Abstract
Hypoxic induction of erythropoietin (Epo) and other oxygen-dependent genes is mediated by the hypoxia-inducible factor-1 (HIF-1), a heterodimeric transactivator consisting of an α and a β subunit. We previously found that the mouse gene encoding HIF-1α harbors two alternative first exons (I.1 and I.2), giving rise to two different HIF-1α mRNA isoforms. Here, we show by RNase protection analysis that the exon I.1-derived mRNA isoform is differentially expressed in mouse tissues, being highest in kidney, tongue, stomach, and testis, but undetectable in liver, whereas the exon I.2 mRNA isoform is ubiquitously expressed. Sequence and methylation analysis showed that, in contrast to exon I.1, exon I.2 resides within a region showing typical features of a CpG island, known to be associated with the 5′ end of housekeeping genes. We identified a 232-bp minimal exon I.2 promoter that strongly induced reporter gene expression in mouse L929 fibroblasts and Hepa1 hepatoma cells. In contrast to L929 cells, the exon I.1 promoter was inactive in Hepa1 cells and hypoxic exposure (1% O2) markedly reduced exon I.2 promoter activity in Hepa1 cells. Prolonged exposure of mice to hypoxia (7.5% O2 for up to 72 hours) also caused a decrease in liver HIF-1α mRNA, whereas aldolase mRNA levels increased. These findings might be related to the relatively low Epo levels in the adult liver.
STIMULATION OF ERYTHROPOIESIS during periods of limited oxygen supply improves the oxygen transport capacity of the blood. This physiologic adaptation to hypoxia is regulated by the glycoprotein hormone erythropoietin (Epo) that is mainly produced in the fetal liver and the adult kidney (reviewed in Jelkmann1). Identification of a hypoxia-responsive element in the 3′ flanking region of the gene encoding Epo led to the discovery of the hypoxia-inducible factor-1α (HIF-1α).2HIF-1α is a basic-helix-loop-helix (bHLH) transcription factor that is activated by exposure of cells to physiologically relevant reductions in oxygen partial pressure (reviewed in Bunn and Poyton,3 Wenger and Gassmann4). This activation involves hypoxic stabilization of the protein that is otherwise ubiquitinated and rapidly degraded in proteasomes under normoxic conditions.5-8 Moreover, the activity of the transactivation domain(s) can also be hypoxically induced.6,9,10 So far, the mechanisms leading to these hypoxic effects are not clearly understood, but there is evidence that redox processes,5,8,11 as well as phosphorylation,12,13 might be involved. After hypoxic exposure, HIF-1α forms a heterodimeric complex with the aryl hydrocarbon receptor nuclear translocator (ARNT), also termed HIF-1β, which activates expression of oxygen-dependent genes.3,4These oxy-genes4 include Epo, transferrin, vascular endothelial growth factor, glycolytic enzymes, inducible nitric oxide synthase, heme oxygenase, as well as other genes involved in the adaptation of an organism to reduced oxygenation at the cellular, local, and systemic level. ARNT also serves as a heterodimerization partner for the aryl hydrocarbon receptor (AhR), also called dioxin receptor, which activates genes involved in xenobiotic metabolism such as the gene encoding cytochrome P450IA1. Besides the common bHLH DNA binding and heterodimerization domain, all of these factors contain a region of amino acid similarity termed PAS (PAS is an acronym for the first described members of this family, namely Per,ARNT, and Sim).4
We previously cloned and characterized the gene encoding mouse HIF-1α (Hif1a) and showed that it contains two different first exons (termed I.1 and I.2).14 Expression of these two mouse HIF-1α mRNA transcripts is regulated by distinct promoters rather than being the product of differential splicing. The two alternative first exons give rise to two distinct mRNA isoforms differing in the composition of their 5′ untranslated regions (UTRs). Moreover, the predicted translation product derived from the exon I.1-containing mRNA isoform is 12 amino acids shorter than the exon I.2-containing isoform. These findings raised the possibilities of differential translational regulation and/or different protein functions.14 So far, no human HIF-1α mRNA isoform has been detected that corresponds to mouse exon I.1. We also analyzed theHif1a exon I.1 promoter and found that it is only moderately active in the cell lines tested, suggesting that additional cis-regulatory elements and/or so far unknown activators (eg, developmental-stage, tissue-specific, or conditional signals) are required for efficient exon I.1 promoter activity.14
Here, we present the sequence analysis, as well as the structural and functional characterization, of Hif1a exon I.2 and flanking regions. In addition, we examined the expression profile of both mRNA isoforms in various adult mouse tissues. Our results suggest thatHif1a exon I.1 regulation exhibits tissue-specific features, whereas the exon I.2 promoter resembles a housekeeping-type promoter.
MATERIALS AND METHODS
Reverse transcription-polymerase chain reaction (RT-PCR) analysis.
Total RNA was isolated from mouse tissues according to the method described by Chomczynski and Sacchi.15 Expression of the two mouse HIF-1α mRNA isoforms was examined as described.14 Briefly, each 2.5 μg of total RNA was reverse transcribed, and equal aliquots of the cDNA were amplified by PCR using specific forward primers for either exon I.1 or exon I.2 and an exon III reverse primer. Equal fractions of the reaction mixes were electrophoresed through 1% agarose gels. Ethidium bromide-stained gels were recorded using a video imaging system (Vilber Lourmat, Inotech, Dottikon, Switzerland).
RNase protection assay.
Hif1a exon I.1 and exon I.2 DNA fragments were subcloned in pBluescript (Stratagene, La Jolla, CA) and antisense cRNA probes were obtained by in vitro transcription with T7 RNA polymerase (MBI Fermentas, Vilinius, Lithuania) in the presence of a32P-uridine triphosphate (UTP). After purification by denaturing urea/polyacrylamide gel electrophoresis, the probes were hybridized with 50 μg total RNA and digested with RNase A and RNase T1 according to the manufacturer's directions (RPA-II Kit; Ambion, Austin, TX). Protected products were resolved by nondenaturing 5% polyacrylamide gel electrophoresis. Radioactive signals were recorded and quantified by phosphorimaging (Molecular Dynamics, Sunnyvale, CA), and the images were displayed using a linear relationship between signal and image intensity. A mouse β-actin probe (Ambion) was used to control for equal RNA amounts between the different samples. These reactions were performed using 2 μg total RNA only.
Cloning and sequencing.
The exon I.2-containing λ phage clone λH30 was cloned from a LambdaGEM-11 (Promega, Madison, WI) genomic library as described previously.14 A 270-bp EcoRI-NcoI (all restriction enzymes were purchased from Fermentas) cDNA fragment containing mouse exon I.2 sequences (kindly provided by A. Damert, Bad Nauheim, Germany) served as hybridization probe. A 2.9-kb XbaI fragment from λH30, containing exon I.2 and flanking sequences, was subcloned into pBluescript SK+ yielding the plasmid pH30X. The insert of this plasmid was sequenced on both strands using a combination of automated and manual sequencing procedures with fluorescently labeled dideoxynucleotides or a 35S-deoxy adenosine triphosphate (dATP) incorporation in cycle sequencing reactions and T7 sequencing reactions, respectively, according to the instructions provided by the manufacturers (Applied Biosystems, Foster City, CA and Pharmacia, Uppsala, Sweden). Sequence analysis was performed using the GCG program package16 or the DNASIS for Windows program (Hitachi, Tokyo, Japan).
Cell culture.
The mouse hepatoma cell line Hepa1 (also termed Hepa1c1c7)17 was a kind gift of L. Poellinger (Huddinge, Sweden). The mouse fibroblast cell line L929 (American Type Culture Collection, Rockville, MD, CCL-1 NCTC clone 929) was a kind gift of V. O'Donnall (Bern, Switzerland). Both cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM, high glucose; Life Technologies, Basel, Switzerland) supplemented with 10% heat-inactivated fetal calf serum (Boehringer, Mannheim, Germany), 100 U/mL penicillin, 100 μg/mL streptomycin, 1 × nonessential amino acids, 2 mmol/L L-glutamine, and 1 mmol/L Na-pyruvate (all purchased from Life Technologies) in a humidified atmosphere containing 5% CO2 at 37°C. Oxygen tensions in the incubator (Forma Scientific, model 3319; Bioblock, Frenkendorf, Switzerland) were either 140 mm Hg (20% O2 vol/vol, normoxia) or 7 mm Hg (1% O2 vol/vol, hypoxia).
Mapping of the exon I.2 transcription initiation site.
The cap site of the mouse HIF-1α exon I.2-containing mRNA was determined by primer extension and mung bean nuclease protection. For primer extension, the oligonucleotide mHIFpex2 (see Fig 2) was radioactively labeled by phosphorylation of the 5′ end with [γ-32P]ATP (Hartmann Analytik, Braunschweig, Germany) and T4 polynucleotide kinase (Fermentas) as described previously.18 Total RNA was extracted from mouse Hepa1 hepatoma cells according to Chomczynski and Sacchi.15Poly(A)+ RNA was prepared using oligo dT cellulose spin columns according to the manufacturer's instructions (Pharmacia). Endlabeled mHIFpex2 (0.4 pmol) was coprecipitated with 5 μg poly(A)+ RNA and resuspended in 10 μL 0.4 mol/L NaCl, 10 mmol/L Pipes [pH 6.4] at 96°C for 2 minutes. After annealing at 56°C for 10 minutes and at room temperature for 15 minutes, primer extension was performed in 50 μL RT buffer (Stratagene), 1 U/μL ribonuclease inhibitor (RNasin, Promega), 0.5 mmol/L deoxynucleotide triphosphates (dNTPs; Promega) containing 250 U reverse transcriptase (Stratascript, Stratagene) at 42°C for 2 hours. The reaction products were ethanol precipitated, resuspended in 50% formamide, electrophoresed through a 6% polyacrylamide/urea sequencing gel, and visualized by autoradiography. A T7 polymerase sequencing reaction, performed with the primer mHIFpex2 and pH30X single-stranded DNA according to the manufacturer's instructions (Pharmacia), served as position marker.
Relative expression levels of Hif1a exon I.1 and exon I.2-containing mRNA isoforms in various mouse tissues. RNase protection assays using each 50 μg total RNA and exon I.1 and I.2-specific antisense probes. A total of 2 μg total RNA was used for the β-actin control reactions. Besides the sole protected 113-bp fragment in Hepa1 cells, a second mRNA species appeared in mouse tissues probably exceeding the 5′ end of the exon I.1 probe (134 nt).
Relative expression levels of Hif1a exon I.1 and exon I.2-containing mRNA isoforms in various mouse tissues. RNase protection assays using each 50 μg total RNA and exon I.1 and I.2-specific antisense probes. A total of 2 μg total RNA was used for the β-actin control reactions. Besides the sole protected 113-bp fragment in Hepa1 cells, a second mRNA species appeared in mouse tissues probably exceeding the 5′ end of the exon I.1 probe (134 nt).
Mung bean nuclease protection was performed essentially as described before.14 Briefly, single-stranded DNA from the plasmid pH30X was prepared using M13K07 helper phages. The 5′ endlabeled mHIFpex2 oligonucleotide was annealed to the template DNA and extended using Klenow fragment of DNA polymerase I (Fermentas). After cleaveage with PvuII, the single-stranded antisense probe was prepared by alkaline agarose gel electrophoresis. This probe (100 kcpm) was hybridized to 50 μg total RNA isolated from Hepa1 cells and digested with 0, 10, 30, or 90 U mung bean nuclease (Life Technologies) for 30 minutes at 30°C. The products were analyzed on a sequencing gel as described above using an unrelated sequencing reaction as length marker.
In vivo methylation analysis.
To isolate genomic DNA, cell lines and mouse tissues were homogenized in 0.2 mol/L NaCl, 50 mmol/L Tris-HCl [pH 8.0], 10 mmol/L EDTA, 1% sodium dodecyl sulfate (SDS), and digested with 0.5 mg/mL Proteinase K (Boehringer Mannheim) at 60°C for 12 hours. After addition of 0.2 mL saturated NaCl and microcentrifugation, the DNA was precipitated from the supernatant by adding 0.45 mL isopropanol. The DNA was resuspended in 10 mmol/L Tris-HCl [pH 7.6], 1 mmol/L EDTA, and cleaved with XbaI, either alone or in combination withCfr42I, SmaI, NotI, HhaI,HpaII, or MspI. After gel electrophoresis through a 0.7% agarose gel, the DNA was transferred to uncharged Biodyne A membranes (Pall Filtron, Northborough, UK) and cross-linked by ultraviolet irradiation (Stratalinker; Stratagene). The membrane was hybridized to a 561-bp Bsp1407I-BstEII fragment derived from pH30X as described previously.14 The signals were recorded by phosphorimaging.
Reporter gene assays.
A firefly luciferase reporter gene construct (pHXN1aluc) containing exon I.2 upstream regulatory regions was obtained by inserting a 1.5-kbKpnI − NheI fragment (KpnI cuts in the polylinker 5′ to the XbaI site shown in Fig 2) into the promoterless luciferase vector pGL3Basic (Promega). Three deletion constructs were made using BspTI, Eco91I, andPvuII, which cleaved 801, 613, and 232 bp, respectively, upstream of the first transcription initiation site. A similar reporter gene construct containing 499 bp of exon I.1 upstream regulatory sequences (pGL499Luc) was described previously.14 Tissue culture cells (1 × 107 in 350 μL medium without fetal calf serum) were coelectroporated with each 25 μg luciferase reporter gene construct and a β-galactosidase reference vector as described previously.14 After incubation for 24 to 38 hours, the cells were lysed in reporter lysis buffer (Promega) and luciferase and β-galactosidase activities were determined according to the manufacturer's instructions (Promega) using a Lumat LB9501 luminometer (Berthold, Bad Wildbad, Germany) and a DigiScan 96-well plate photometer (ASYS, Eugendorf, Austria), respectively. Differences in the transfection efficiency and extract preparation were corrected by normalization to the corresponding β-galactosidase activities.
Hypoxic exposure of mice.
Animals were exposed to a gas mixture containing 35% air and 65% N2 (7.5% O2 final concentration) in an airtight chamber containing water and nutrients ad libidum. After exposure, the mice were killed by cervical dislocation and the organs were withdrawn and frozen in liquid N2. Liver RNA was extracted and analyzed by Northern blotting as described previously.19 The filters were subsequently hybridized with probes for mouse HIF-1α,20 aldolase A,19 and ribosomal protein L28,19 the latter used to normalize for differences in loading and blotting efficiency.
RESULTS
Differential tissue expression of the two mouse HIF-1α mRNA isoforms.
We previously reported that the two mRNA isoforms coding for HIF-1α are coexpressed in L929 and Hepa1 mouse cell lines.14 Here, we examined the expression pattern of the two HIF-1α mRNAs in several mouse tissues. To distinguish between the two isoforms, RT-PCR reactions were performed using primers specific for either exon I.1 or exon I.2 in combination with an exon III primer. As shown in Fig 1, this yielded specific PCR products of 371 bp and 472 bp for exon I.1 and exon I.2, respectively. The specificity of these products was further confirmed by restriction digestions and Southern blotting (data not shown). Interestingly, while the exon I.2 mRNA isoform was ubiquitously expressed with less than fourfold variation in signal intensity between the different tissues, the exon I.1 mRNA isoform showed a distinct expression pattern. Exon I.1 mRNA levels were highest in kidney, spleen, thymus, tongue, and the reproductive organs (testis, ovary, and uterus); moderate in brain, heart, lung, bone marrow, and skeletal muscle; and no exon I.1 mRNA isoform could be detected in any liver of five mice analyzed. Exposure of the mice to 0.1% carbon monoxide for 4 hours, causing functional anemia,21 did not significantly alter the expression pattern of the two HIF-1α mRNA isoforms (data not shown), whereas Epo mRNA in kidney and liver was increased under the same conditions.21 In agreement with our previous observations using various cell lines,14 the present RT-PCR analysis of mouse tissues did not yield any evidence for the existence of an mRNA isoform containing exon I.2 as an alternative splice variant, further supporting the notion that two independent promoters drive expression of the two alternative first exons.
Hif1a exon I.1 and exon I.2-containing mRNA isoforms are detectable in most mouse tissues. RT-PCR analysis of total RNA isolated from the indicated mouse tissue. PCR was performed using forward primers unique to either exon I.1 or exon I.2 and a common reverse primer specific for exon III. Single PCR products were indicative for the exon I.1 (371 bp) or the exon I.2 (472 bp) mRNA isoform. Representative results of two to five independent experiments are shown. Note that the exon I.1 mRNA isoform could not be detected in liver.
Hif1a exon I.1 and exon I.2-containing mRNA isoforms are detectable in most mouse tissues. RT-PCR analysis of total RNA isolated from the indicated mouse tissue. PCR was performed using forward primers unique to either exon I.1 or exon I.2 and a common reverse primer specific for exon III. Single PCR products were indicative for the exon I.1 (371 bp) or the exon I.2 (472 bp) mRNA isoform. Representative results of two to five independent experiments are shown. Note that the exon I.1 mRNA isoform could not be detected in liver.
Due to the distinct base compositions and lengths of exons I.1 and I.2, this RT-PCR approach did not allow to determine the relative expression levels of the two mouse HIF-1α mRNA isoforms. To circumvent this problem, an RNase protection assay was established using labeled antisense probes specific for exons I.1 and I.2, respectively (Fig 2). RNase protection showed that the exon I.1 mRNA isoform was detectable only in kidney, tongue, testis, stomach, and embryonic tissue (embryonic day E18.5). Interestingly, whereas the previously mapped14 113-bp protected fragment represented the sole exon I.1 mRNA species in Hepa1 cells, an additional mRNA species might exist in mouse tissues whose 5′ end probably extends the length of the 134 nt cRNA probe (Fig 2), indicating that a second transcriptional start site might exist in mouse tissues upstream to that mapped in Hepa1 cells. In contrast to the exon I.1-derived mRNA isoform, the I.2 isoform was ubiquitously expressed. Taking into consideration the differences in probe length, the exon I.2 mRNA isoform was estimated to be at least sevenfold more abundant (testis) than the exon I.1 mRNA isoform.
DNA sequence of mouse Hif1a exon I.2 and flanking regions.
The ubiquitous expression of the exon I.2-derived HIF-1α mRNA isoform implied that, unlike exon I.1, exon I.2 is expressed from a housekeeping-type promoter. To test this hypothesis directly, we subcloned and sequenced exon I.2 and flanking regions (Fig 3) using the previously isolated λ phage λH30 that bridged the gap between exon I.1 and exon III-containing phage clones.14 Exon I.2 was 96.8% identical to the corresponding sequence of the mouse HIF-1α cDNA 5′ end reported by Li et al,9 containing three mismatches and seven insertions. In contrast to exon I.1, exon I.2 had an ATG translation initiation codon in frame with the ATG on exon II, leading to a predicted translation product that was 12 amino acids longer than the one derived from the exon I.1 mRNA isoform (Fig 3). These 12 N-terminal amino acids were identical to those predicted from the cDNA reported by Li et al.9 The observed translation initiation site (TTCGCCATGG) matched the consensus reported by Kozak (GCCRCCATGG).22 Furthermore, the exon-intron splice junction conformed to the consensus sequence.23
Hif1a exon I.2 and flanking regions. The sequence of exon I.2 is in bold, repetitive elements and restriction enzyme recognition sites are underlined, and putative transcription factor consensus binding sites are double underlined. The transcription initiation sites mapped by primer extension and mung bean nuclease protection (see Fig 4) are indicated by filled arrows and the start of intron 1 by an open arrow. The predicted translation initiation codon and the first 12 amino acids of the exon I.2 isoform are indicated. The location of the oligonucleotide mHIFpex2 is depicted with a line over the sequence.
Hif1a exon I.2 and flanking regions. The sequence of exon I.2 is in bold, repetitive elements and restriction enzyme recognition sites are underlined, and putative transcription factor consensus binding sites are double underlined. The transcription initiation sites mapped by primer extension and mung bean nuclease protection (see Fig 4) are indicated by filled arrows and the start of intron 1 by an open arrow. The predicted translation initiation codon and the first 12 amino acids of the exon I.2 isoform are indicated. The location of the oligonucleotide mHIFpex2 is depicted with a line over the sequence.
Exon I.2 was 75% identical to the 5′ UTR of the human HIF-1α cDNA and the predicted amino acid sequence differed in two of the 12 positions (Glu7 to Ala and Glu9 to Asp). Thus, exon I.2 encoded the mouse homologue of the human HIF-1α 5′ end. On the other hand, no human homologue for exon I.1 has been reported so far. Because the in-frame ATG codon on the second exon of mouse Hif1a is not present in the human gene, we consider that a putative human homologue would contain the ATG initiation codon on the alternative first exon rather than on the second exon. Alternatively, a human homologue of mouse exon I.1 might not exist.
Mapping of the transcription initiation site of Hif1a exon I.2.
To determine the cap site of the HIF-1α exon I.2 mRNA isoform, primer extension analysis was performed using poly(A)+ RNA isolated from mouse Hepa1 hepatoma cells primed with the exon I.2-specific oligonucleotide mHIFpex2 (see Fig 3). As depicted in Fig 4, two major bands were obtained, corresponding to C1390 and C1397 of the sequence shown in Fig 3. To confirm this result, we also applied a nuclease protection assay using an endlabeled antisense DNA probe and total Hepa1 RNA. After hybridization of the probe to the RNA, increasing amounts of mung bean nuclease were added to digest protruding ends of the DNA-RNA hybrids, as well as excess single-stranded antisense probe. In this case, two major bands were identified corresponding to C1390 and C1395 (Fig 3). Thus, the longest Hif1a exon I.2-derived transcription product was 325 bp (corresponding to a 5′ UTR of 290 bp), which was 16 bp longer than the cDNA reported by Li et al.9 All three start sites (indicated by arrowheads in Fig 3) conformed to the CA rule for eukaryotic transcription initiation sites.24 25 However, as reported by these investigators, the preferred cap site corresponds to an adenosine rather than to a cytosine. The reason for this discrepancy is currently unknown, but might be related to the lack of a canonical TATA box, known to be associated with multiple transcriptional start sites.
Mapping of the Hif1a exon I.2 transcription initiation sites. (A) Primer extension. Poly(A)+ mRNA was isolated from mouse Hepa1 cells and annealed to the endlabeled, complementary oligonucleotide mHIFpex2 (see Fig 3). The primer was extended with reverse transcriptase and the products were resolved on a 6% denaturing polyacrylamide gel together with a sequencing reaction performed with the same oligonucleotide as primer and a plasmid containing the sequence shown in Fig 3 as template. (B) Nuclease protection. Total RNA derived from Hepa1 cells was hybridized to an endlabeled single-stranded antisense probe prepared as described in Materials and Methods. The DNA-RNA hybrids were treated with the indicated amounts of mung bean nuclease and separated on a sequencing gel along with an unrelated sequencing ladder that served as length marker. Numbers indicate the lengths of reaction products including the mHIFpex2 primer.
Mapping of the Hif1a exon I.2 transcription initiation sites. (A) Primer extension. Poly(A)+ mRNA was isolated from mouse Hepa1 cells and annealed to the endlabeled, complementary oligonucleotide mHIFpex2 (see Fig 3). The primer was extended with reverse transcriptase and the products were resolved on a 6% denaturing polyacrylamide gel together with a sequencing reaction performed with the same oligonucleotide as primer and a plasmid containing the sequence shown in Fig 3 as template. (B) Nuclease protection. Total RNA derived from Hepa1 cells was hybridized to an endlabeled single-stranded antisense probe prepared as described in Materials and Methods. The DNA-RNA hybrids were treated with the indicated amounts of mung bean nuclease and separated on a sequencing gel along with an unrelated sequencing ladder that served as length marker. Numbers indicate the lengths of reaction products including the mHIFpex2 primer.
Hif1a exon I.2 is located within an essentially methylation-free CpG island.
One intriguing feature of mouse Hif1a exon I.2 was the finding that it is located within a G+C rich region. Figure 5 shows a comparison of the G+C content of the exon I.1 and I.2 loci. Whereas the exon I.1 locus displayed an average G+C content of approximately 43%, comparable to that of the bulk genome, exon I.2 and upstream regions had a G+C content of 76%. After a short oligo T repeat at the beginning of intron 1 (Fig 3), a second G+C rich (81%) stretch was found. Because G+C rich regions associated with 5′ ends of genes are indicative for the presence of methylation-free CpG islands (reviewed in Cross and Bird,26 Gardiner-Garden and Frommer27), we analyzed the frequency of CpG dinucleotides, as well as the in vivo CpG methylation pattern.
Hif1a exon I.2 is located within a CpG island. G+C content across the exon I.1 (A) and exon I.2 (B) regions. A window of 100 bp shifted in steps of 1 bp was used in the computer-assisted analysis. The positions of exon I.1 and exon I.2 are indicated. The CpG/GpC ratio is given for each DNA segment that could be distinguished from adjacent segments by the difference in the G+C content.
Hif1a exon I.2 is located within a CpG island. G+C content across the exon I.1 (A) and exon I.2 (B) regions. A window of 100 bp shifted in steps of 1 bp was used in the computer-assisted analysis. The positions of exon I.1 and exon I.2 are indicated. The CpG/GpC ratio is given for each DNA segment that could be distinguished from adjacent segments by the difference in the G+C content.
CpG dinucleotide frequencies are usually estimated as the ratio of CpG (ie, observed) versus GpC (ie, expected) over a certain nucleotide region. As shown in Fig 5A, the occurrence of the CpG dinucleotide was suppressed in the exon I.1 locus (CpG to GpC ratio of 0.3). By contrast, the frequency of the CpG dinucleotide in the G+C rich exon I.2 locus was similar to the GpC frequency (Fig 5B).
CpG methylation was assessed in several mouse cell lines and tissues by Southern blotting using CpG methylation-sensitive restriction enzymes. As shown in Fig 6A, genomic DNA was cleaved with XbaI, either alone or in combination with a second enzyme recognizing selected sites in exon I.2 or in the 5′ and 3′ flanking regions. A 561-bp 5′ fragment derived from a region outside of the G+C rich CpG island served as hybridization probe. As shown in Fig 6B, the SmaI site in exon I.2 and the NotI site in the downstream region were entirely methylation-free. Interestingly, methylation of the Cfr42I site in the upstream region was about 50% in all cell lines and tissues tested. To date, we have no conclusive explanation for this result, but it might be related to the observation of Matsuo et al,28 who showed that the mouse experiences more accidental CpG island methylation than man, resulting in erosion of mouse CpG islands during evolution. Because this feature was rather unexpected for a CpG island-type promoter, we further analyzed the 5′ region using the CpG methylation-sensitive restriction enzymes HhaI andHpaII together with the methylation-insensitiveHpaII-isoschizomer MspI. As shown in Fig 6C, the upstream CpG dinucleotides of the G+C rich region were methylation-free, whereas one CpG outside of the CpG island was mostly methylated. Of note, an HpaII/MspI restriction fragment length polymorphism was detected in these experiments (Fig 6C). Taken together, the exon I.2 locus fulfills the criteria for a CpG island, as it is G+C rich, shows unsuppressed CpG frequency, and is essentially CpG methylation-free.
Methylation pattern of the Hif1a exon I.2 region. (A) Map of the CpG methylation-sensitive restriction enzyme recognition sites selected to assess the methylation pattern of distinct CpG dinucleotides in the exon I.2 region. The methylation status of a particular restriction enzyme site is indicated by open and partially filled circles. (B) Southern blot analysis of genomic DNA isolated from various mouse cell lines and tissues. The DNA was cleaved either withXbaI alone, or in combination with CpG methylation-sensitive restriction enzymes cutting 5′ (Cfr42I, C), within (SmaI, S), or 3′ (NotI, N) of exon I.2. (C) Detailed exon I.2 upstream Southern blot analysis using the CpG methylation-sensitive restriction enzymes Cfr42I (C),HhaI (H) and HpaII (P), or the methylation-insensitiveHpaII-isoschizomer MspI (M). Note that anHpaII/MspI restriction fragment length polymorphism (RFLP) was detected.
Methylation pattern of the Hif1a exon I.2 region. (A) Map of the CpG methylation-sensitive restriction enzyme recognition sites selected to assess the methylation pattern of distinct CpG dinucleotides in the exon I.2 region. The methylation status of a particular restriction enzyme site is indicated by open and partially filled circles. (B) Southern blot analysis of genomic DNA isolated from various mouse cell lines and tissues. The DNA was cleaved either withXbaI alone, or in combination with CpG methylation-sensitive restriction enzymes cutting 5′ (Cfr42I, C), within (SmaI, S), or 3′ (NotI, N) of exon I.2. (C) Detailed exon I.2 upstream Southern blot analysis using the CpG methylation-sensitive restriction enzymes Cfr42I (C),HhaI (H) and HpaII (P), or the methylation-insensitiveHpaII-isoschizomer MspI (M). Note that anHpaII/MspI restriction fragment length polymorphism (RFLP) was detected.
As often observed in CpG islands, we noted the lack of a canonical TATA box and a high number of putative Sp1 binding sites (Fig 3). It has been reported that Sp1 binding is implicated in maintaining CpG islands methylation-free.26 Furthermore, two putative HIF-1α binding sites were identified, which perfectly matched the consensus sequence we reported previously.4 However, because HIF-1α mRNA levels in cell lines29 and mice20 were not upregulated by hypoxia, these putative sites are probably not functional. Notably, also a putative HIF-1 binding site in the exon I.1 promoter was found to be nonfunctional.14
Comparison of the Hif1a exon I.1 and I.2 promoter activities.
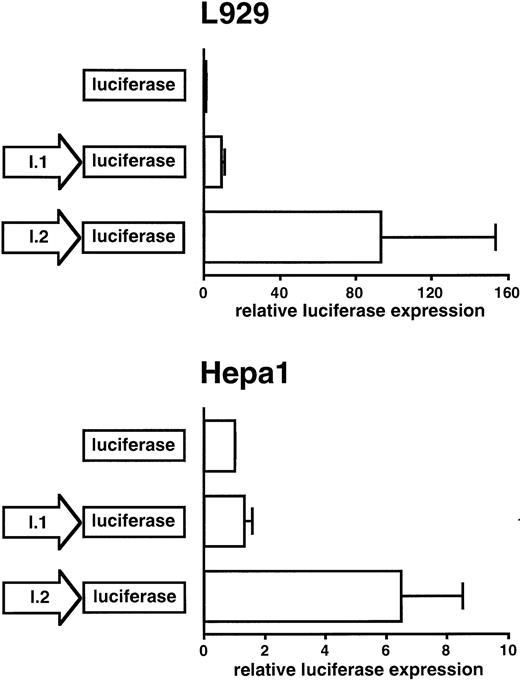
To analyze the presumed exon I.2 promoter activity, a 1.4-kb fragment of the exon I.2 upstream region was inserted into a promoterless firefly luciferase reporter gene vector. To directly compare the respective promoter activities of exon I.1 and exon I.2 upstream sequences, a 499-bp exon I.1 promoter-containing luciferase construct was also analyzed. As reported previously,14 the exon I.1 promoter activity using either a 0.5-kb or a 1.0-kb fragment was about equal. Mouse L929 fibroblasts and Hepa1 hepatoma cells were transiently transfected with both constructs and incubated for 24 to 28 hours at normoxic conditions. A cotransfected β-galactosidase expression vector served as a reference to correct for differences in transfection efficiency and extract preparation. As shown in Fig 7, the exon I.2 promoter construct was 10-fold and fivefold more active than the exon I.1 promoter construct in L929 and Hepa1 cells, respectively. Interestingly, while in these experiments the exon I.1 promoter stimulated basal luciferase expression ninefold in L929 fibroblasts, only very low promoter activity could be detected in Hepa1 hepatoma cells, suggesting that the exon I.1 promoter was not active in hepatoma cells. This finding is in line with the lack of detectable exon I.1 mRNA isoform expression in mouse liver (see above).
Comparison of the Hif1a exon I.1 and exon I.2 promoter activities. The empty parental vector pGL3Basic or the same vector containing either the exon I.1 promoter or the exon I.2 promoter were transiently transfected into mouse L929 fibroblast and Hepa1 hepatoma cells. Luciferase activity was determined after 24 to 28 hours of normoxic incubation. A cotransfected β-galactosidase expression vector served as internal control for transfection efficiency and extract preparation. All values were normalized to the respective luciferase activity obtained with the empty vector pGL3Basic, which was arbitrarily defined as 1. Mean ± standard deviation (SD) of three independent experiments are shown. Note the different scales.
Comparison of the Hif1a exon I.1 and exon I.2 promoter activities. The empty parental vector pGL3Basic or the same vector containing either the exon I.1 promoter or the exon I.2 promoter were transiently transfected into mouse L929 fibroblast and Hepa1 hepatoma cells. Luciferase activity was determined after 24 to 28 hours of normoxic incubation. A cotransfected β-galactosidase expression vector served as internal control for transfection efficiency and extract preparation. All values were normalized to the respective luciferase activity obtained with the empty vector pGL3Basic, which was arbitrarily defined as 1. Mean ± standard deviation (SD) of three independent experiments are shown. Note the different scales.
Hypoxia decreases Hif1a exon I.2 promoter activity in mouse hepatoma cells.
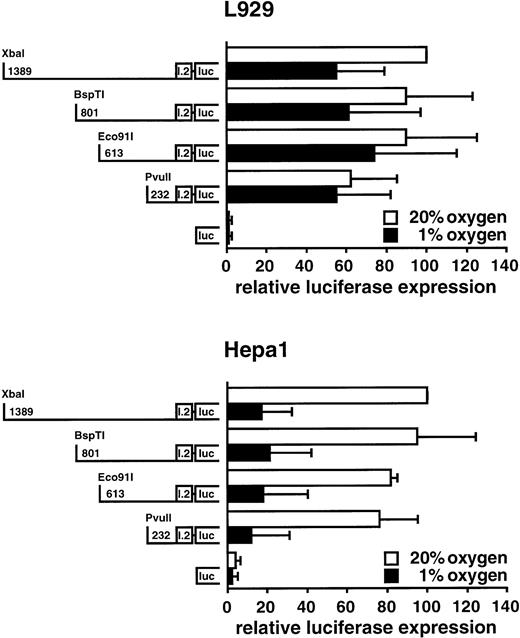
To further analyze the exon I.2 promoter, several deletion constructs were prepared containing 1389, 801, 613, or 232 bp of exon I.2 upstream regulatory sequences. After transient transfection, L929 and Hepa1 cells were split and exposed to normoxia (20% O2) or hypoxia (1% O2) for 36 to 38 hours. As shown in Fig 8, deletion of most of the 1389-bp fragment only marginally reduced exon I.2 promoter activity in normoxic cells, and the 232-bp fragment was almost equally active as the 1389-bp fragment. Interestingly, exposure of the cells to 1% oxygen reduced luciferase expression about fivefold in Hepa1 cells, but only marginally in L929 cells. Conversely, parallel transfections with an SV40 promoter-driven luciferase vector containing three concatamerized Epo-derived HIF-1 binding sites17,18,30 showed about twofold hypoxic induction (data not shown). These results imply that the previously observed17 30 time-dependent hypoxic decrease in HIF-1α steady-state mRNA levels in Hepa1 cells is due to a concomitant reduction in exon I.2 promoter activity.
Hypoxic downregulation of Hif1a exon I.2 promoter activity. Exon I.2 upstream sequences of various length as indicated were placed in front of the firefly luciferase reporter gene. These constructs were transiently transfected into mouse L929 fibroblasts or Hepa1 hepatoma cells, followed by exposure to normoxia or hypoxia for 36 to 38 hours. Subsequently, luciferase expression was determined as described in Fig 7. All values were normalized to the respective normoxic luciferase activity obtained with the construct containing the longest 5′ region, which was arbitrarily defined as 100. Mean ± SD of three independent experiments are shown.
Hypoxic downregulation of Hif1a exon I.2 promoter activity. Exon I.2 upstream sequences of various length as indicated were placed in front of the firefly luciferase reporter gene. These constructs were transiently transfected into mouse L929 fibroblasts or Hepa1 hepatoma cells, followed by exposure to normoxia or hypoxia for 36 to 38 hours. Subsequently, luciferase expression was determined as described in Fig 7. All values were normalized to the respective normoxic luciferase activity obtained with the construct containing the longest 5′ region, which was arbitrarily defined as 100. Mean ± SD of three independent experiments are shown.
Hypoxic reduction of HIF-1α mRNA in mouse liver.
The hypoxic decrease of exon I.2 mRNA levels and promoter activity in mouse hepatoma cells and the lack of the exon I.1 mRNA expression in mouse liver implied that chronic hypoxia might result in reduced hepatic HIF-1α mRNA levels. To test this hypothesis, mice were exposed in triplicates to hypoxic hypoxia (7.5% O2) for up to 3 days. After exposure, liver RNA was isolated and analyzed by Northern blotting. Figure 9 depicts the ratio between HIF-1α mRNA and the ribosomal protein L28 mRNA, which was used to normalize for differences in loading and blotting efficiency. Interestingly, the HIF-α to L28 ratio slightly decreased in hypoxic mouse liver by 20% to 30%, whereas the hypoxia-inducible glycolytic enzyme aldolase A mRNA to L28 ration transiently increased 2.2-fold after 24 hours and decreased again to 1.6-fold over normoxic controls after 48 to 72 hours.
Hypoxic reduction of HIF-1α mRNA levels in mouse liver. Three mice each were exposed to 7.5% O2 for 0 to 72 hours and liver RNA was analyzed by Northern blotting. Shown are the ratios of HIF-1α mRNA and aldolase A mRNA, respectively, and the ribosomal protein L28 mRNA (mean ± SD).
Hypoxic reduction of HIF-1α mRNA levels in mouse liver. Three mice each were exposed to 7.5% O2 for 0 to 72 hours and liver RNA was analyzed by Northern blotting. Shown are the ratios of HIF-1α mRNA and aldolase A mRNA, respectively, and the ribosomal protein L28 mRNA (mean ± SD).
DISCUSSION
In this work, we showed that the mouse HIF-1α exon I.1-containing mRNA isoform is tissue-specifically expressed, whereas the exon I.2 isoform displays an ubiquitous expression pattern in all mouse tissues examined. Ubiquitous HIF-1α expression correlates with the previous observation that HIF-1 DNA binding activity and HIF-1–mediated transactivation of reporter genes is widespread in mammalian cells.31,32 We sequenced and functionally characterized the alternative mouse Hif1a exon I.2 and 5′ flanking regions that we previously mapped approximately 6 kb downstream of theHif1a exon I.1.14 As predicted from the G+C rich 5′ UTR of human and mouse HIF-1α mRNA,14 exon I.2 is located within a 1.3-kb G+C rich, mostly methylation-free CpG island, known to be associated with the promoters of housekeeping genes. This observation is in agreement with the ubiquitous expression of the exon I.2 mRNA isoform. However, because about 40% of all tissue-specifically expressed genes also contain CpG island-type promoters,26,27 we cannot exclude the possibility that theHif1a exon I.2 promoter works in a cell type-specific manner in vivo. The RT-PCR and RNase protection approaches used in this work were not suitable to identify the cell type expressing a particular mRNA. Even if the exon I.2 promoter is ubiquitously active in cell culture, in situ hybridization experiments of mouse tissues will be necessary to unambigously determine the HIF-1α mRNA levels in a particular cell type. In view of the fact that adaptation to different oxygenation is likely to be mandatory for every single cell, a tissue-specific HIF-1 expression seems to be rather improbable. However, tissue-specific expression of the alternative mouse HIF-1α mRNA isoforms, as well as the recently discovered hypoxia-inducible HIF-2α4 (also termed EPAS1,33HLF,34 or HRF35) might complement each other to ensure ubiquitous HIF-1–like activity. Of note, also two tissue- and developmental stage–specifically expressed ARNT relatives (ARNT236,37 and BMAL138), as well as several other bHLH-PAS proteins, have recently been detected, implying a complex network of spatial and temporal formation of heterodimeric bHLH-PAS transcription factors.
A remarkable finding of our HIF-1α mRNA expression analysis in various mouse tissues is the lack of detectable exon I.1 mRNA isoform and a slight hypoxic reduction of the exon I.2 mRNA isoform in mouse liver. In mouse Hepa1 hepatoma cells in culture, reporter gene experiments showed that the Hif1a exon I.1 promoter is inactive and hypoxic exposure reduced exon I.2 promoter activity. These data are consistent with our previous finding that hypoxia time-dependently decreases HIF-1α (but not ARNT) mRNA levels in Hepa1 cells,17,30 and they might also explain why reporter gene experiments using hypoxia-responsive luciferase constructs consistently gave lower induction levels in Hepa1 cells than in other cell lines.18,30 39 Both promoters were much more active in L929 cells compared with Hepa1 cells and the hypoxic downregulation could neither be found in L929 nor in any other human hepatoma (Hep3B and HepG2) or nonhepatoma cell line examined (unpublished observation, June 1997).
Overall, our in vitro data provide an explanation for the in vivo findings and it is tempting to speculate that altered HIF-1α mRNA levels might also influence target gene expression. For example, endogenous transferrin mRNA was less induced by hypoxia in Hepa1 compared with the human hepatoma cell lines Hep3B and HepG2,40 and oxygen-regulated Epo expression is also exclusively found in Hep3B and HepG2,3 but was undetectable in Hepa1 cells (our unpublished observations, August 1995). One hallmark of Epo expression is the switch during development from the fetal liver to the adult kidney as the main source of Epo synthesis. In mammals, this switch takes place in the third trimester of gestation.1 However, the molecular mechanism(s) underlying this phenomenon are currently unknown. At least in the adult mouse, our results on HIF-1α mRNA expression might provide a (partial) explanation of why hypoxia-dependent Epo production is lowered in the adult liver. Clearly, examination of HIF-1α mRNA isoform expression in murine liver and kidney during development will be required to elucidate a possible role of HIF-1 in tissue-specific and developmental stage–specific Epo expression.
The kinetics of hypoxic HIF-1 protein activation appears different from that of HIF-1α mRNA downregulation in Hepa1 cells. Four hours of 1% oxygen are sufficient to induce HIF-1 DNA binding activity in Hepa1 cells.17,29,30 Four hours of 0.1% carbon monoxide treatment are also typically used to elicit a hypoxic response in mice.21 However, 4 hours of hypoxia does not alter expression of the two HIF-1α mRNA isoforms in Hepa1 cells17,29,30 or in mice,20 but rather, at least 8 hours hypoxia are required to downregulate HIF-1α mRNA in Hepa1 cells.17,30 For transient expression experiments, we typically induced the cells for 36 to 38 hours, and 24 hours of hypoxic exposure was also necessary to see a change in mouse liver HIF-1α mRNA. Thus, hypoxic HIF-1α protein activation precedes its mRNA downregulation. The presence of a putative HIF-1 binding site in theHif1a exon I.2 5′ flanking region, which perfectly matched the tentative consensus sequence,4 opens the possibility that HIF-1 could downregulate HIF-1α expression in an autoregulatory loop. Indeed, in the ARNT-deficient subline Hepa1C4, known to be incapable of activating hypoxia-responsive reporter genes,17,30 we observed no hypoxic reduction in HIF-1α mRNA levels,30 supporting the hypothesis of a feedback inhibition. However, deletion of this putative HIF-1 binding site in the 232-bp exon I.2 upstream reporter gene construct did not alter the hypoxic downregulation of luciferase expression, suggesting that this putative HIF-1 binding site is not functional. Of note, a nonfunctional HIF-1 binding site is also present in the exon I.1 promoter,14 confirming that one such element in isolation is not sufficient to convey hypoxic activation of gene expression.4 However, this result cannot exclude that HIF-1 might be involved in the regulation of its own expression.
In summary, the present report completes our work on the two alternative mouse Hif1a promoters, establishes a housekeeping-type major HIF-1α mRNA isoform corresponding to the one known in humans, and opens intriguing questions on the biological function(s) of the tissue-specific alternative mRNA isoform.
ACKNOWLEDGMENT
We are grateful to A. Damert, V. O'Donnall, S. Kozlov, U. Müller, and L. Poellinger for the gifts of material; H. Marti and A. Görlach for helpful discussions and critically reading the manuscript; R. Städeli for technical help; C. Gasser for the artwork; and C. Bauer for support.
Supported by the Swiss National Science Foundation (Grant No. 31-47111.96). R.H.W. is a recipient of the Sondermassnahmen des Bundes zur Förderung des akademischen Nachwuchses.
The novel nucleotide sequence reported in this paper has been deposited with the EMBL/GenBank/DDBJ data bases and is available under accession number Y13656.
Address reprint requests to Roland H. Wenger, PhD, Physiologisches Institut der Universität Zürich-Irchel, Winterthurerstrasse 190, CH-8057 Zürich, Switzerland.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.









This feature is available to Subscribers Only
Sign In or Create an Account Close Modal