer autoradiographic exposure shown to the left.
Recently, Stuart et al36 have proposed on the basis of transient cell transfection data that Pax-5 (BSAP) is a potent repressor of p53 gene transcription. According to this hypothesis, Pax-5 should mediate its oncogenic effect through inactivation of the tumor-suppressor gene p53. However, the malignant B lymphocytes of patient MZL-1 expressed normal levels ofp53 mRNA compared with the control cell lines Raji, BJA-B, and HeLa (Fig 9B). No intact p53 transcripts could be detected in KIS-1 cells. However, a truncated p53 mRNA was expressed at a low level, suggesting that both alleles of the p53 gene are mutated in the KIS-1 cell line. Moreover, a medulloblastoma tumor, MB-238, also contained normal levels of p53 mRNA (Fig 9B) despite the fact that this tumor was previously shown to highly overexpress the PAX-5 gene.20 Hence, these data do not support a role for Pax-5 (BSAP) in p53 gene repression.
DISCUSSION
The transcription factor Pax-5 (BSAP) is known to play an important role in early B-cell development as shown by loss-of-function mutation in the mouse.11,14 The chromosomal t(9;14)(p13;q32) translocation has now been shown to correspond to a gain-of-function mutation of the human PAX-5 gene, pointing to a critical role of this transcription factor in late B-cell differentiation and in the pathogenesis of NHL. As summarized in Fig10, to date, three t(9;14) translocations have been molecularly characterized, all of which affect the 5′ region of thePAX-5 gene. Recently, we have shown that the Eμ enhancer of the IgH locus is juxtaposed next to the exon 1A promoter ofPAX-5 in the diffuse large-cell lymphoma KIS-1.8Here, we have shown that an antisense promoter in the switch Sμ region of the IgH gene is linked to the downstream exon 1B ofPAX-5 in a case of MZL. Interestingly, the translocation breakpoint of the B-SLL case 10529 was also located inPAX-5 exon 1B just three base pairs upstream of that of patient MZL-1 (Fig 5A and 10). The corresponding breakpoint on the IgHlocus was, however, only approximately mapped to the Sμ region, and the effect of the t(9;14) translocation on PAX-5 expression was not analyzed.9 Hence, it is unclear at present whether the translocation in case 1052 occurred upstream or downstream of the antisense Sμ promoter used in patient MZL-1. The same Sμ promoter was previously shown to drive expression of a translocated c-MYC gene in Burkitt’s lymphoma.28 In this tumor, the coding region of the affected c-MYC gene is known to be frequently altered by somatic mutations.37 Sequence analysis of cloned PAX-5 cDNA from the MNC of patient MZL-1 demonstrated, however, that the translocated PAX-5 gene codes for a wild-type BSAP protein (data not shown). Furthermore, expression analyses directly showed an increase in the initiation of PAX-5transcription from the Sμ promoter in the MZL-1 lymphoma and from the upstream exon 1A promoter in the KIS-1 cells. These data indicate therefore that the t(9;14) translocation corresponds to a regulatory mutation that increases PAX-5 expression as a result of either promoter replacement or enhancer insertion. In this regard, the oncogenic activation of PAX-5 clearly differs from that ofPAX-3 and PAX-7 in alveolar rhabdomyosarcoma. A novel fusion gene coding for a more potent transcription factor is generated in this pediatric muscle tumor by a specific translocation involving one of the two PAX loci and the fork head domain geneFKHR.38-40
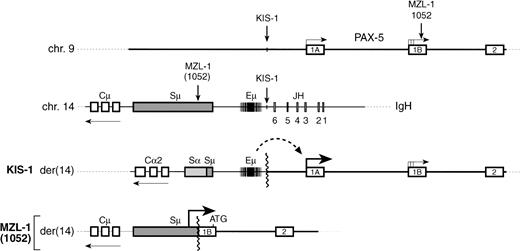
t(9:14) translocations involving the PAX-5 andIgH loci. A schematic diagram depicts the 5′ region of the PAX-5 gene, the JH-to-Cμ region of the IgH locus and the corresponding translocation breakpoints present on the derivative chromosme 14 in the lymphomas of patients KIS-1,8 1052,9 and MZL-1 (this study). The PAX-5 expression level and the translocation breakpoint relative to the antisense promoters in the immunoglobulin Sμ region have not been mapped in patient 1025,9 and hence it is not known at present whether promoter replacement as described for patient MZL-1 may be a molecular mechanism for PAX-5 gene deregulation in this case. The position of the translation start codon at the very 3′ end of PAX-5 exon 1B is indicated by ATG.
t(9:14) translocations involving the PAX-5 andIgH loci. A schematic diagram depicts the 5′ region of the PAX-5 gene, the JH-to-Cμ region of the IgH locus and the corresponding translocation breakpoints present on the derivative chromosme 14 in the lymphomas of patients KIS-1,8 1052,9 and MZL-1 (this study). The PAX-5 expression level and the translocation breakpoint relative to the antisense promoters in the immunoglobulin Sμ region have not been mapped in patient 1025,9 and hence it is not known at present whether promoter replacement as described for patient MZL-1 may be a molecular mechanism for PAX-5 gene deregulation in this case. The position of the translation start codon at the very 3′ end of PAX-5 exon 1B is indicated by ATG.
Southern blot analysis of 18 B-SLL tumors with plasmacytoid differentiation did not show any further translocation in the promoter regions of the PAX-5 gene. Hence, our analysis indicates, in agreement with the study of Iida et al,9 that the immediate 5′ region of PAX-5 is not a mutational hotspot in B-SLL tumors. However, long-range FISH analysis has shown a more consistent involvement of PAX-5 in t(9;14) translocations of these tumors,9 suggesting that activation from distantly locatedIgH enhancers, as exemplified by the MAL translocation,41 may be the most common mechanism for deregulating the PAX-5 gene. As shown in the present report, oncogenic activation of PAX-5 is not only observed in B-SLL tumors with plasmacytoid differentiation, but also at least in one case of MZL, a related non-Hodgkin’s disease. The tumor of patient MZL-1 was classified as splenic MZL based on the observed splenomegaly, the characteristic bone marrow infiltation pattern,16 the tumor involvement of peripheral blood, the immunophenotype and villous morphology of the malignant B lymphocytes,5 and the expression of the DBA.44 antigen that is normally present only on cells of the mantle and marginal zones of lymphoid follicles.25Because the marginal zone is known to be enriched in memory B cells with the potential of differentiating into plasma cells,42,43 it has been postulated that transformation of these cells may give rise to MZL.5
Recently, it has been suggested that Pax proteins exert their oncogenic effect by inactivating the transcription of the tumor suppressor genep53.36 This hypothesis is primarily based on protein-DNA binding studies and transient cell transfection experiments that suggested that Pax-5 (BSAP) may repress the p53 gene by binding to its 5′ untranslated sequence.36 However, we have been unable to confirm these data either in vitro or in vivo. In contrast to the prediction of the hypothesis, the level ofp53 transcription was totally unaffected in early pro-B cells lacking Pax-5.15 Second, we have not observed any inverse correlation between PAX-5 and p53 expression in a large panel of medulloblastoma tumors (Fig 9B; Z. Kozmik and M.B., unpublished data), which were previously shown to overexpress the PAX-5 gene.20 Third, thep53 expression level was also normal in the malignant B lympocytes of patient MZL-1 (Fig 9B). Moreover, the previously reported decrease of p53 expression in the large-cell lymphoma KIS-19 was shown to be caused by a truncation of thep53 transcript (Fig 9B) in agreement with the consistent involvement of p53 gene mutations in disease progression to large-cell lymphoma.44,45 All of these data therefore strongly argue against the hypothesis that p53 is a target gene of the transcription factor BSAP (Pax-5). In this context it is interesting to note that the immunoglobulin J-chain gene was expressed in all human B-cell lines analyzed except in the KIS-1 and MZL-1 B lymphocytes. Hence, it is conceivable that overexpression ofPAX-5 may interfere with the regulation of the human J-chain gene in analogy to the murine homologue that is considered to be under negative control by BSAP.33
How can we explain the oncogenic activation of PAX-5 by the t(9;14) translocation now that our data rule out an involvement ofPAX-5 in p53 gene regulation? In principle, we can distinguish between a quantitative and temporal effect of PAX-5deregulation on late B-cell differentiation. It has been well documented that the function of PAX genes is highly sensitive to gene dosage. This sensitivity is reflected by the frequent association of heterozygous PAX gene mutations with human disease syndromes46 as is best illustrated forPAX-6. Inactivation of one PAX-6 allele causes aniridia in humans and the Small eye phenotype in mice,47whereas even a moderate increase in gene-copy number also causes aSmall eye-like phenotype referred to as microphthalmia.48 By analogy, it is therefore possible that the small, but consistent increase in BSAP (Pax-5) protein synthesis could also alter the B-cell phenotype. For instance, antisense oligonucleotide inhibition experiments suggested a role for BSAP in the proliferation control of mature B cells.49 Interestingly, the proliferation rate of these cells was even slightly increased by transient overexpression of BSAP.49 Hence, it is conceivable that deregulation of PAX-5 perturbs the cell-cycle regulation of mature B cells, may prevent them from entering the quiescent state, and could thus contribute to lymphomagenesis. On the other hand, it is also possible that the proximity of immunoglobulin control elements overrules the correct temporal regulation ofPAX-5 during the terminal phase of B-cell differentiation. ThePAX-5 gene is known to be downregulated during plasma cell differentiation based on the following evidence. First, PAX-5expression is detected in neither plasmacytoma nor myeloma cells of mouse and human origin.12,50 Second, in vitro differentiation experiments with mouse splenic B lymphocytes have shown that Pax-5 expression is repressed on stimulation of mature B cells to undergo plasma cell differentiation.51,52 In contrast, the IgH locus is transcriptionally most active in immunoglobulin-secreting plasma cells.53 Hence, the insertion of IgH control elements by the t(9;14) translocation may force the translocated PAX-5 allele to remain active at a time when the endogenous PAX-5 gene is usually switched off. In agreement with this hypothesis, B lymphocytes carrying the t(9;14) translocation frequently exhibit features of plasmacytoid differentiation.4,9 Furthermore, ectopic expression ofPAX-5 in late B cells has recently been shown to interfere with immunoglobulin secretion that is normally induced on terminal differentiation.54 Hence, the t(9;14) translocation could contribute to tumorigenesis by interfering with the downregulation ofPAX-5 transcription, thereby preventing completion of the plasma cell differentiation program. This hypothesis can now be experimentally verified by recreating the human t(9;14) translocation breakpoint in transgenic mice.
ACKNOWLEDGMENT
We thank H. Ohno for providing the KIS-1 cell line, H. Pirc-Danoewinata for cytogenetic analysis, R. Thalhammer for help with photography, R. Kurzbauer for DNA sequencing, P. Steinlein for cell sorting, and G. Christofori for critical reading of the manuscript.
Supported by the Research Institute of Molecular Pathology, Vienna, by a grant from the Austrian Industrial Research Promotion Fund, Vienna, and by Grant No. NB5829 from the Austrian National Bank.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Meinrad Busslinger, PhD, Research Institute of Molecular Pathology, Dr. Bohr-Gasse 7, A-1030, Vienna, Austria; email: Busslinger@nt.imp.univie.ac.at.

![Fig. 2. Morphological characterization of the malignant B lymphocytes in the bone marrow and blood of patient MZL-1. (A,B) Immunohistochemical analysis of bone marrow biopsies. Sections were stained with anti-CD79a [Ig-] (A) and anti-DBA.44 (B) antibodies. Both antibodies stained lymphoid cells that were predominantly located within the sinusoids of the bone marrow. The sinusoids are indicated by arrowheads (cross-section) or arrows (oblique [A] and longitudinal [B] sections), respectively. The clear spheroidal areas are occupied by fat cells. (C) Staining of CD5+CD19+peripheral blood cells. CD5+CD19+ cells were sorted from mononuclear cells of the peripheral blood and stained with the anti-DBA.44 antibody.25 About 70% of all sorted cells were strongly positive for the DBA.44 antigen. (D) Morphology of the malignant B lymphocytes. Blood smears of patient MZL-1 were analyzed by the modified Wright staining. Four representative B cells are shown at high magnification together with adjacent erythrocytes. These B cells contain a round or ovoid nucleus that is surrounded by a moderately increased amount of basophilic cytoplasm. Most malignant B cells have the appearance of villous lymphocytes as evidenced by the presence of cytoplasmic projections or short thin villi, which are often asymmetrically located on one side of the cell.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/92/10/10.1182_blood.v92.10.3865/4/m_blod422mor02z.jpeg?Expires=1770144582&Signature=T5~sq3keSMKDnbGklWxvAkkn5xfm~npoEpX4uL33n8x5Ex08tdsAiNqcQsAn1AJUHe7F6EBl4esK2aopjWyZXUyJo5LPYyCLiPGfdTU-Lk4ouXXZe92wKe7vJeAhLcy30iKO0iapC2r-l08KOeLssfOS46vCgx8cVmTXMBjH-0cHhyeuZUU-SgqJOjZElNWbwXtnhWNYeA4v9FOKUlkWcPnNc056qrvspVZE5fV-PNwi4e1JxFvMfiigDDUrG1771MTBxVQz3D0-ZMj-ZCJioavg3n4H0~agc6fL7dp~j5LIwNI1M7xgwDAO19aZjvAlloXTadxl7mD3GJvTWyfnlg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 6. Characterization of exon 1B of PAX-5. (A) Heterogeneous transcription initiation. Total RNA (20 μg) isolated from the B-lymphoid BJA-B and KIS-1 cell lines as well as from control HeLa cells was analyzed by S1 nuclease protection assay with a DNA probe labeled at the EagI site in exon 1B. The S1-resistant DNA fragments were separated by electrophoresis together with a G+A sequence ladder obtained by partial depurination of the same end-labeled DNA probe. Arrowheads point to the most abundant transcription start sites. (B) DNA sequence of exon 1B. The most frequently used transcription start sites are denoted by arrows. Exon 1B sequences are shown in capital letters, whereas the flanking promoter and intron sequences are shown in lowercase letters. The numbers to the left refer to the nucleotide position relative to the first prominent transcription start site. The deduced amino acids of exon 1B, the reciprocal breakpoints [der(9) and der(14)] of the MLZ-1 translocation and the restriction sites relevant for probe design are indicated together with a consensus recognition sequence for the transcription factor Sp155 that was shown, by EMSA assay, to bind to this −40 promoter region in B-cell nuclear extract (data not shown). The invariant GT dinucleotide of the 5′ splice junction is underlined.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/92/10/10.1182_blood.v92.10.3865/4/m_blod42201006w.jpeg?Expires=1770144582&Signature=TPsEyIy0gLmHnUtaOhoH~Mz8fofcY6ZtY7~OmoK78qWbZElE5i9rOODcyHR2ppenN9vWvHRSYhmK9iDOHbj8fDpmF7MTEfP-2ZeSX-ZdvhnwVP4FTQW~-6ALnD1X3hElDbmpOcEbFWdVvMfvOOfX9ObeYkrno2VbFSi6C-aw2OdB1fBzD6E4D09oMkdfYySyz3Y3q5dCsSEXldepFObBUepBIL9QUc~4dG6W5K5P7UP2m5uLu6xKzKqVMWC4hdIGX0UNFusrk8cCWPdydq23c6Mkvsj5M9UoiElgb6g4j80wuczsMgo4zgPQXX5Esnu6E1s5o8x2ah33DhEY51PtCQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal