Abstract
Two new members of human β-chemokine cDNA were isolated based on structural and functional similarities to human leukotactin-1. One of these clones was identical to the previously isolated human β-chemokine, CKβ8, whereas the other is a splicing variant of CKβ8, therefore named CKβ8-1. CKβ8 was short in 51 nucleotides (17 amino acids) compared with CKβ8-1. The mature proteins of CKβ8-1 and CKβ8 consisted of 116 and 99 amino acids with calculated molecular weights of 12,500 and 10,950, respectively. Both CKβ8-1 and CKβ8 were potent agonists at CCR1. These chemokines chemoattracted neutrophils, monocytes, and lymphocytes. They also significantly suppressed colony formation by human bone marrow, granulocyte-macrophage, erythroid, and multipotential progenitor cells stimulated by combinations of growth factors. To our knowledge, this is the first example that an alternative splicing produces two active β-chemokines from a single gene.
THE CHEMOKINES ARE a family of small cytokines that consist of basic, heparin-binding small molecular-weight proteins. Four subfamilies of chemokines have been discovered to date: CXC (α), CC (β), γ (C), and CX3C.1-3 The classification of the chemokine subfamilies is based on the distance between the first two of four-to-six conserved cysteine residues. The CX3C chemokine module is tethered to the surface of the cell membrane by a long mucine-like stalk.3
Reported bioactivities include human immunodeficiency virus (HIV)-suppression, immunoregulation, leukocyte migration, and suppression of hematopoietic stem/progenitor cell proliferation.4-7 Chemokines activate leukocytes by binding to seven-transmembrane domain G protein-coupled receptors, several of which also act pathologically as HIV-1 coreceptors. Nine CC chemokine receptor subtypes, CCR1-9, have previously been identified.8-17 They are all expressed on leukocytes, and together they account for binding sites for most of the known CC chemokines. Some of these receptors, such as CCR1, CCR2, CCR3, CCR4, CCR5, and CCR9 exhibit a promiscuous binding property, whereas one high-affinity ligand has been identified for CCR6, CCR7, and CCR8.
Among the β-chemokines reported to date, murine (m)MRP-1 (also called C10),18 mMRP-2,19 and leukotactin-1 (Lkn-1)20 are distinct from the rest of this group in that they contain an extra two cysteines, which may form a third disulfide bond, and the NH2-terminal regions are fairly extended. MRPs and Lkn-1 can be classified as C6 β-chemokines because they include six conserved cysteines, whereas only four cysteines are aligned in other β-chemokines. These structural features may suggest that C6 β-chemokines constitute a subgroup of β-chemokines which are associated with a distinct bioactivity. Here, we report an additional two members of C6 β-chemokine family, which are created by alternative splicing.
MATERIALS AND METHODS
Cell lines.
Human monocyte/macrophage cell line THP-1, human melanoma cell line OCM-1, and human osteogenic sarcoma cell line HOS were cultured in Dulbecco's modified Eagle's medium with 10% fetal calf serum and antibiotics.
Production of THP-1 cDNA library.
Two micrograms of poly(A)+ RNA from interleukin-4 (IL-4)-stimulated THP-1 cells were converted into double-stranded cDNA using a Time Saver cDNA kit (Pharmacia Biotech, Piscataway, NJ).BstX I adaptors (Invitrogen, San Diego, CA) were added to the cDNA. cDNA larger than 0.5 kb was selected by agarose gel fractionation and cloned into PRc/CMV (Invitrogen) that had been cleaved with BstXI.
DNA sequencing.
CKβ8-1 and CKβ8 cDNAs were sequenced by the dideoxy chain termination method using Sequenase version 2.0 (US Biochemical, Cleveland, OH). CKβ8-1/CKβ8 genomic sequence was determined by an automatic DNA sequencer (ABI).
Northern blot.
The multiple tissue Northern blot and human master blot were purchased from Clonetech (Palo Alto, CA). To investigate differential tissue expression of Lkn-1, CKβ8-1, and CKβ8, each chemokine-specific oligonucleotide probe was generated. For the Lkn-1–specific probe, a COOH-terminal region corresponding to the nucleotide position +292 through +342 (19) was used, whereas the CKβ8-1–specific probe was derived from the nucleotide position +141 to +189 corresponding to 17 amino acids created by alternative splicing. For the CKβ8-1 and CKβ8 common probe, a COOH-terminal region encompassing +316 to +366 was used. Each oligonucleotide was synthesized in antisense manner, kinased in the presence of 100 μCi [γ-32p] adenosine triphosphate with T4 polynucleotide kinase, and purified through G-50 column. Hybridization was performed at 45°C in 5 mL of ExpressHyb solution (Clonetech) with 5 × 106 cpm/mL. Filters were washed according to the instruction provided and were exposed for different time periods.
Chromosomal mapping and determination of exon-intron organization of CKβ8-1/CKβ8 using bacterial artificial chromosome (BAC).
Two human BAC libraries were screened with the CKβ8-1 probe encompassing 5′ untranslated region (UTR) and 103 nucleotides of open reading frame. This region had a 97% identity with Lkn-1 at nucleotide level. Thus, it was hybridized to both Lkn-1 and CKβ8-1 even at high stringent hybridization and washing conditions. Seven positive clones were isolated (Research Genetics, Huntsville, AL). Among these, three clones (9-0-20, 23-1-14, and 110-F-6) were further analyzed. BAC DNAs were purified with either standard protocol omitting phenol-chloroform extraction or QIAGEN-tip 100 (Chatsworth, CA), digested with HindIII, and subjected to Southern blot analysis. In parallel with these BAC DNA, the human genomic DNA was prepared from three different human cell lines (OCM1, a human melanoma cell line; and GM02430 and GM03365, human B-cell lymphoma cell lines) and was subjected to Southern blot analysis. Hybridization was performed at 65°C using QuickHyb solution (Stratagene, LaJolla, CA) for 1 hour, and final wash was at 65°C in the presence of 2 × standard saline citrate (SSC) for 20 minutes. For Southern blot analysis, Lkn-1 and CKβ8-1 cDNA fragments encompassing nucleotide position 103 through respective 3′ UTR were released with HindIII and Xba1 located at nucleotide position 103 and vector, respectively, labeled, and used for hybridization.
To determine exon-intron organization and their nucleotide sequences, a 3-kb HindIII fragment was isolated from BAC 110-F-6 and the entire nucleotide sequence was determined. This provided sequence information on a portion of exons 2, 3, and 4 and introns 2 and 3. The remaining exon 2 sequence was determined from another BAC clone (125-F-14).
Recombinant CKβ8-1 and CKβ8.
The open reading frame of CKβ8-1 and CKβ8 excluding the putative signal sequence was amplified with the following set of polymerase chain reaction (PCR) primers: forward primers (5′-CGAATTCCATATGCA GTTCACAAA TGATGCAGAG-3′) and reverse primers (5′-CGCCGCTCGAGTTGAGTA GGGCTTCAGC-3′). Pfu polymerase (Stratagene, La Jolla, CA) was used for the amplification, and the amplified fragments were digested with NdeI/XhoI and cloned into pET21a (Novagene, Madison, WI) that had been digested withNdeI/XhoI. After the sequence was confirmed, the recombinant plasmid was introduced into expression hostEscherichia coli HMS 174 (Novagen). The construct will produce recombinant CKβ8-1 and CKβ8 to which one amino acid (Met) is added at its amino terminus and six histidines added at its carboxyl terminus.
Because most of the CKβ8-1 and CKβ8 were found in the inclusion body, denaturation and refolding procedures were conducted. The inclusion bodies were dissolved in 20 mL denaturation buffer (6 mol/L guanidine-HCl; 20 mmol/L Tris-HCl, pH 7.9; 500 mmol/L NaCl; and 4 mmol/L n-octylglucopyranoside), and spun down at 15,000 rpm for 30 minutes. The supernatant containing the denatured rCKβ8-1 and rCKβ8 was applied to the activated Ni-column. After binding, the column was washed once with 25 mL buffer containing 6 mmol/L urea; 20 mmol/L Tris-HCl, pH 7.9; 500 mmol/L NaCl; and 20 mmol/L imidazole, followed by solubilization with the addition of 25 mL buffer containing 20 mmol/L Tris-HCl, pH 7.9; and 150 mmol/L NaCl. The refolded rCKβ8-1 and rCKβ8 were eluted with 6 mL elution buffer (20 mmol/L Tris-HCl, pH 7.9; 150 mmol/L NaCl; and 50 mmol/L EDTA). The eluted rCKβ8-1 and rCKβ8 were diluted 10-fold with distilled water, applied to a 1-mL heparin agarose column (Pharmacia Fine Chemicals, Piscataway, NJ), and eluted with a linear salt gradient. The active fractions were obtained between 300 mmol/L and 800 mmol/L NaCl.
Colony assays.
Human bone marrow cells were collected from donors after obtaining informed consent. Low-density human bone marrow cells retrieved after density cut on Ficoll-Hypaque gradients (1.070 g/cm3) were plated at 5 × 104 cells/mL in 0.3% agar culture medium with 10% fetal bovine serum (FBS) for colony formation by granulocyte-macrophage progenitors (CFU-GM), stimulated by recombinant human (rhu) granulocyte-macrophage colony-stimulating factor (GM-CSF; 100 U/mL) plus rhu steel factor (SLF; 50 ng/mL), or were plated in 1% methylcellulose culture medium with 30% FBS for colony formation by CFU-GM, erythroid progenitors (burst forming unit-erythroid; BFU-E), and multipotential progenitors (CFU granulocyte-erythroid-macrophage-megakaryocyte; CFU-GEMM) stimulated by recombinant rhu preparations of erythropoietin (Epo; 1 U/mL), IL-3 (100 U/mL), and SLF (50 ng/mL). Epo was purchased from Amgen Corporation (Thousand Oaks, CA), and GM-CSF, IL-3, and SLF were kind gifts from Immunex Corporation (Seattle, WA). CKβ8-1 or CKβ8 were added to each of the plates at 50 ng/mL. Rabbit polyclonal antibodies to CKβ8-1 and CKβ8 were added to neutralize the chemokines in certain experiments. Cultures were incubated at 5% CO2 and lowered (5%) O2 in a BNP-210 incubator (Tabai ESPEC Corp, South Plainfield, NJ). Colonies were scored after 14 days of incubation.
Cell preparation and chemotaxis assay.
Human peripheral blood mononuclear cells (PBMC) were obtained from healthy donors by gradient centrifugation on Ficoll-paque (Pharmacia Biotech). Monocytes were isolated from PBMC by exploiting their ability to adhere to plastic surface. Lymphocytes were obtained by two-step removal of monocytes from PBMC. The resulting purities of monocytes and lymphocytes were 90% and 88%, respectively, as determined by microscopic examination. Human neutrophils were prepared by erythrocyte sedimentation with 3% Dextran T500 (Sigma Chemical Co, St Louis, MO) followed by Ficoll-Histopaque (Sigma) separation and hypotonic lysis. The purity of neutrophils assessed by morphology was greater than 95%. Purified monocytes and lymphocytes were washed, counted, and resuspended at 2 × 106, 8 × 106, and 2 × 106 cells/mL, respectively, in RPMI 1640 supplemented with 0.5% bovine serum albumin and 20 mmol/L Hepes. Human neutrophils were suspended in Hanks' Balanced Salt Solution (HBSS) at 1 × 106 cells/mL.
Migration of cells was assessed in a microchamber (Neuroprobe, Cabin John, MD) as described previously. Briefly, the lower wells of the microchamber were filled with dilutions of chemokines or with control, and the upper wells with 50 μL of cell suspension. The two compartments were separated by a polyvinylpyrrolidone-free filter with 3-μm pores for neutrophils and lymphocytes and 5-μm pores for monocytes. After incubation at 37°C for 1 hour (neutrophils), 2 hours (monocytes), or 4 hours (lymphocytes), the filters were removed from the chambers, washed, fixed, and stained with Diff-Quick (Dade Diagnostics Inc, Aquada, PR). Finally, the cells of 5 random oil-immersion fields were counted. The chemotactic index was calculated from the number of cells migrated to the test samples divided by the number of cells migrated to the control. Human recombinant IL-8 and hMIP-1α were purchased from R & D Systems (Minneapolis, MN) and used for positive controls.
Calcium flux assay.
Receptor activation was assessed by real time measurement of (Ca2+)i changes using a MSIII flurometer (Photon Technology International, South Brunswick, NJ) in the HOS cell lines which expressed recombinant CCR1, CCR2, CCR3, CCR4, CCR5, and CXCR4 and purified peripheral blood leukocyte subsets. The HOS cell lines were obtained through the AIDS Research and Reference Reagent Program (Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health) from Dr Nathaniel R. Landau. Briefly, 2 × 107 cells were detached with the addition of 5 mL of warm 1 mmol/L EDTA in Dulbecco's phosphate-buffered saline followed by incubation at 37°C for 10 minutes, and washed twice with 10 mmol/L HEPES buffer, pH 7.4, supplemented with 1 × HBSS. The HOS cells and leukocytes were loaded with 5 μmol/L fura-2AM in 2 mL of the above buffer supplemented with 0.8 mmol/L MgCl2, 1.8 mmol/L CaCl2, and 20 mmol/L glucose at 37°C for 30 minutes, washed twice, and resuspended at 1 × 106cells/mL in the same buffer. Two milliliters of the cell suspension was placed in a stirred, water-jacked cuvette at 37°C. Excitation scans between 300 and 400 nm were performed to determine whether fura-2AM was appropriately loaded. Excitation ratios at 340 and 380 nm were measured. rhMIP-1α was purchased from R & D Systems.
RESULTS
Cloning of the gene encoding CKβ8-1 and CKβ8 and alternative splicing.
While we were screening THP.1 cDNA library with a probe representing a portion of exon 4 of Lkn-1, a related cDNA was isolated. The cDNA clone was termed CKβ8-1 because this cDNA was highly homologous to CKβ8.21 Its open reading frame has a 73% amino acid identity with Lkn-1. The nucleotide sequence of CKβ8-1 signal sequence was 95% identical to that of Lkn-1. To determine whether Lkn-1 and CKβ8-1 were unique messages in THP.1 cells, we conducted a reverse transcriptase–PCR using the forward primer (common primer) derived from 5′ UTR and the reverse primers derived from 3′ UTR of Lkn-1 or CKβ8-1. We were able to detect unique Lkn-1 and CKβ8-1 messages. We also found that the CKβ8-1 PCR products involved two more related bands, which had shorter sizes and appeared to be minor species (our unpublished observation, May 1996). Using the CKβ8-1 DNA probe, we again screened the same cDNA library. We obtained ten positive clones, among which seven clones represented CKβ8-1 and two were a short version of CKβ8-1 with identical 5′ UTR and 3′ UTR, indicating that these might be alternative splicing variants. These cDNAs were identical to previously published CKβ8. The remaining cDNA clone was a faulty splicing variant in which exon 3 and a portion of exon 4 were deleted, resulting in a frame-shift mutation.
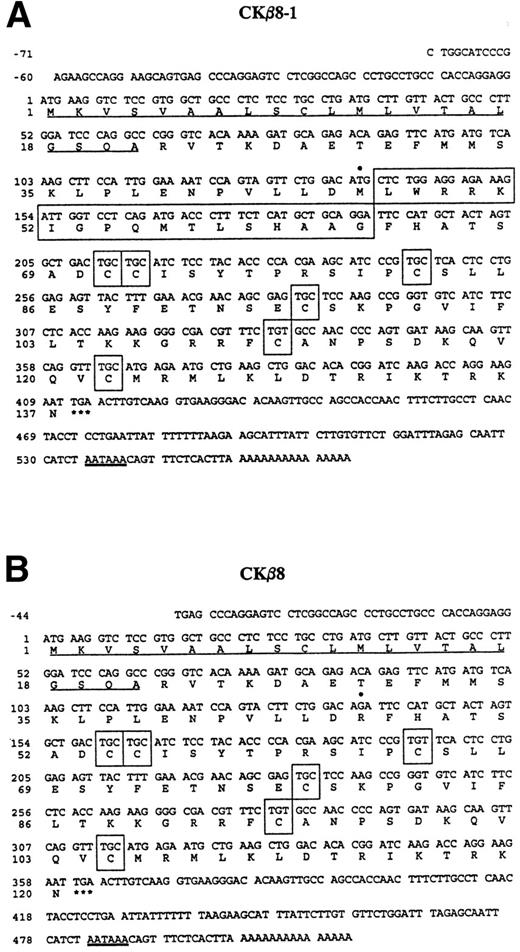
As shown in Fig 1A and B, CKβ8-1 is composed of 137 amino acids, of which the first 21 amino acids constitute a putative signal peptide. The first cysteine is preceded by 49 amino acids. CKβ8 has an identical signal peptide, which is followed by 32 amino acids before the first cysteine. The predicted signal peptides are underlined. Six conserved cysteine motifs are shown in boxes. Seventeen amino acids in a large box were spliced out in CKβ8.
Nucleotide sequence of cDNAs encoding CKβ8-1 and CKβ8 and the deduced amino acid sequence. The nucleotide sequence of the message strand is numbered in the 5′ to 3′ direction. The 5′ UTR sequence is indicated by negative numbers. The predicted amino acid sequence is shown below the nucleotide sequence. Underlined are the putative signal peptides. The stop codons are indicated by a star (*). The six cysteine residues are depicted in boxes. These sequences have been deposited in the GenBank data base (Accession No. U58913 andU67128 for CKβ8-1 and CKβ8, respectively). (A) The CKβ8-1 cDNA sequence is shown. Seventeen amino acids (Leu47 to Gly63) created by alternative splicing are represented by a large box. In addition, Met46 is denoted by a filled circle (•) that was converted to Arg46 in CKβ8 due to the alternative splicing. (B) The CKβ8 cDNA sequence is shown. Depicted by a filled circle (•) is Arg46, which was derived fro m Met46 of CKβ8-1.
Nucleotide sequence of cDNAs encoding CKβ8-1 and CKβ8 and the deduced amino acid sequence. The nucleotide sequence of the message strand is numbered in the 5′ to 3′ direction. The 5′ UTR sequence is indicated by negative numbers. The predicted amino acid sequence is shown below the nucleotide sequence. Underlined are the putative signal peptides. The stop codons are indicated by a star (*). The six cysteine residues are depicted in boxes. These sequences have been deposited in the GenBank data base (Accession No. U58913 andU67128 for CKβ8-1 and CKβ8, respectively). (A) The CKβ8-1 cDNA sequence is shown. Seventeen amino acids (Leu47 to Gly63) created by alternative splicing are represented by a large box. In addition, Met46 is denoted by a filled circle (•) that was converted to Arg46 in CKβ8 due to the alternative splicing. (B) The CKβ8 cDNA sequence is shown. Depicted by a filled circle (•) is Arg46, which was derived fro m Met46 of CKβ8-1.
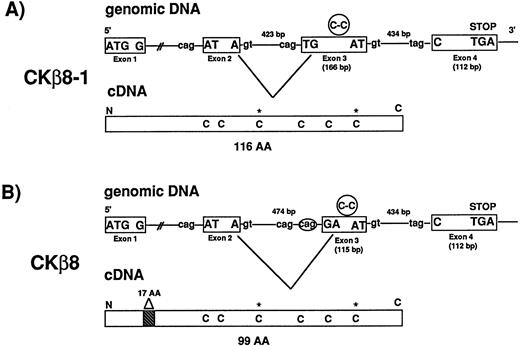
We isolated seven BAC clones representing CKβ8-1 and CKβ8. Exon-intron organization of the CKβ8-1/CKβ8 gene was determined. As seen in Fig 2, the CKβ8-1/CKβ8 gene is comprised of four exons whereas most C-C chemokine genes contain three exons.22 Fig 2A and B show how CKβ8-1 and CKβ8 are created by alternative splicing. Alternative splicing occurred at a region of exon 3 before C-C motif in such a way that CKβ8 was shortened by 17 amino acids at the NH2-terminus. Among these 17 amino acids, one amino acid was created as a unique residue (arginine), which was not found in CKβ8-1.
CKβ8 is an alternative splicing form of CKβ8-1. (A) Two BAC clones (110-F-6 and 125-F-14) representing the CKβ8-1 gene were isolated and further analyzed for determination of exon and intron organization of the gene. A 3.0-kb Hind III fragment was isolated from 110F-6 and the entire nucleotide sequence covering a portion of exons 2, 3, and 4 and introns 2 and 3 was determined. A 1.3-kb HindIII fragment was also isolated from 125-F-14 and the entire nucleotide sequence was determined. This sequence included portions of exon 2 and intron 1. Two independent cDNA clones representing CKβ8-1 and CKβ8 were isolated from THP.1 cDNA library. Their sequences were determined. Exons are shown in boxes. The putative splicing donor (gt) or acceptor (cag or tag) consensus sequences are also shown. The Cys-Cys motifs are represented in circles in exon 3, whereas the extra two cysteines are denoted by stars (*) in cDNAs. (B) Alternative splicing deleted 57 bp from exon 3 in CKβ8-1 in such a way that CKβ8 was formed. A 17–amino acid deletion point is indicated in CKβ8 cDNA.
CKβ8 is an alternative splicing form of CKβ8-1. (A) Two BAC clones (110-F-6 and 125-F-14) representing the CKβ8-1 gene were isolated and further analyzed for determination of exon and intron organization of the gene. A 3.0-kb Hind III fragment was isolated from 110F-6 and the entire nucleotide sequence covering a portion of exons 2, 3, and 4 and introns 2 and 3 was determined. A 1.3-kb HindIII fragment was also isolated from 125-F-14 and the entire nucleotide sequence was determined. This sequence included portions of exon 2 and intron 1. Two independent cDNA clones representing CKβ8-1 and CKβ8 were isolated from THP.1 cDNA library. Their sequences were determined. Exons are shown in boxes. The putative splicing donor (gt) or acceptor (cag or tag) consensus sequences are also shown. The Cys-Cys motifs are represented in circles in exon 3, whereas the extra two cysteines are denoted by stars (*) in cDNAs. (B) Alternative splicing deleted 57 bp from exon 3 in CKβ8-1 in such a way that CKβ8 was formed. A 17–amino acid deletion point is indicated in CKβ8 cDNA.
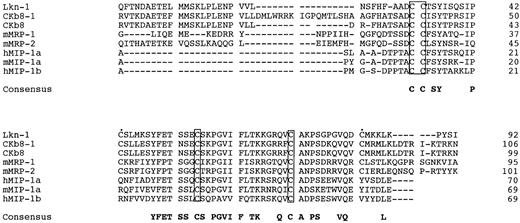
Figure 3 shows an amino acid alignment among three Lkn-1, CKβ8, CKβ8-1, mMRPs, MIP-1α, and MIP-1β, to show two features of Lkn-1, CKβ8s, and MRPs, distinct from the rest of the β-chemokines; Lkn-1, CKβ8-1, CKβ8, and mMRPs constitute a long amino-terminus to the first cysteine and contain six conserved cysteines. The additional two cysteines conserved among Lkn-1, CKβ8-1, CKβ8, and mMRPs may form an extra disulfide bond. Therefore, we refer to this subgroup of β-chemokines as C6 β-chemokines.
Alignment of CKβ8-1 and CKβ with Lkn-1, mMRPs, MIP-1α, and MIP-1β. The putative signal peptide sequence is not shown. Shown in boxes are four conserved cysteine residues, whereas two extra cysteines conserved in MRP families are indicated by filled circles (•). Gaps were introduced for optimum alignment.
Alignment of CKβ8-1 and CKβ with Lkn-1, mMRPs, MIP-1α, and MIP-1β. The putative signal peptide sequence is not shown. Shown in boxes are four conserved cysteine residues, whereas two extra cysteines conserved in MRP families are indicated by filled circles (•). Gaps were introduced for optimum alignment.
Chromosomal localization.
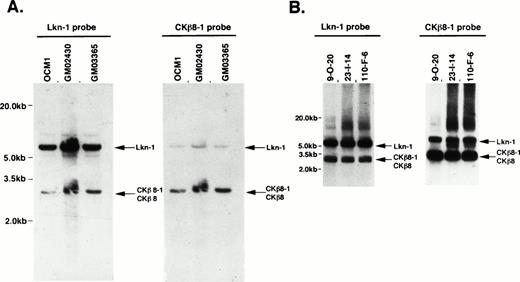
We investigated the location of the gene encoding CKβ8-1 and CKβ8. Figure 4A shows a genomic southern pattern using three different cell lines in which the genes encoding Lkn-1 and CKβ8-1/CKβ8 exist as a single-copy gene. Because there is a 73% identity between these genes, we observed some degree of cross-hybridization in a high stringent hybridization and washing condition (final at 65°C with 2 × SSC), but each probe was preferentially hybridized to its own gene. We isolated several BAC clones representing CKβ8-1/CKβ8 genes. These BAC clones carried approximately 200-kb inserts. Of these, three were subjected to further analysis. As shown in Fig 4B, the genes encoding Lkn-1 and CKβ8-1/CKβ8 were detected from the individual BAC clone by hybridization with either the Lkn-1 or CKβ8-1 probe, suggesting that these two genes might be located on the same chromosome. We determined the sequences of both the 7.0-kb Lkn-1 and 3.0-kb CKβ8-1/CKβ8 bands. Because Lkn-1 gene has been mapped to human chromosome 17,20 we concluded that the gene encoding CKβ8-1 and CKβ8 would be mapped to human chromosome 17, in which the Lkn-1 and CKβ8-1/CKβ8 genes were separated within 200 kb of each other, at the most.
Chromosomal localization of the CKβ8-1/CKβ8 gene. (A) Genomic Southern blots revealed that the genes encoding Lkn-1 and CKβ8-1/CKβ8 are single copy, but related genes. Twenty micrograms of genomic DNA prepared from a melanoma cell line (OCM1) and two B-cell lymphoma cell lines (GM02430 and GM03365) were digested withHindIII and were transferred to a nylon filter membrane. The Lkn-1 or CKβ8-1 probe was hybridized to the membrane, which was finally washed with 2 × SSC at 65°C for 30 minutes. Indicated by arrows are the genes encoding Lkn-1 and CKβ8-1/CKβ8. (B) The genes encoding Lkn-1 and CKβ8-1/CKβ8 are located at the same chromosome. Three BAC clones (9-0-20, 23-1-14, 110-F-8) were isolated and 1 μg of each DNA was digested with HindIII. Southern blot was performed as described above.
Chromosomal localization of the CKβ8-1/CKβ8 gene. (A) Genomic Southern blots revealed that the genes encoding Lkn-1 and CKβ8-1/CKβ8 are single copy, but related genes. Twenty micrograms of genomic DNA prepared from a melanoma cell line (OCM1) and two B-cell lymphoma cell lines (GM02430 and GM03365) were digested withHindIII and were transferred to a nylon filter membrane. The Lkn-1 or CKβ8-1 probe was hybridized to the membrane, which was finally washed with 2 × SSC at 65°C for 30 minutes. Indicated by arrows are the genes encoding Lkn-1 and CKβ8-1/CKβ8. (B) The genes encoding Lkn-1 and CKβ8-1/CKβ8 are located at the same chromosome. Three BAC clones (9-0-20, 23-1-14, 110-F-8) were isolated and 1 μg of each DNA was digested with HindIII. Southern blot was performed as described above.
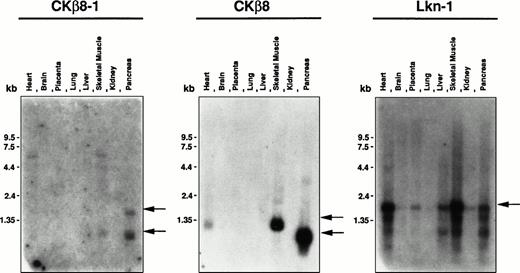
Tissue distribution of Lkn-1, CKβ8-1, and CKβ8.
To investigate the tissue distribution of Lkn-1, CKβ8-1, and CKβ8 mRNA, Lkn-1–, and CKβ8-1–specific, or CKβ8-1/CKβ8 common oligonucleotides were labeled and hybridized to the human multiple tissue blot and the human master blot containing poly (A)+RNAs derived from a variety of tissues which had been normalized. These oligonucleotide probes were specific to the corresponding cDNA according to the Southern hybridization to Lkn-1, CKβ8-1, and CKβ8 cDNAs (data not shown). As shown in Fig 5,the 1.0-kb and 2.0-kb CKβ8-1 mRNAs were detected in the pancreas, and 1.0-kb mRNA was found in skeletal muscle. When the CKβ8-1/CKβ8 common probe was used, the 1.0-kb mRNA was abundantly detected in the pancreas, whereas 1.2-kb mRNA was detected in heart and skeletal muscle, suggesting that CKβ8 may be a major species in these tissues. The reason why pancreas produces a different size of CKβ8-1 or CKβ8 remains unclear. Even after a more stringent washing, these messages remained (data not shown), suggesting that they may be true signals. Lkn-1–specific probe hybridized to a 2.0-kb mRNA species that is the most abundant in the heart and skeletal muscle. The Lkn-1 mRNA was also detectable in placenta, liver, and pancreas at a lower level. By using the human master blot, the expression of the three chemokines was also examined. The human master blot well correlated to the human multiple tissue blot shown in Fig 5 (data not shown). An abundant expression of Lkn-1 was detected in the adrenal gland. Interestingly, expression for all three chemokines was not seen in peripheral leukocyte and spleen, whereas a moderate expression of Lkn-1 was found in thymus. A low expression of Lkn-1 or CKβ8 was detected in bone marrow.
Tissue distribution of Lkn-1, CKβ8-1, and CKβ8 mRNA. The human multiple tissue blot (first panel) was purchased from Clonetech. Each oligonucleotide probe (5 × 106 cpm/mL) was hybridized to the membrane at 45°C and finally washed with 2 × SSC at 45°C for 20 minutes. Indicated by arrows are CKβ8-1, CKβ8, and Lkn-1 mRNAs.
Tissue distribution of Lkn-1, CKβ8-1, and CKβ8 mRNA. The human multiple tissue blot (first panel) was purchased from Clonetech. Each oligonucleotide probe (5 × 106 cpm/mL) was hybridized to the membrane at 45°C and finally washed with 2 × SSC at 45°C for 20 minutes. Indicated by arrows are CKβ8-1, CKβ8, and Lkn-1 mRNAs.
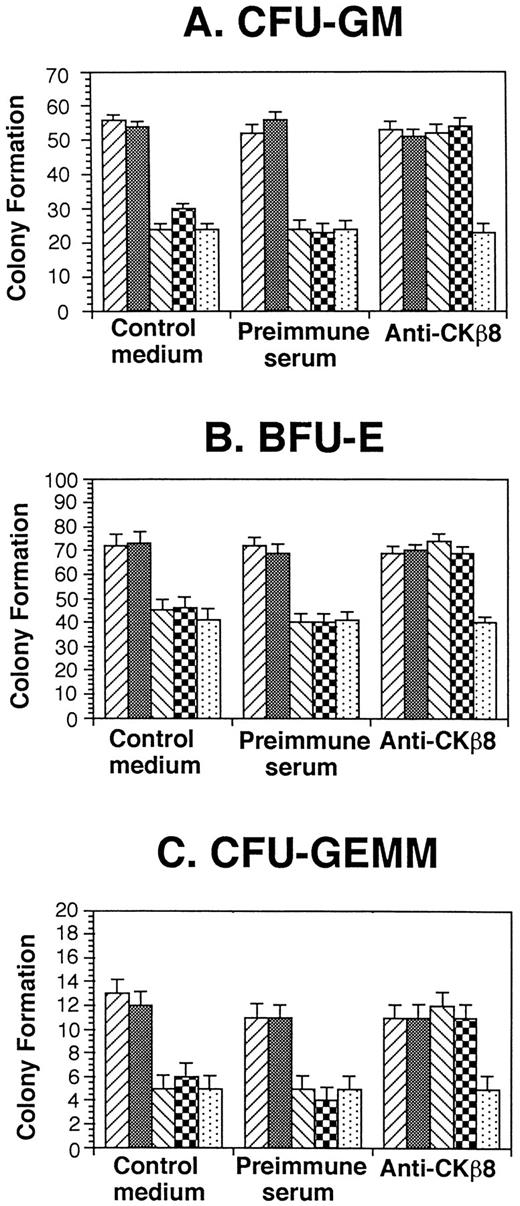
CKβ8-1 and CKβ8 suppress progenitor cell proliferation.
Because a number of chemokines have suppressive effects on proliferation of myeloid progenitor cells,7,9-11 we assessed the effects of CKβ8-1 and CKβ8 in comparison with that of hMIP-1α on colony formation by myeloid progenitor present in a low-density fraction of normal human bone marrow. As shown in Fig6, 50 ng/mL CKβ8-1 and CKβ8 significantly suppressed colony formation (P < .001) to a level equivalent to that of hMIP-1α, a known suppressive chemokine.7 19 Moreover, antiserum specific for CKβ8-1 and CKβ8 neutralized the suppressive effects of CKβ8-1 and CKβ8, but not that of MIP-1α. Preimmune serum had no effect on the suppressive activities of CKβ8-1, CKβ8, or MIP-1α.
Effect of CKβ8-1 and CKβ8 on colony formation by low-density human marrow cells. Low-density human bone marrow cells were plated at 5 × 104 cells/mL with 10% to 30% FBS growth factors, and chemokines (50 ng/mL) in a 0.3% agar or 1% methylcellulose culture medium. Colony formation was scored 14 days after incubation in 5% CO2 and lowered (5%) O2. Results are representative of three separate samples. (▨), Control medium; (▩) control diluent; (▧), CKβ8-1; (▩), CKβ8; (▧), MIP-1α.
Effect of CKβ8-1 and CKβ8 on colony formation by low-density human marrow cells. Low-density human bone marrow cells were plated at 5 × 104 cells/mL with 10% to 30% FBS growth factors, and chemokines (50 ng/mL) in a 0.3% agar or 1% methylcellulose culture medium. Colony formation was scored 14 days after incubation in 5% CO2 and lowered (5%) O2. Results are representative of three separate samples. (▨), Control medium; (▩) control diluent; (▧), CKβ8-1; (▩), CKβ8; (▧), MIP-1α.
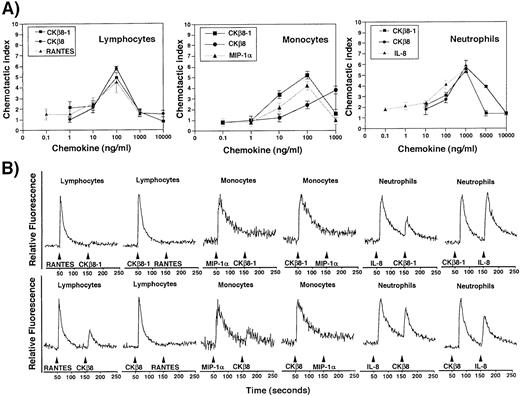
CKβ8-1 and CKβ8 induce chemotaxis and calcium flux in neutrophils, monocytes, and lymphocytes.
The chemotactic activities of CKβ8-1 and CKβ8 were evaluated in comparison with other known chemokines on human leukocyte subsets. As shown in Fig 7, these chemokines chemoattracted peripheral blood lymphocytes, monocytes, and neutrophils. Chemotactic effects of CKβ8-1 and CKβ8 were comparable to regulated on activation, normal T-cell expressed and secreted (RANTES) in lymphocytes, and to IL-8 in neutrophils, showing peak activities at 100 ng/mL and 1,000 ng/mL, respectively, and typical bell-type curves. It is interesting that CKβ8-1 and CKβ8 showed a strong chemotactic activity for neutrophils. IL-8 chemoattracted neutrophils at 0.1 ng/mL, whereas CKβ8-1 and CKβ8 chemoattracted neutrophils at 10 ng/mL. MIP-1α did not chemoattract neutrophils (data not shown). CKβ8 differed from CKβ8-1 in the monocyte chemoattraction. CKβ8-1 showed a peak at 100 ng/mL, whereas CKβ8 exhibited a weaker chemotactic activity at a lower concentration, which continued to increase up to 1,000 ng/mL. MIP-1α and CKβ8-1 showed a typical bell-shaped curve. This suggests that CKβ8-1 and CKβ8 could possibly have different kinetics of chemotactic function in vivo.
(A) Chemotactic activities of the recombinant CKβ8-1 and CKβ8. Lymphocytes, monocytes, and neutrophils were isolated from human peripheral blood buffy coat. Chemotaxis assays were performed using a Boyden chamber kit (Neuro Probe) with varying concentrations of CKβ8-1 and CKβ8. Microscopic observation at 100 × was used for counting each cell type. Three independent regions were selected and cell numbers were counted. The chemotactic index was driven by the following formula: (the number of cells chemoattracted by each concentration of CKβ8-1 or CKβ8)/(the number of cells chemoattracted by medium alone). (B) Each leukocyte population was isolated and examined for its response to CKβ8-1 and CKβ8, along with RANTES and MIP-1α. Two million cells were loaded with fura-2AM (Molecular Probes, Inc, Eugene, OR), stimulated with 25 nmol/L each chemokine, and used for Ca2+ flux assay.
(A) Chemotactic activities of the recombinant CKβ8-1 and CKβ8. Lymphocytes, monocytes, and neutrophils were isolated from human peripheral blood buffy coat. Chemotaxis assays were performed using a Boyden chamber kit (Neuro Probe) with varying concentrations of CKβ8-1 and CKβ8. Microscopic observation at 100 × was used for counting each cell type. Three independent regions were selected and cell numbers were counted. The chemotactic index was driven by the following formula: (the number of cells chemoattracted by each concentration of CKβ8-1 or CKβ8)/(the number of cells chemoattracted by medium alone). (B) Each leukocyte population was isolated and examined for its response to CKβ8-1 and CKβ8, along with RANTES and MIP-1α. Two million cells were loaded with fura-2AM (Molecular Probes, Inc, Eugene, OR), stimulated with 25 nmol/L each chemokine, and used for Ca2+ flux assay.
The capacity of CKβ8-1 and CKβ8 to induce a rapid Ca2+flux was assessed on lymphocytes, monocytes, and neutrophils. CKβ8-1 and CKβ8 induced a Ca2+ flux at 25 nmol/L in these cells. Although MIP-1α was not able to chemoattract neutrophils, it induced a Ca2+ flux, which is in line with the observation by McCall et al.23 CKβ8-1 and CKβ8 were able to desensitize lymphocytes to a subsequent stimulation with RANTES and desensitize monocytes to a subsequent stimulation with MIP-1α. RANTES and MIP-1α were able to desensitize lymphocytes and monocytes to subsequent stimuli with CKβ8-1, whereas the secondary stimulation of these cells with CKβ8 produced a weak Ca2+ flux. These data indicate that CKβ8-1 and CKβ8 share receptors with RANTES and MIP-1α, and CKβ8 seems to have a stronger desensitizing capability than RANTES and MIP-1α.
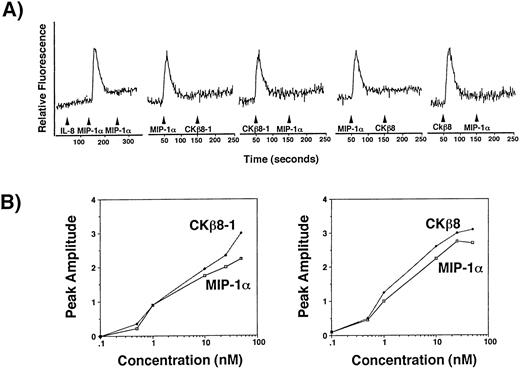
CCR1 is a functional receptor for CKβ8-1 and CKβ8.
To determine a receptor for CKβ8-1 and CKβ8, we performed transient Ca2+ flux assays using a human osteogenic sarcoma cell line, HOS cells expressing the human CD4 alone or CD4 along with one of CCR1, CCR2B, CCR3, CCR4, CCR5, or CXCR4. The HOS cell line expressing the human CD4 alone did not respond to CKβ8-1, CKβ8, MIP-1α, and RANTES (data not shown). A robust Ca2+ flux was detected only through CCR-1 at 25 nmol/L of CKβ8-1 and CKβ8. Figure8A shows the specificity of MIP-1α–mediated Ca2+ signal through CCR1. It also shows that the CKβ8-1 and CKβ8-mediated Ca2+ signal completely desensitized that of MIP-1α, suggesting that MIP-1α, CKβ8-1, and CKβ8 use CCR1 as a common receptor. We also showed similar desensitization patterns from neutrophils, monocytes, and lymphocytes in Fig 7. These data suggest that CCR1 may be a physiological receptor for CKβ8-1 and CKβ8.
(A) CCR1 is a receptor for CKβ8-1 and CKβ8. HOS cells (2 × 107) expressing CCR1 were loaded with fura-2AM at 5 μmol/L for 30 minutes and washed. Cells (2 × 106) in 2 mL were used for Ca2+ flux assay. For this assay, 25 nmol/L of each chemokine was used. CCR1 expressing HOS cells were treated with 25 nmol/L of IL-8, MIP-1α, and MIP-1α, successively. Their Ca2+ response was measured. For desensitization experiments, the HOS cells were treated with 25 nmol/L of MIP-1α and CKβ8-1 or CKβ8-1 and MIP-1α, or were treated with 25 nmol/L of MIP-1α and CKβ8 or CKβ8 and MIP-1α. (B) Potency of Ca2+ flux. Fura-2AM-loaded CCR1-HOS cells were stimulated with the indicated concentrations (0.1 to 50 nmol/L) of chemokines, and relative fluorescence was measured. The peak amplitude of Ca2+ response versus chemokine concentration was plotted in a semilog scale.
(A) CCR1 is a receptor for CKβ8-1 and CKβ8. HOS cells (2 × 107) expressing CCR1 were loaded with fura-2AM at 5 μmol/L for 30 minutes and washed. Cells (2 × 106) in 2 mL were used for Ca2+ flux assay. For this assay, 25 nmol/L of each chemokine was used. CCR1 expressing HOS cells were treated with 25 nmol/L of IL-8, MIP-1α, and MIP-1α, successively. Their Ca2+ response was measured. For desensitization experiments, the HOS cells were treated with 25 nmol/L of MIP-1α and CKβ8-1 or CKβ8-1 and MIP-1α, or were treated with 25 nmol/L of MIP-1α and CKβ8 or CKβ8 and MIP-1α. (B) Potency of Ca2+ flux. Fura-2AM-loaded CCR1-HOS cells were stimulated with the indicated concentrations (0.1 to 50 nmol/L) of chemokines, and relative fluorescence was measured. The peak amplitude of Ca2+ response versus chemokine concentration was plotted in a semilog scale.
To compare agonistic potential between CKβ8-1, CKβ8, and MIP-1α, dose-response experiments were conducted using the CCR1-expressing HOS cells. Figure 8B contains representative data of two independent experiments. CKβ8-1 and CKβ8 exhibited comparable responses to MIP-1α at 10 nmol/L.
DISCUSSION
We have isolated two more members of the C6 β-chemokine subfamily, CKβ8-1 and previously published CKβ8,24 which were created by alternative splicing. CKβ8-1 and CKβ8 are highly homologous to Lkn-1 (about 73% identity) and distantly related to MIP-1α (about 50% identity). Because the NH2-terminal regions of β-chemokines may play a critical role in the interaction with receptors, the alternative splicing of CKβ8-1 and CKβ8, which leads to the different size of the NH2-terminal region, may have biological significance. An additional splice variant of CKβ8 has been deposited in the Swiss-Prot data base under Accession Number P55773. Interestingly, the splicing occurred at the sequence representing the signal peptide. Because the genes encoding the signal peptides of β-chemokines have been shown to reside in a single exon,22 the nature of this variant remains unclear at this time. Regarding alternative splicing of chemokines, there are two precedent examples: one is stromal cell-derived factor-1 (SDF-1) in which an alternative splicing occurred at the COOH-terminal region,25 and the other is HCC-1, whose sequence was deposited in the public data base under Accession NumberZ79293. However, functional comparisons have not been made, thus our report is the first example showing that alternative splicing produces biologically active chemokines. To date, we do not have strong evidence that CKβ8-1 and CKβ8 are functionally different: they have similar Ca2+ flux patterns, chemotaxis, and myelosuppressive activities.
Although Lkn-1 and CKβ8-1/CKβ8 are structurally related and located in proximity, their gene expression patterns seem to be different. Lkn-1 is expressed in a wide range of tissues, whereas CKβ8-1 and CKβ8 are largely expressed in skeletal muscle and the pancreas. Although a significant expression of Lkn-1 was detected in the skeletal muscle, there was very low Lkn-1 expression in pancreas. The highest Lkn-1 expression was found in the adrenal gland, whereas only marginal expression of CKβ8 was detected. We did not see the differential expression of CKβ8-1 and CKβ8. Whereas CKβ8 is a major species in tissues tested, CKβ8-1 was detected in very low abundancy. However, because CKβ8-1 was a major species in THP-1 cells, monocytes need to be tested. Forssmann et al21 were able to detect the expression of CKβ8, which were upregulated by IL-1β or interferon-γ.
The Ca2+ flux patterns of CKβ8-1 and CKβ8 correlated to chemotactic activities in neutrophils, monocytes, and lymphocytes. CCR1 is known to be a receptor expressed in these leukocyte populations.8,23,26 Interestingly, even though MIP-1α induced robust Ca2+ flux in lymphocytes, monocytes, and neutrophils, it was not able to chemoattract neutrophils. Gao et al26 recently showed that neutrophils derived from normal mice migrated in response to MIP-1α but neutrophils from CCR1 knockout mice exhibited an impaired chemotaxis toward MIP-1α, suggesting that the CCR1-mediated signals may be responsible for neutrophil chemotaxis. On the other hand, McCall et al23found that MIP-1α elicited a robust Ca2+ flux in human neutrophils but failed to induce neutrophil chemotaxes. We also observed similar phenomena with MIP-1α. One plausible interpretation could be that MIP-1α signaling through the human CCR1 may not be sufficient for leading to chemotaxis in comparison with the mouse CCR-1. This is possibly achieved by coupling of different Gα proteins to CCR1 between mice and humans. MIP-1α–mediated neutrophil chemoattraction awaits further study. However, CKβ8-1 and CKβ8 induced a rapid Ca2+ flux and chemoattraction in neutrophils as well.
CKβ8-1 and CKβ8 are structurally related to Lkn-1. However, Lkn-1 binds to CCR1 and CCR3, whereas CKβ8-1 and CKβ8 bind to CCR1, suggesting that in vivo function of Lkn-1, CKβ8-1, and CKβ8 may not be redundant. CCR1-ligands, MIP-1α, RANTES, MCP-3, and Lkn-1 also bind to another receptor such as CCR5,12CCR2B,9 or CCR3.10 However, CKβ8-1 and CKβ8 are unique CCR1 ligands identified to date in that they do not have overlapping receptor specificity. Because Forssmann et al21showed desensitization of CKβ8 by MCP-1, 3, and 4, it is still possible that there may exist another receptor for CKβ8-1 and CKβ8. In addition to this, four more CC chemokine receptors CCR6, 7, 8, and 9 have been recently identified.14-17 Whether CKα8-1 or CKα8 bind to any of these receptors needs to be tested.
CKβ8-1 and CKβ8 genes were mapped to human chromosome 17q in which a human β-chemokine cluster is located.27 Based on the southern patterns using BAC and human genomic DNA, the Lkn-1 gene and CKβ8-1 and CKβ8 genes are located in proximity, at most within 200 kb. Others reported that a YAC contig containing around 1,500 kb included MIP-1α (LD78α), MIP-1β (LD78β), RANTES, NCC-2 (an expression sequence tag [EST] of HCC-1), and a novel chemokine-like gene (NCC-4). Therefore, CKβ8-1 and CKβ8 could be included in the YAC contig.
Because of their broad chemotactic specificities, β-chemokines could play a central role in the development and maintenance of the leukocyte infiltration found in many diseases, such as allergic inflammation, arthritis, nephritis, and experimental autoimmune encephalomyelitis.28 Because CKβ8-1 and CKβ8 chemoattract most leukocytes subsets, these chemokines could be associated with these diseases.
Chemokines have been implicated in regulation of hematopoiesis.6,7,29 It is of interest that all C6 β-chemokines so far analyzed,6,18 including CKβ8-1 and CKβ8 as shown in the present study, have shown myelosuppressive activity on hCFU-GM, BFU-E, and CFU-GEMM, whereas some non-C6 β-chemokines, such as MIP-1β, RANTES, MCP-2, MCP-3, and eotaxin29 do not manifest suppressive activity. Perhaps the C6 β-chemokine family members will give us a hint as to why some chemokines suppress myelopoiesis, whereas others do not.
Although CKβ8-1 has not been published previously, two groups of investigators reported characterization of MPIF-130 or CKβ8.21 CKβ8 was considered chemotactic only for monocytes, but inactive for IL-2–conditioned T cells and neutrophils.21 MPIF-1 was considered chemotactic to monocytes, resting T cells, and neutrophils.30 The difference of T-cell chemoattraction between these studies suggests that IL-2 signaling or T-cell activation may regulate the response of T cells to CKβ8 or myeloid progenitor inhibitory factor-1 (MPIF-1). Given the difference of neutrophil chemotaxis, it may be easily explained by the difference in assay conditions, the methods used to isolate neutrophils, or the relative activation state of the neutrophils isolated by the various techniques. We have shown that CKβ8 is a strong chemoattractant for peripheral blood monocytes, lymphocytes, and neutrophils. The chemotactic results were further strengthened by calcium flux assays using the purified leukocyte subsets. Therefore, our current data agree more closely with the studies with MPIF-1.
CKβ821 cross-desensitized only the calcium signal produced by MIP-1β, but did not desensitize MIP-1α–mediated calcium flux. MPIF-130 was able to desensitize MIP-1α response. Our current data showed that CKβ8 desensitized MIP-1α and RANTES signals in monocytes and lymphocytes, but did not desensitize IL-8 signals in neutrophils. Our data again more closely support the data with MPIF-1 and this information is now extended to neutrophils. MPIF-130 was shown to selectively inhibit colony formation of CFU-GM and CFU-GEMM, but not BFU-E, whereas our data showed that CKβ8 inhibited colony formation of all early progenitors. There may be two possible explanations of this difference, and one is the difference between human and mouse. We used human bone marrow cells, whereas Patel et al30 used mouse bone marrow for much of their work. The other is different assay conditions such as the combinations of growth factors. Furthermore, we showed specific blocking of the colony formation with antibodies specific for CKβ8. The specificity of the antibodies was demonstrated by showing that they did not neutralize the MIP-1α activities.
ACKNOWLEDGMENT
The authors thank Sister Mary Etta Kiefer for editing, and Audrey Carson for typing this manuscript.
Supported by U.S. Public Health Service Grants RO1 AI 28125 and DE 12156 (B.S.K.) and RO1 HL 54037, RO1 DK 53674, HL 56416, and a project in PO1 HL 53586 (H.E.B.) from the NIH.
Address reprint requests to Byoung S. Kwon, PhD, Indiana University School of Medicine, Department of Microbiology & Immunology, 635 Barnhill Dr, Indianapolis, IN 46202-5120.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal