Abstract
The t(8;21)-encoded AML1-ETO chimeric product is believed to be causally involved in up to 15% of acute myelogenous leukemias through an as yet unknown mechanism. To directly investigate the role of AML1-ETO in leukemogenesis, we used gene targeting to create anAML1-ETO “knock-in” allele that mimics the t(8;21). Unexpectedly, embryos heterozygous for AML1-ETO(AML1-ETO/+) died around E13.5 from a complete absence of normal fetal liver–derived definitive hematopoiesis and lethal hemorrhages. This phenotype was similar to that seen following homozygous disruption of either AML1 orCBFβ. However, in contrast to AML1- or CBFβ-deficient embryos, fetal livers from AML1-ETO/+ embryos contained dysplastic multilineage hematopoietic progenitors that had an abnormally high self-renewal capacity in vitro. To further document the role of AML1-ETO in these growth abnormalities, we used retroviral transduction to express AML1-ETO in murine adult bone marrow–derived hematopoietic progenitors. AML1-ETO–expressing cells were again found to have an increased self-renewal capacity and could be readily established into immortalized cell lines in vitro. Taken together, these studies suggest that AML1-ETO not only neutralizes the normal biologic activity of AML1 but also directly induces aberrant hematopoietic cell proliferation.
THE (8;21)(q22;q22) TRANSLOCATION is one of the most frequent karyotypic abnormalities detected in acute myelogenous leukemia (AML), accounting for approximately 40% of de novo AML cases that have M2 morphology in the French-American-British (FAB) classification.1 As a result of this translocation, the gene encoding the AML1 (CBFA2) transcription factor from chromosome 21 is fused to the eight twenty-one gene (ETO,also known as MTG8 and CBFA2T1) on chromosome 8.2-5This fusion gene encodes a chimeric AML1-ETO protein consisting of the N-terminus of AML1 fused in frame to the C-terminus of ETO. The fact that the t(8;21) is the sole cytogenetic abnormality in the majority of these cases suggests that AML1-ETO plays a critical role in the establishment of the leukemia clone.
One of the targets of this translocation, AML1, is the DNA-binding subunit of the AML1/CBFβ transcription factor complex and binds the enhancer core motif, TGT/cGGT.6,7 AML1's DNA-binding affinity is increased through heterodimerization with CBFβ, and both its DNA-binding and interaction with CBFβ are mediated through a central domain with high homology to the Drosophilasegmentation gene, runt.6,8,9 The AML1-ETO chimeric product retains this domain and therefore also binds the core enhancer sequence and interacts with CBFβ.6,10 Compared with AML1, relatively little is known about ETO, the other target of this translocation. Although ETO contains a zinc finger motif and appears to be the mammalian homologue of the Drosophila genenervy,11 no direct DNA-binding activity has been detected, nor has its function been identified.12
Transcriptional regulation by AML1 through the enhancer core motif has been shown to be important for the tissue specific expression of a number of hematopoietic specific genes including interleukin-3 (IL-3),13 granulocyte-macrophage colony-stimulating factor (GM-CSF),14 the receptor for CSF-1 (CSF-1R),15myeloperoxidase,16 and subunits of the T-cell antigen receptor (TCR).8,17 By creating mice deficient in AML1/CBFβ, we18 as well as others19-21 have shown that the AML1/CBFβ transcription factor complex is essential for the establishment of definitive hematopoiesis.
Consistent with a critical role in hematopoiesis, the genes encoding the AML1/CBFβ transcription factor complex are the most frequent targets of chromosomal abnormalities in human leukemia. In addition to t(8;21), AML1 fusion products are generated as a result of the t(3;21) (AML1-EVI1) in myelodysplasia and blast crisis of chronic myelogenous leukemia,22-24 and the t(12;21) (TEL-AML1), the most frequent translocation in pediatric acute lymphoblastic leukemia.25-28 Similarly, the CBFβgene is fused to the smooth muscle myosin heavy chain gene,MYH11, as a result of the inv(16) or t(16;16) in the majority of AML cases with M4Eo FAB morphology.29,30 Biochemical studies of these translocation-encoded fusion products suggest that both AML1 and CBFβ chimeric products function, at least in part, to dominantly interfere with normal AML1/CBFβ-mediated transcription.10,31-34 For example, AML1-ETO represses transcription of reporter genes driven by theTCRβ enhancer10 or theGM-CSF promoter,31 and this activity is dependent on a putative repression domain within the C-terminus of ETO.32
To directly investigate the in vivo mechanism through which AML1-ETO contributes to leukemogenesis, we designed a “knock-in” strategy to generate mice containing a single allele of AML1-ETO(AML1-ETO/+) whose expression is regulated by the endogenous transcriptional regulatory elements of murine AML1. Our analysis shows that AML1-ETO both neutralizes normal AML1/CBFβ activity and directly generates signals that lead to the generation of abnormal hematopoietic progenitors.
MATERIALS AND METHODS
Construction of AML1-ETO knock-in targeting vector.
A silent A → G mutation at codon 122 was introduced by site-directed mutagenesis into the human AML1-ETO cDNA (a gift from Dr S. Hiebert, Vanderbilt University, Nashville, TN) to create aSacII cleavage site that is in an identical position to aSacII site within exon 4 of murine AML1. A 1.9-kbSacII fragment from this cDNA, which included 62 bp ofAML1 exon 4, all of exon 5, and the entire fused portion ofETO, was then inserted into the murine AML1 exon 4SacII site within a 12-kb murine genomic AML1clone.18 This resulted in an in frame fusion of the first 95 bp of murine AML1 exon 4 to the remaining portion of the human AML1-ETO cDNA. A 470-bp fragment containing the polyadenylation signal from the rabbit globin gene excised from pEμSR,35 followed by a 1.7-kb HSV-tk promoter-neomycin resistance cassette,18 inserted in the opposite transcriptional orientation was inserted immediately 3′ of the 1.9-kb cDNA fragment. Finally, a 1.2-kb HSV-tk promoter-diphtheria toxin-A negative selection cassette was inserted at the 3′ end of the construct. The vector was linearized at a unique Not I site located at the 3′ end of the construct and introduced into embryonic stem (ES) cells.
Production of chimeric mice.
The linearized targeting vector (50 μg) was electroporated into E14 ES cells as previously described.18 G418-resistant clones were analyzed for homologous recombination by Southern analysis ofXba I–digested DNA hybridized with 5′ (0.4 kb) or 3′ (0.5 kb)AML1 fragments derived from murine genomic sequences outside the vector. Clones with homologous recombination of the targeting vector were then assessed by reverse transcriptase-polymerase chain reaction (RT-PCR) for expression of the fusion gene. Oligonucleotide primers from murine AML1 exon 3 (5′-CCAGCAAGCTGAGGAGCGGCG-3′) and human ETO (5′-AGGCTGTAGGAGAATGG-3′) were used for PCR amplification, and the products were analyzed by Southern analysis using a murine AML1 exon 4 specific oligonucleotide probe (5′-GTGGTGGCACTGGGGGACGT-3′) for detection. AML1-ETO/+ clones with undifferentiated morphology and normal karyotypes were injected into C57BL/6 blastocysts. Male chimeras were bred with C57BL/6 females and tail biopsy specimens of agouti offspring were screened for the presence of the knock-in allele by Southern analysis.
Histology.
Embryos were removed from the uterus, dissected free from the fetal membranes, and inspected under a dissecting microscope for evidence of gross abnormalities. Fetal tissues were then obtained for genotyping and cells from yolk sacs or fetal livers were isolated under sterile conditions and used for in vitro hematopoietic colony assays. Embryos were fixed in Bouin's solution, embedded in paraffin, and sections stained with hematoxylin and eosin. Peripheral blood was collected in 10 mmol/L EDTA and smears stained with Wright-Giemsa.
In vitro culture of hematopoietic cells.
Cells from yolk sacs, fetal livers, or bone marrows were obtained and cultured in methylcellulose semisolid media containing a combination of colony stimulating factors as previously described.18 Cell aggregates containing more than 50 cells were counted as colonies. Cytocentrifuge preparations of hematopoietic colonies were stained with Wright-Giemsa for morphologic examination or for the presence of nonspecific esterase (alpha naphthyl butyrate) cytochemical activity. In replating experiments, either individual hematopoietic colonies or all of the cells from the primary culture were collected, washed, and then replated at 1 to 3 × 104 cells/plate into new methylcellulose cultures under conditions identical to those used for the primary cultures. Colonies were scored as above and subsequent replatings were performed in an identical fashion. Cell lines were generated by harvesting colonies from a single plate and growing the cells in liquid cultures containing RPMI 1640 media containing 10% fetal calf serum, 2 mmol/L L-glutamine, 50 U/mL penicillin G, 50 μg/mL streptomycin, and 10 ng/mL of IL-6 and SCF and 2 ng/mL of IL-3.
Western blot and immunophenotypic analysis.
Cells were lysed in boiling RIPA lysis buffer, electrophoretically separated on a 10% denaturing polyacrylamide gel, and transferred to a nitrocellulose membrane. Proteins were detected using affinity purified AML-1 N-terminal peptide antiserum6 or affinity purified ETO N-terminal peptide antiserum10 and visualized with supersignal ULTRA chemiluminescence substrate (Pierce, Rockford, IL). Cell surface antigens were detected by a standard direct immunofluorescence assay using phycoerythrin-conjugated monoclonal antibodies (MoAb) from PharMigen (San Diego, CA) to Ly-6G (Gr-1), CD116 (MAC-1), c-kit, Sca-1, and Thy-1. Fluorescence activity was analyzed on an FACScan (Becton Dickson, San Jose, CA). Isotypically matched MoAbs at the same protein concentration were used as negative controls in all experiments.
Production of retroviral stocks and infection of murine bone marrow cells.
A human AML1-ETO cDNA10 was inserted into the retroviral vector MSCVneo36 to generate MSCV/AML1-ETOneo. Helper-free retrovirus was generated by transfecting the Bosc23 packaging cell line37 with MSCVneo or MSCV/AML1-ETOneo DNA, and retroviral containing supernatants were collected and stored at −80°C. Supernatants were titered on NIH3T3 cells in G418 containing media as described previously.38
Murine bone marrow cells were obtained from the femur and tibia of 8- to 10-week-old female BALB/cByJ mice 2 days following an intraperitoneal injection of 150 mg/kg of 5-FU (SoloPak Laboratories, Elk Grove Village, IL). Harvested cells were prestimulated for 48 hours with IL-6 and SCF as previously described38 and then infected on fibronectin fragment CH-296-(Takara Shuzo, Otsu, Japan) coated bacterial dishes with 10 mL of viral supernatant supplemented with 100 ng/mL SCF and 100 ng/mL IL-6. Fresh viral supernatant was added after 2 hours and again after 22 hours, and the infection continued for a total of 48 hours. Infected cells were then plated directly in methycellulose medium containing G418 at a concentration of 1.0 mg/mL, and the hematopoietic growth factors IL-3, IL-6, and SCF. Serial replating of hematopoietic colonies was performed as described above. Cell lines were established by growing cells in liquid cultures containing Iscove's media containing 15% fetal calf serum (FCS), 2 mmol/L glutamine, 10 ng/mL of IL-3 and IL-6, 50 ng/mL SCF, 0.1 mmol/L 2-mercaptoenthanol, 10 μg/mL insulin, 200 μg/mL transferrin, and 3 U/mL erythropoietin.
RESULTS
Embryonic lethality, absence of definitive fetal liver hematopoiesis, and lethal hemorrhages in mice heterozygous for the AML1-ETOknock-in allele.
To create an AML1-ETO chimeric gene that mimics that formed by the t(8;21), we fused human AML1-ETO sequences in frame to murine AML1 exon 4 (Fig 1). This targeting strategy resulted in the generation of a murine/human hybridAML1-ETO gene whose expression is controlled by endogenous murine AML1 regulatory sequences. Three independentAML1-ETO/+ ES cell clones with normal ploidy were analyzed and shown by RT-PCR to express AML1-ETO (Fig 1). In undifferentiated ES cells, endogenous murine AML1 is expressed and can be detected by RT-PCR (data not shown).
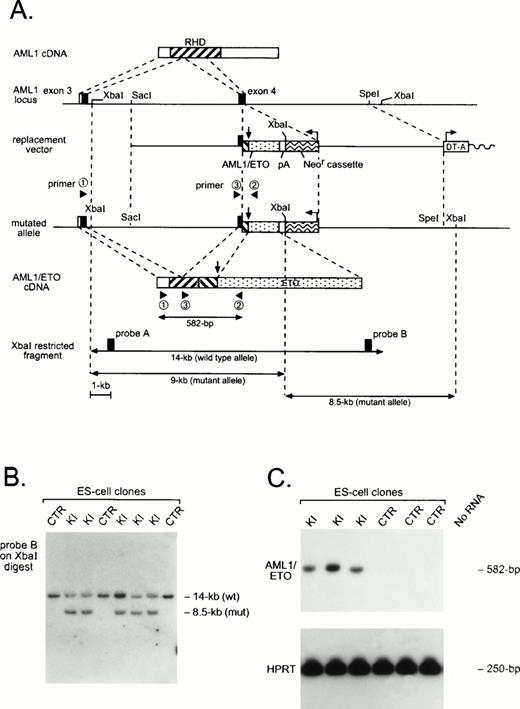
Generation of an AML1-ETO chimeric gene by homologous recombination. (A) Schematic of AML1 cDNA, partial murine AML1 genomic locus, and replacement vector containing a partial human AML1-ETO cDNA, polyadenylation signal (pA), and the positive selection neomycin resistance cassette (Neor) and negative selection diphtheria toxin-A cassette (DT-A). Arrows indicate the position of the AML1-ETO fusion and the transcriptional orientation of selection cassettes. The structure of the targeted allele and the chimeric AML1-ETO cDNA is shown, as are the primers used for RT-PCR amplification and detection of theAML1-ETO fusion transcript. Use of AML1 genomic probes A or B on Xba I–digested DNA allows resolution of wild-type and targeted alleles. (B) Southern analysis of control (CTR) and AML1-ETO knock-in (KI) ES cell clones. (C) RT-PCR analysis of CTR and KI ES cell clones. AML1-ETO mRNA was amplified using primers 1 and 2, and products were hybridized with the murine AML1-specific oligonucleotide 3. Amplification was also performed for HPRTmRNA as a control for the presence of amplifiable RNA.
Generation of an AML1-ETO chimeric gene by homologous recombination. (A) Schematic of AML1 cDNA, partial murine AML1 genomic locus, and replacement vector containing a partial human AML1-ETO cDNA, polyadenylation signal (pA), and the positive selection neomycin resistance cassette (Neor) and negative selection diphtheria toxin-A cassette (DT-A). Arrows indicate the position of the AML1-ETO fusion and the transcriptional orientation of selection cassettes. The structure of the targeted allele and the chimeric AML1-ETO cDNA is shown, as are the primers used for RT-PCR amplification and detection of theAML1-ETO fusion transcript. Use of AML1 genomic probes A or B on Xba I–digested DNA allows resolution of wild-type and targeted alleles. (B) Southern analysis of control (CTR) and AML1-ETO knock-in (KI) ES cell clones. (C) RT-PCR analysis of CTR and KI ES cell clones. AML1-ETO mRNA was amplified using primers 1 and 2, and products were hybridized with the murine AML1-specific oligonucleotide 3. Amplification was also performed for HPRTmRNA as a control for the presence of amplifiable RNA.
Chimeric male mice generated from two independent AML1-ETO/+ ES cells were bred with wild-type C57BL/6 females, and agouti pups were obtained, showing germline transmission of the ES cell–derived genome. Of the 94 live agouti pups genotyped, none contained theAML1-ETO knock-in allele, suggesting that expression of AML1-ETO was embryonic lethal (Table 1). To define the stage of embryonic development at which expression of AML1-ETO is lethal, we analyzed embryos between 10.5 and 14.5 days of gestation. At E10.5 and E11.5 the majority of AML1-ETO/+ embryos were viable and showed no significant morphologic abnormalities when compared with their normal litter mates (Table 1 and data not shown). However, by E12.5 a significant proportion of theAML1-ETO/+ embryos were dead, and by E14.5 no viable embryos with the knock-in allele were detected (Table 1). From E12.5 to E13.5 the AML1-ETO/+ embryos showed normal overall organ development and were equal in size to their control litter mates; however, theAML1-ETO/+ embryos were identifiable by marked fetal liver pallor and by massive hemorrhages within the ventricles of the central nervous system (CNS) and within the pericardial cavity and the soft tissues of the back (Fig 2).
Genotype and Phenotype of Embryos Derived From AML1-ETO Chimeric Males Mated with Normal Females
| Stages . | Number of Pups . | Genotype . | Phenotype . | ||
|---|---|---|---|---|---|
| +/+ . | +/KI* . | Hemorrhage . | Dead . | ||
| E10.5 | 48 | 34 | 14 | 0 | 0 |
| E11.5 | 58 | 38 | 20 | 1 | 1 |
| E12.5 | 62 | 37 | 25 | 21 | 5 |
| E13.5 | 60 | 39 | 21 | 21 | 10 |
| E14.5 | 32 | 18 | 14 | 14 | 14 |
| At birth | 94 | 94 | 0 | — | — |
| Stages . | Number of Pups . | Genotype . | Phenotype . | ||
|---|---|---|---|---|---|
| +/+ . | +/KI* . | Hemorrhage . | Dead . | ||
| E10.5 | 48 | 34 | 14 | 0 | 0 |
| E11.5 | 58 | 38 | 20 | 1 | 1 |
| E12.5 | 62 | 37 | 25 | 21 | 5 |
| E13.5 | 60 | 39 | 21 | 21 | 10 |
| E14.5 | 32 | 18 | 14 | 14 | 14 |
| At birth | 94 | 94 | 0 | — | — |
*KI, AML1-ETO knock-in allele.
Phenotype of E12.5 AML1-ETO/+ and wild-type (WT) embryos. (Top panels) AML1-ETO/+ embryos were identical in size to wild-type littermates, but were easily identifiable by the presence of fetal liver pallor and massive hemorrhages within the ventricles of the CNS and the soft tissues of the back. (Second panels) Sections showing hemorrhages within the dorsal root ganglia ofAML1-ETO/+ embryos. (Third panels) Sections of the fetal liver from AML1-ETO/+ embryos showing a complete absence of hematopoietic precursors with only rare primitive nucleated erythrocytes seen within hepatic sinusoids. By contrast, sections of the fetal liver from control littermates show numerous erythroblasts and scattered myeloblasts and megakaryocytes. (Bottom panels) Smears of peripheral blood show the absence of definitive erythrocytes and platelets in the AML1-ETO/+ embryos. By contrast, numerous enucleated definitive erythrocytes and platelets are seen in the peripheral blood from wild-type embryos.
Phenotype of E12.5 AML1-ETO/+ and wild-type (WT) embryos. (Top panels) AML1-ETO/+ embryos were identical in size to wild-type littermates, but were easily identifiable by the presence of fetal liver pallor and massive hemorrhages within the ventricles of the CNS and the soft tissues of the back. (Second panels) Sections showing hemorrhages within the dorsal root ganglia ofAML1-ETO/+ embryos. (Third panels) Sections of the fetal liver from AML1-ETO/+ embryos showing a complete absence of hematopoietic precursors with only rare primitive nucleated erythrocytes seen within hepatic sinusoids. By contrast, sections of the fetal liver from control littermates show numerous erythroblasts and scattered myeloblasts and megakaryocytes. (Bottom panels) Smears of peripheral blood show the absence of definitive erythrocytes and platelets in the AML1-ETO/+ embryos. By contrast, numerous enucleated definitive erythrocytes and platelets are seen in the peripheral blood from wild-type embryos.
On microscopic examination, the CNS hemorrhages appeared to originate within midbrain parenchyma, the dorsal root ganglia, and the ganglia of the VII/VIII cranial nerve complex (Fig 2). Focal hemorrhages were detected within these structures as early as E11.5. Apoptotic neural cells were also identified within these ganglia; however, no differences in the number of apoptotic cells were noted between normal and AML1-ETO/+ embryos. In addition, we saw no evidence of necrosis in the developing neural structures before the onset of bleeds.
The major developmental defect identified in the AML1-ETO/+ embryos was the complete absence of definitive fetal liver–derived hematopoiesis. Primitive yolk sac–derived hematopoiesis was intact, as assessed by the absence of overt anemia and by normal morphology of primitive nucleated erythrocytes. By contrast, microscopic sections of the fetal liver revealed the complete absence of definitive fetal liver–derived hematopoiesis (Fig 2). Within the liver no erythroid, myeloid, or megakaryocytic progenitors were identified, and only scattered primitive nucleated erythrocytes were seen. Peripheral blood smears from E11.5-E13.5 AML1-ETO/+ embryos contained only primitive nucleated erythrocytes and lacked visible platelets (Fig 2). By contrast, platelets and definitive erythrocytes were easily identified within smears from control litter mates. In addition, neutrophils and monocytes were seen upon scanning the smears from normal E11.5-E13.5 embryos but were absent from AML1-ETO/+ smears (data not shown).
Expression of AML1-ETO leads to the generation of dysplastic hematopoietic progenitors.
To further characterize the hematopoietic defect identified inAML1-ETO/+ embryos, we analyzed cells from yolk sacs and fetal livers for in vitro hematopoietic colony forming activity. Cells from the yolk sacs of viable E10.5 embryos were plated in methycellulose media under conditions optimal for the development of myeloid, erythroid, and mixed colonies. At this stage of development no primitive hematopoietic progenitors are detected under the in vitro culture conditions used,39 and thus all hematopoietic colonies detected in these assays are of definitive origin. Numerous colonies grew in cultures of yolk sacs from E10.5 wild-type embryos, whereas only rare granulocytic or granulocytic-monocytic colonies were identified in yolk sac cultures from E10.5 AML1-ETO/+ embryos (Table 2). Similarly, numerous definitive hematopoietic colonies were detected in cultures of fetal liver cells from E11.5 wild-type embryos. By contrast, 20- to 30-fold fewer cells were recovered from fetal livers of E11.5 AML1-ETO/+ embryos, and when these cells were plated in methycellulose at numbers equal to that used in wild-type cultures, only rare hematopoietic colonies were identified (Table 2). AML1-ETO expression was confirmed within these colonies by RT-PCR analysis (data not shown). Taken together, these data show that expression of AML1-ETO leads to almost a complete absence of definitive hematopoiesis; however, in contrast to AML1-deficient embryos,18 19 a few progenitors could be identified.
Hematopoietic Progenitors in Yolk Sac and Fetal Liver
| Source . | Age . | Genotype . | Number of Embryos . | Erythroid . | Myeloid . | Mixed . |
|---|---|---|---|---|---|---|
| Yolk sac | E10.5 | +/+ | 10 | 30 ± 14* | 179 ± 60 | 59 ± 43 |
| +/KI | 10 | 0 | 5 ± 5 | 0 | ||
| Fetal livers | E11.5 | +/+ | 23 | 6 ± 6† | 61 ± 29 | 13 ± 13 |
| +/KI | 9 | 0 | 4 ± 4 | 4 ± 4 | ||
| E12.5 | +/+ | 7 | 9 ± 5 | 62 ± 36 | 12 ± 7 | |
| +/KI | 12 | 0 | 0 | 82 ± 53‡ | ||
| E13.5 | +/+ | 7 | 2 ± 1 | 61 ± 11 | 4 ± 3 | |
| +/KI | 3 | 0 | 0 | 37 ± 13 |
| Source . | Age . | Genotype . | Number of Embryos . | Erythroid . | Myeloid . | Mixed . |
|---|---|---|---|---|---|---|
| Yolk sac | E10.5 | +/+ | 10 | 30 ± 14* | 179 ± 60 | 59 ± 43 |
| +/KI | 10 | 0 | 5 ± 5 | 0 | ||
| Fetal livers | E11.5 | +/+ | 23 | 6 ± 6† | 61 ± 29 | 13 ± 13 |
| +/KI | 9 | 0 | 4 ± 4 | 4 ± 4 | ||
| E12.5 | +/+ | 7 | 9 ± 5 | 62 ± 36 | 12 ± 7 | |
| +/KI | 12 | 0 | 0 | 82 ± 53‡ | ||
| E13.5 | +/+ | 7 | 2 ± 1 | 61 ± 11 | 4 ± 3 | |
| +/KI | 3 | 0 | 0 | 37 ± 13 |
*Mean ± SD per yolk sac.
Mean ± SD per 10,000 fetal liver cells.
Abnormal colonies with dysplastic morphology.
Similar to the results obtained with E11.5 fetal livers, the total number of cells recovered from fetal livers of viable E12.5 and E13.5AML1-ETO/+ embryos was also 20- to 30-fold less than that recovered from wild-type embryos. In contrast to the results obtained with E11.5 embryos, however, cultures of fetal livers cells from viable E12.5 and E13.5 AML1-ETO/+ embryos contained numerous abnormal multilineage colonies (Table 2 and Fig 3).These colonies consisted of large, tightly packed aggregates of differentiating erythroid precursors, monocytes, megakaryoblasts, and hypergranular cells of the myeloid lineage. Each lineage showed evidence of dysplastic morphology with numerous abnormal erythroblast, megakaryoblast, and multinucleated myeloid cells identified (Fig 3).
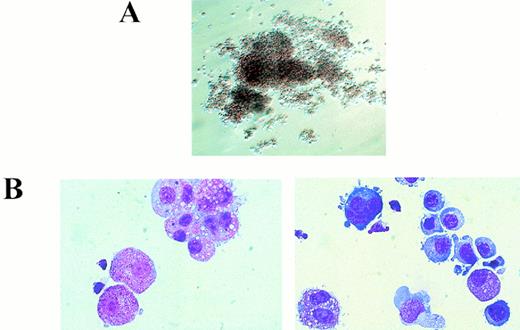
Morphology of dysplastic AML1-ETO/+ mixed hematopoietic colonies. (A) Abnormal mixed colonies derived fromAML1-ETO/+ fetal liver cells. (B) Cytocentrifuge preparations of cells contained within these colonies. (Left) Numerous hypergranular myeloid cells are seen with frequent abnormal binucleated and trinucleated cells detected. (Right) Illustration of the mixed nature of the colony with maturing erythroid cells, a monocyte, binucleated myeloid precursors, and a dysplastic megakaryoblast.
Morphology of dysplastic AML1-ETO/+ mixed hematopoietic colonies. (A) Abnormal mixed colonies derived fromAML1-ETO/+ fetal liver cells. (B) Cytocentrifuge preparations of cells contained within these colonies. (Left) Numerous hypergranular myeloid cells are seen with frequent abnormal binucleated and trinucleated cells detected. (Right) Illustration of the mixed nature of the colony with maturing erythroid cells, a monocyte, binucleated myeloid precursors, and a dysplastic megakaryoblast.
To further characterize these abnormal colonies, we collected cells from individual AML1-ETO/+ or wild-type fetal liver cultures after 7 to 10 days of growth in methylcellulose, disaggregated the cells, and then grew them in liquid cultures either in the presence or absence of IL-3, IL-6, and SCF. Under these conditions, cellular growth and survival was dependent on the presence of hematopoietic growth factors, with rapid cell death observed in their absence. To confirm expression of AML1-ETO within the expanding hematopoietic population, Western blot analysis was performed using an affinity purified ETO-specific rabbit antisera.10 As shown in Fig4, the AML1-ETO chimeric protein was not observed in cells from control cultures but was easily detected within the hematopoietic cells derived from each of the AML1-ETO/+ fetal liver cultures (representative results from six cultures presented). The level of AML1-ETO expression was similar to that of endogenous AML1 as detected with an AML1-specific antiserum (data not shown).
AML1-ETO expression in the dysplastic hematopoietic cells. Western blot analysis of total cell lysates prepared from wild-type (Wt.) or AML1-ETO/+ knock-in (KI) cells. Cell lysates transferred to nitrocellulose membranes were blotted with affinity purified ETO N-terminal peptide antisera and visualized with supersignal ULTRA chemiluminescence substrate.
AML1-ETO expression in the dysplastic hematopoietic cells. Western blot analysis of total cell lysates prepared from wild-type (Wt.) or AML1-ETO/+ knock-in (KI) cells. Cell lysates transferred to nitrocellulose membranes were blotted with affinity purified ETO N-terminal peptide antisera and visualized with supersignal ULTRA chemiluminescence substrate.
To examine the self-renewal capacity of the cells within the fetal liver-derived colonies, we replated either single colonies or bulk cell populations in secondary methylcellulose cultures. When these cells were replated under conditions optimal for the development of multipotential hematopoietic progenitors, similar numbers of colonies were observed from both the AML1-ETO/+ and the control cells (Table3). However, in contrast to the wild-type cells, the AML1-ETO/+ cells formed relatively large dysplastic granulocyte-monocyte and multilineage colonies identical to those seen in the primary cultures (Fig 5A). Moreover, upon subsequent replating the AML1-ETO/+ cells continued to form dysplastic mixed colonies at a high efficiency. Cells from control cultures failed to form colonies after the seventh replating, whereas AML1-ETO/+ cells continued to generate colony-forming progenitors in methylcellulose, without any loss in efficiency of colony formation beyond 20 passages (results from the initial 10 passages are shown in Table 3). Although the colony-forming efficiency for the AML1-ETO/+ cells remained relatively constant, the frequency of multilineage colonies containing erythroid cells and megakaryocytes decreased with each passage. Remarkably, however, definitive multilineage colonies remained through passage 16 (Fig 5A). The immature dysplastic myeloid and monocytic cells from the replated colonies readily expanded in liquid cultures containing IL-3, IL-6, and SCF and coexpressed the myeloid and monocytic markers GR1 and MAC1 (Fig 5B). Identical results were obtained from AML1-ETO–expressing fetal liver hematopoietic progenitors derived from two independently targeted ES cell clones. Taken together, these data suggest that expression of AML1-ETO during the establishment of definitive hematopoiesis directly leads to the generation of multipotential progenitors that have a high self-renewal capacity and dysplastic maturation.
Serial In Vitro Assays of Fetal Liver Hematopoietic Progenitors
| Genotype . | In Vitro Passage* . | |||||
|---|---|---|---|---|---|---|
| 1° . | 2° . | 4° . | 6° . | 8° . | 10° . | |
| Wild-type | 198 ± 25† | 2,112 ± 219 | 2,533 ± 525 | 13 ± 19 | 0 | 0 |
| AML1-ETO/+ | 135 ± 60 | 900 ± 154 | 1,321 ± 407 | 702 ± 278 | 600 ± 188 | 650 ± 153 |
| Genotype . | In Vitro Passage* . | |||||
|---|---|---|---|---|---|---|
| 1° . | 2° . | 4° . | 6° . | 8° . | 10° . | |
| Wild-type | 198 ± 25† | 2,112 ± 219 | 2,533 ± 525 | 13 ± 19 | 0 | 0 |
| AML1-ETO/+ | 135 ± 60 | 900 ± 154 | 1,321 ± 407 | 702 ± 278 | 600 ± 188 | 650 ± 153 |
*Hematopoietic cells from methylcellulose cultures were disaggregated, washed, and 3 × 104 viable cells were replated in triplicate into fresh 35-mm methylocellulose cultures.
Numbers represent myeloid and mixed colonies per 3 × 104 viable cells ± SD.
Morphology and immunophenotype of AML1-ETO/+ hematopoietic cells. (A, left-to-right) Typical dysplastic multilineage AML1-ETO/+ colony from passage 16 with prominent hemaglobinization. Wright-Giemsa–stained cytocentrifuge preparation of this colony showing erythroid, monocytic, and megakaryocytic elements. Prominent alpha naphthyl butyrate cytochemical activity in a cluster of myeloid and monocytic cells from within this colony. (B) Flow cytometeric analysis of surface antigen expression in AML1-ETO/+ hematopoietic cells expanded in liquid cultures. Solid black line represents staining obtained with antibodies specific for the indicated hematopoietic antigen. Dashed line corresponds to the signal obtained with an isotype matched control antibody.
Morphology and immunophenotype of AML1-ETO/+ hematopoietic cells. (A, left-to-right) Typical dysplastic multilineage AML1-ETO/+ colony from passage 16 with prominent hemaglobinization. Wright-Giemsa–stained cytocentrifuge preparation of this colony showing erythroid, monocytic, and megakaryocytic elements. Prominent alpha naphthyl butyrate cytochemical activity in a cluster of myeloid and monocytic cells from within this colony. (B) Flow cytometeric analysis of surface antigen expression in AML1-ETO/+ hematopoietic cells expanded in liquid cultures. Solid black line represents staining obtained with antibodies specific for the indicated hematopoietic antigen. Dashed line corresponds to the signal obtained with an isotype matched control antibody.
Murine adult hematopoietic cells transduced with AML1-ETO exhibit a high self-renewal capacity and readily establish cell lines in vitro.
To further define the role of AML1-ETO in the growth abnormalities observed in hematopoietic progenitors, we targeted its expression to adult bone marrow–derived progenitors by retroviral infection (Fig6). A human AML1-ETO cDNA was inserted into the murine stem cell retroviral vector (MSCV), which contains a neomycin resistance gene (neo) under the control of the phosphoglycerate kinase promoter.36 This vector yields high titer virus capable of efficiently transducing and expressing genes in murine hematopoietic stem cells and their progeny and allows direct selection of infected cells based on resistance to G418. In two independent experiments bone marrow cells obtained from 5-fluorouracil (5-FU)–treated mice were infected with helper-free retroviral supernatants containing MSCV/AML1-ETOneo or MSCVneo.Infected cells were then plated directly in G418-containing methylcellulose medium under conditions optimal for the growth of multilineage hematopoietic colonies (Fig 6A and B). Using viral supernatants with titers of 1 to 5 × 105 G418-resistant cfu/mL, between 5% and 10% of colony forming progenitors acquired G418 resistance. Although resistant granulocyte, granulocyte-macrophage, macrophage, and mixed colonies were identified in all cultures, fewer colonies were generated following infection with MSCV/AML1-ETOneo. The cause of this difference is not immediately apparent.
The effects of AML1-ETO expression on bone marrow–derived hematopoietic progenitors. (A and B) Replating efficiencies in two independent experiments. The open circles with red line depict the results of cell infected with MSCVneo, whereas the solid squares with blue line represent the results from cell infected with MSCV/AML1-ETOneo. (C) Flow cytometric analysis of surface antigen expression on MSCV/AML1-ETOneo–infected hematopoietic progenitors derived from the 13th replating. These cells were carried in liquid cultures for 3 weeks before immunophenotypic analysis. The solid line represents the staining obtained with the antibody specific for the indicated antigen. The dashed line represents the signal obtained with an isotype matched control antibody. (D) Wright-Giemsa–stained cytocentrifuge preparation of cells obtained from the 13th replating. (E) Western blot analysis of cell lysates prepared from MSCV/AML1-ETOneo–infected hematopoietic cells using affinity purified antibodies raised against an AML1–N-terminal peptide.
The effects of AML1-ETO expression on bone marrow–derived hematopoietic progenitors. (A and B) Replating efficiencies in two independent experiments. The open circles with red line depict the results of cell infected with MSCVneo, whereas the solid squares with blue line represent the results from cell infected with MSCV/AML1-ETOneo. (C) Flow cytometric analysis of surface antigen expression on MSCV/AML1-ETOneo–infected hematopoietic progenitors derived from the 13th replating. These cells were carried in liquid cultures for 3 weeks before immunophenotypic analysis. The solid line represents the staining obtained with the antibody specific for the indicated antigen. The dashed line represents the signal obtained with an isotype matched control antibody. (D) Wright-Giemsa–stained cytocentrifuge preparation of cells obtained from the 13th replating. (E) Western blot analysis of cell lysates prepared from MSCV/AML1-ETOneo–infected hematopoietic cells using affinity purified antibodies raised against an AML1–N-terminal peptide.
Cells were obtained from primary cultures 8 to 10 days after plating and replated in secondary methycellulose cultures in the presence of G418. This process was serially repeated and the number and morphology of colonies analyzed after each passage. As shown in Fig 6A and B, a slight increase in the efficiency of colony formation was noted through the first four passages for both MSCVneo- and MSCV/AML1-ETOneo–infected cells. Following passage 4, however, the number of colony forming progenitors rapidly declined in the MSCVneo population, with no colonies detected following the 6th passage. By contrast, MSCV/AML1-ETOneo cells continued to show an increase in the efficiency of colony formation through subsequent passages. In the first experiment (Fig 6A), MSCV/AML1-ETOneo–infected cells formed colonies at a high efficiency through 22 generations (results from 13 passages shown). Similarly, in a second more recent experiment that has to date been carried through a shorter number of generations (Fig 6B), MSCVneo cells failed to form colonies after the 5th replating, whereas MSCV/AML1-ETOneo cells continued to efficiently form colonies through 13 passages (data from 6 passages shown).
The replated colonies from MSCV/AML1-ETOneo cells were composed primarily of immature blasts like cells that readily expanded into cell lines in liquid cultures in the presence of IL-3, IL-6, and SCF. These cells expressed c-kit but were negative for GR-1, Mac-1, Sca-1, and Thy-1 (Fig 6C). In addition, the cells had the morphology of immature hematopoietic cells with only rare mid-mature myeloid and monocytic cells identified (Fig 6D). Western blot analysis of cell lysates from MSCV/AML1-ETOneo–infected populations revealed AML1-ETO expression (Fig 6E). Taken together, these data further show that expression of AML1-ETO can directly lead to the generation of abnormal hematopoietic progenitors with a high self-renewal capacity.
DISCUSSION
To directly investigate the mechanistic role of the t(8;21)-encoded AML1-ETO chimeric product in leukemogenesis, we used gene targeting to create mice with an AML1-ETO “knock-in” allele that mimics the t(8;21). We now show that murine embryos heterozygous for the AML1-ETO knock-in allele have normal yolk sac–derived primitive hematopoiesis but have a near total absence of fetal liver-derived definitive hematopoiesis and die at embryonic day 13.5 from massive bleeding in the CNS, pericardial sac, and soft tissue. This bleeding diathesis appeared to result, at least in part, from the absence of circulating platelets compounded by an evolving anemia due to the inability to make definitive erythrocytes. Thus, the phenotype that resulted from AML1-ETO expression was nearly identical to the embryonic lethal phenotype seen following homozygous disruption of eitherAML118,19 orCBFβ.20,21 This similarity in phenotypes suggests that AML1-ETO effectively neutralizes the normal biologic activity of the AML1/CBFβ transcriptional factor complex and thus supports published data that suggested that AML1-ETO represses AML1-mediated transcriptional activity through a dominant-negative mechanism.10 31-34
Despite the overall similarity between the phenotypes resulting from the loss of AML1/CBFβ and expression of AML1-ETO, two significant differences were observed. First, AML1-ETO–expressing embryos lived 1 day longer than AML1- or CBFβ-deficient embryos. Second, fetal livers from AML1-ETO–expressing embryos contained rare hematopoietic progenitors, whereas no progenitors were detected in the fetal livers of AML1- or CBFβ-deficient embryos. The presence of these cells in AML1-ETO/+ embryos could, in part, be responsible for the longer survival of these embryos, given that these cells have the capacity to differentiate into megakaryocytes and presumably functional platelets, hematopoietic elements capable of providing protection from bleeding. These progenitors, however, failed to establish a functional hematopoietic system within the developing embryos. Despite this apparent inability to expand in vivo, these cells readily grew in cultures where they displayed significant morphologic dysplasia. Moreover, these AML1-ETO–expressing cells had an abnormally high self-renewal capacity, a property not seen in normal fetal liver–derived progenitors but more typical of leukemic cells. The identical hematopoietic abnormalities were observed in AML1-ETO/+ heterozygous mice derived from two independent ES cell lines, suggesting that the phenotype was a direct result of AML1-ETO expression. This was further confirmed by expressing AML1-ETO from a retroviral LTR in adult bone marrow–derived hematopoietic progenitors. In these experiments, immature hematopoietic colony forming progenitors were generated that had a high self-renewal capacity, and again could be readily established into cell lines when grown in liquid cultures in the presence of hematopoietic growth factors. Taken together, these data suggest that AML1-ETO suppresses normal AML1/CBFβ activity and leads to the direct generation of signals that contribute to the initiation of aberrant hematopoietic cell proliferation.
Although our data do not directly address the mechanism by which AML1-ETO mediates the expansion of abnormal hematopoietic progenitors, several possibilities are suggested by our observations: (1) the AML1-ETO chimeric protein may cause an incomplete repression of normal AML1 transcriptional activity and thereby result in dysregulated expression of target genes critical for hematopoiesis; (2) expression of AML1-ETO may result in novel gain-of-function activities that alter the transcription of genes normally regulated by AML1/CBFβ or other runt homology domain-containing transcription factors; (3) novel AML1-ETO–mediated activities may affect the expression of genes that are not normally regulated by this class of transcription factors; or (4) a combination of these effects. Irrespective of the underlying mechanism, our data provide direct evidence that expression of AML1-ETO contributes to the expansion of abnormal hematopoietic progenitors by increasing the self-renewal capacity of multipotential hematopoietic progenitors.
Somewhat surprising was the observation that the dysplasticAML1-ETO/+ hematopoietic cells failed to expand to appreciable levels within the embryos. Similarly, in preliminary experiments theAML1-ETO/+ cells fail to induce leukemia in sublethally irradiated syngeneic or severe combined immunodeficiency disease (SCID) mice (Downing and Cai, unpublished data, March 1998). Despite this lack of growth in vivo, these cells readily expanded in vitro when cultured in the presence of IL-3, IL-6, and SCF. Moreover, the cells are growth factor–dependent both for survival and proliferation. One possible explanation for these results is that aberrant expression of AML1-ETO may lead to an altered sensitivity to growth factor–induced proliferation or differentiation, with high growth factor concentrations such as those used for in vitro cultures required for the expansion of the abnormal AML1-ETO progenitors. This possibility is strengthened by the known role that AML1/CBFβ plays in the transcriptional regulation of growth factors such as GM-CSF and IL-313,14 and growth factor receptors such as CSF-1R.15 The role of growth factor signaling pathways in the expansion of this abnormal hematopoietic population remains to be determined.
Although expression of AML1-ETO resulted in an abnormally high self-renewal capacity in both fetal liver– and adult bone marrow–derived hematopoietic progenitors, the phenotypes of the expanding cells differed between these populations. Immature myelomonocytic cells were obtained following expression of AML1-ETO in fetal liver–derived progenitors, whereas more undifferentiated hematopoietic progenitors were obtained following expression of AML1-ETO in bone marrow–derived progenitors. These differences may result from expression of AML1-ETO in distinct stem or progenitor cell populations. Alternatively, the difference may result from subtle differences in the pattern of expression of AML1-ETO in the expanding hematopoietic population that result from the different promoters used to drive its expression. Nevertheless, despite the observed phenotypic differences both populations showed some evidence of terminal differentiation. Significantly, in the fetal liver–derived cells, terminal hematopoietic differentiation was observed along the myeloid, monocytic, erythroid, and megakaryocytic lineages. Thus, these data clearly show that although AML1-ETO expression leads to abnormal proliferation and maturation, it does not block terminal hematopoietic differentiation.
A number of investigators have recently used knock-in approaches to replace one gene with another40 or to create chimeric oncogenes.41-43 Yergeau and colleagues43generated mice heterozygous for an AML1-ETO knock-in allele by using a strategy that was slightly different from ours. Their data also showed that expression of AML1-ETO resulted in the death of embryos at midgestation from CNS hemorrhages and a severe impairment of fetal liver–derived hematopoiesis. In contrast to our results, however, they did not observe dysplastic hematopoietic progenitors within the fetal liver but instead identified rare apparently normal macrophage colonies in yolk sac cultures. Several differences exist between the targeting strategies used by the two groups, including the exon targeted and the transcriptional orientation of the neomycin resistance gene in relationship to AML1. These differences could result in significant changes in the level of expression of the chimeric gene and thus may be responsible for the phenotypic differences observed. Although the level of AML1-ETO expression was not documented in Yergeau studies, using our strategy we consistently observed levels of AML1-ETO that were similar to that of endogenous AML1. Moreover, easily detectable levels of AML1-ETO were obtained following retroviral mediated expression in bone marrow–derived progenitors, and again this level of expression resulted in an increase in the self-renewal capacity and in vitro immortilization of these cells.
In summary, we have presented data that suggest that expression of AML1-ETO results in both the neutralization of normal AML1/CBFβ-mediated activities required for definitive hematopoiesis and the direct generation of signals that result in the expansion of dysplastic hematopoietic progenitors. These AML1-ETO–expressing progenitors are likely to represent the immediate precursors to full blown leukemic populations. An investigation of the growth properties and leukemic potential of the AML1-ETO/+ cells derived through these experiments should provide valuable insights into the pathways through which AML1-ETO contributes to human acute myeloid leukemia.
ACKNOWLEDGMENT
The authors thank W. Paul Conn, W. Kent Williams, Cristy Nagy, and John Swift for excellent technical assistance and Drs Scott Hiebert, Darin O'Brien, and Gerard Grosveld for helpful discussions and critical reading of the manuscript.
Supported by National Instituted of Health (NIH) Grant P01 CA71907-01, NIH Cancer Center CORE Grant CA-21765, and the American Lebanese and Syrian Associated Charities of St Jude Children's Research Hospital.
Address reprint requests to James R. Downing, MD, Department of Pathology and Laboratory Medicine, St Jude Children's Research Hospital, 332 N Lauderdale, Memphis, TN 38105.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal