Abstract
A characteristic of normal neutrophil maturation is the induction of secondary granule protein (SGP) mRNA expression. Several leukemic human cell lines mimic normal morphologic neutrophil differentiation but fail to express SGPs, such as lactoferrin (LF) and neutrophil gelatinase (NG). In contrast, two murine cell lines (32D C13 and MPRO) are able to differentiate into neutrophils and induce expression of LF and NG. Therefore, to study the normal regulation and function of these genes, the corresponding murine homologs must be isolated. Using cDNA representational difference analysis (RDA) to compare a committed myeloid progenitor cell line (EPRO) with the multipotent stem cell line from which it was derived (EML), we isolated a fragment bearing homology to human neutrophil collagenase (hNC). Here, we describe the cloning and characterization of a full-length (∼2 kb) clone that exhibits nearly 65% nucleotide and 73% amino acid identity to hNC. Ribonuclease protection analysis (RPA) of the tissues and cell lines shows that mouse NC (mNC) is expressed only in cell lines exhibiting neutrophilic characteristics, further confirming its identity as the mouse homolog of hNC. Furthermore, we have demonstrated a shared negative regulatory pathway for this and other SGP genes. We have previously shown that CCAAT displacement protein (CDP/cut) binds to a specific region of the LF promoter, and overexpression of CDP blocks G-CSF–induced upregulation of LF gene expression in 32D C13 cells. We show here that in these cells, upregulation of both NC and NG is also blocked. CDP is thus the first identified transcription factor that is a candidate for mediating the shared regulation of neutrophil SGP protein genes.
MATRIX METALLOPROTEINASES (MMPs) are responsible for the turnover of extracellular matrix associated with a variety of biologic processes, including tissue regeneration and wound healing, and may play a role in the acquisition of metastatic potential in transformed cells.1 A neutrophil-specific MMP, human neutrophil collagenase (hNC), has been identified and is distinct from the closely related fibroblast collagenase with respect to peptide and collagen specificity.2 hNC has been shown to colocalize with lactoferrin (LF) in specific granules within polymorphonuclear neutrophils,3 and its expression is thought to be coordinately regulated with other secondary granule protein (SGP) transcripts such as LF and neutrophil gelatinase (NG).4 5
Cell lines used for the study of neutrophil differentiation include HL60 and NB4 cells, both of which were derived from patients with acute promyelocytic leukemia.6,7 The NB4 cell line is characterized by t(15;17) involving the retinoic acid receptor α (RARα) and the PML gene,8 and both HL60 and NB4 cells exhibit neutrophil differentiation upon induction with all-trans retinoic acid (ATRA).7,9 Although morphologic maturation into neutrophils is apparent, neither HL60 nor NB4 cell lines express SGP transcripts characteristic of normal neutrophils.2,10-13 In contrast, the murine cell line 32D C1314 is able to undergo complete neutrophil maturation with stage-specific expression of SGP transcripts upon induction with granulocyte colony-stimulating factor (G-CSF).4 Therefore, to study the normal regulation of these genes during neutrophil maturation and the potential cause of SGP gene dysregulation in leukemic cells, it is necessary to isolate their murine homologs.
Two murine cell lines have recently been established using a dominant-negative form of RARα.15-18 The stem cell factor (SCF)-dependent EML cell line is arrested at an early point in hematopoiesis and is characterized by the ability to differentiate along multiple lineages while exhibiting a specific block in neutrophil maturation that can be overcome by superphysiologic concentrations of ATRA in the presence of granulocyte-macrophage CSF (GM-CSF).17 The MPRO cell line is GM-CSF–dependent and consists of neutrophilic promyelocytes that terminally differentiate in response to ATRA.16 We have shown that both of these cell lines are able to express mouse NG (mNG) and mLF upon induction with ATRA. Furthermore, a more mature myeloid cell line can be derived from EML cells17; it is referred to as EPRO and possesses characteristics similar to the MPRO cell line, including neutrophil maturation17 and SGP gene expression.
In an attempt to isolate factors involved in myeloid commitment, we used cDNA representational difference analysis (RDA)19 to compare EML and EPRO cells (manuscript in preparation). Among the differentially expressed genes that were isolated was a fragment that exhibited a high degree of homology to hNC. Here, we describe the cloning of the full-length cDNA for mNC using this fragment. We show that the expression of this transcript is neutrophil-specific and exhibits regulation similar to that for the other SGP transcripts, including repression by CCAAT displacement protein (CDP). CDP is a homeodomain protein with extensive homology toDrosophila cut protein and has been shown to inhibit expression of the myeloid-specific cytochrome heavy-chain gene gp91-phox, among others.20 We have previously demonstrated CDP-mediated repression of the SGP gene LF during G-CSF–induced maturation of 32D C13 cells overexpressing CDP.21 Repression of both gp91-phox22 and LF is associated with CDP binding within their respective promoters, although no consensus binding site for CDP has been described.20 22 Here, we demonstrate that mNC and mNG expression are also inhibited at the mRNA level in 32D C13 cells overexpressing CDP, a pattern similar to that of LF. CDP is thus the first candidate for a transcriptional regulator mediating the shared regulation of neutrophil SGP genes.
MATERIALS AND METHODS
Cell lines.
EML, MPRO, BHK/MKL, and HM-5 cells were kindly provided by Schickwann Tsai (Fred Hutchinson Cancer Research Center, Seattle, WA). EML cells were maintained in Iscove's modified Dulbecco's medium (IMDM) containing 20% heat-inactivated horse serum and supplemented with 10% conditioned medium from the BHK/MKL cell line as a source of rat SCF. MPRO cells were maintained in AIM V serum-free medium (Life Technologies Inc, Bethesda, MD) supplemented with 10% conditioned medium from the HM-5 cell line as a source of GM-CSF. 32D C13 and 32D/CDP cells were grown in IMDM with 10% heat-inactivated fetal bovine serum and 10% WEHI-conditioned medium as a source of interleukin-3 (IL-3). The establishment of 32D C13/CDP cells is described elsewhere.21 To derive the EPRO cell line, EML cells were induced with IL-3 and 10 μmol/L ATRA (Sigma, St Louis, MO) in the presence of SCF for 72 hours, followed by culture in IMDM containing 20% heat-inactivated horse serum supplemented with 10% HM-5–conditioned medium.
For induction of the 32D cell lines, cells were washed twice with phosphate-buffered saline and placed in IMDM with 10% fetal bovine serum and 3 × 103 U/mL recombinant human G-CSF (Amgen, Thousand Oaks, CA). MPRO cells were induced by adding ATRA to a final concentration of 10 μmol/L in the presence of normal growth medium as already described. All cell lines were grown at 37°C and 5% CO2 in a humidified incubator.
Alignment of mouse and human NC. Box 1 outlines the cysteine switch region, and box 2 is the zinc binding domain. The proteins share 73% identity overall. Arrow 3, signal peptidase cleavage site; arrow 4, autolytic activation cleavage site; arrow 5, degradation site.
Alignment of mouse and human NC. Box 1 outlines the cysteine switch region, and box 2 is the zinc binding domain. The proteins share 73% identity overall. Arrow 3, signal peptidase cleavage site; arrow 4, autolytic activation cleavage site; arrow 5, degradation site.
Library construction.
Total RNA was isolated from uninduced EPRO cells using TRIzol reagent (Life Technologies), and mRNA was purified using an oligo(dT) column (Life Technologies). First-strand synthesis was initiated using an oligo(dT) primer containing a Xho I site. After second-strand synthesis, the cDNA was blunt-ended and ligated to anEcoRI adapter followed by directional cloning into a phosphatased ZAP Express λ vector (Stratagene, La Jolla, CA) cut withEcoRI and Xho I and packaged using Gigapack Gold extracts (Stratagene).
Approximately 1.5 × 105 clones were plated and transferred to duplicate BioTrace NT nitrocellulose membranes (Gelman Sciences, Ann Arbor, MI). The membranes were hybridized with the fragment EPRO 6-2 labeled with [α-32P]dCTP by random prime (Boehringer Mannheim, Indianapolis, IN). Secondary and tertiary screens were performed as usual,23 and phagemids were excised from isolated tertiary clones according to the manufacturer's protocol (Stratagene). Clones were analyzed by restriction mapping and sequenced using the dideoxy chain-termination method.24
RNA isolation and Northern analysis.
Total RNA was isolated from the cells using TRIzol reagent. Northern analysis was performed by electrophoresis of 10 μg RNA in a 1% agarose/formaldehyde gel, followed by transfer to nitrocellulose membrane (Gelman Sciences). Blots were probed with cDNA fragments labeled with [α-32P]dCTP by random prime (Boehringer Mannheim). Hybridization was performed overnight at 42°C in the presence of 50% formamide and washed with 2× SSC (300 mmol/L NaCl and 60 mmol/L sodium citrate)/0.1% sodium dodecyl sulfate (SDS) twice at room temperature for 10 minutes, followed by two high-stringency washes in 0.1× SSC (15 mmol/L NaCl and 3 mmol/L sodium citrate)/0.1% SDS at 55° to 60°C for 15 minutes.
Probes.
Probes for mLF, mNG, and β-actin were prepared as previously described.4 A probe for human p47 was kindly provided by Steven Chanock (National Cancer Institute, Bethesda, MD). The probe designated EPRO 6-2 was isolated from a RDA screen for differential gene expression and cloned into the BamHI site of pBluescript II (Stratagene). The EPRO 6-2 fragment was excised usingEcoRI and Xba I followed by agarose gel purification and labeled as before.
RPA.
Riboprobes corresponding to β-actin and mNC were prepared using the MAXIscript transcription kit (Ambion, Austin, TX). Before transcription, a plasmid containing the full-length mNC cDNA (pBK-mNC 4-1) was linearized with BstZ17 (New England Biolabs Inc, Beverly, MA) followed by digestion with proteinase K (final concentration, 200 μg/mL) for 30 minutes at 50°C. The linearized plasmid was then phenol/chloroform-extracted and precipitated with ammonium acetate and ethanol. For β-actin, the pTRI-actin plasmid was used (Ambion). A typical transcription reaction contained 1 μg linearized template and 0.5 mmol/L final concentration each for ATP, GTP, and UTP; a final CTP concentration of 25 μmol/L was used with 1 μmol/L32P-CTP, and the reaction was incubated at 25°C for 1 hour followed by digestion with DNAse I at 37°C for 15 minutes. Probes were analyzed on a 6% polyacrylamide/TBE gel, exposed to film, cut from the gel, and eluted from gel slices by soaking in 0.5 mol/L NH4OAc/1% SDS overnight at 37°C.
For the RPA, approximately 10 μg total RNA was simultaneously precipitated with 1 × 104 cpm β-actin and 1 × 105 cpm mNC probes, and the pellets were resuspended in 10 μL HybSpeed buffer (Ambion) at 95°C. All samples were normalized to 50 μg RNA with yeast RNA. As a control, yeast RNA alone was hybridized to the indicated amounts of probe. The probe and RNA were hybridized at 68°C for 10 minutes followed by digestion with a mixture of RNAse A and T1 at a dilution of 1:100 for 20 minutes at 37°C. Digested samples were precipitated, and the pellets were resuspended in loading buffer and separated on a 6% polyacrylamide/TBE gel for 2.5 hours at 250V. The gel was then exposed to film at −70°C with an intensifying screen.
RESULTS
Isolation of mNC cDNA.
Current studies in our laboratory have used the technique of cDNA RDA19 in an attempt to isolate factors involved in myelopoiesis. While using RDA to characterize the EPRO cell line, we isolated a number of cDNA fragments that are differentially expressed with respect to the multipotent cell line (EML) from which it was derived. Sequence analysis of one of these fragments (EPRO 6-2) revealed a high degree of homology to hNC. Therefore, we sought to clone and characterize a full-length cDNA clone for mNC.
An oligo(dT)-primed cDNA library derived from EPRO cells was constructed as described earlier. Approximately 1.5 × 105plaques were screened with EPRO 6-2, resulting in 57 positive clones. From these, five clones were purified by secondary and tertiary screens, after which four of the five clones remained positive and three were identical in size (2 kb) while the fourth was shorter (about 1.5 kb), as determined by restriction digestion analysis (data not shown).
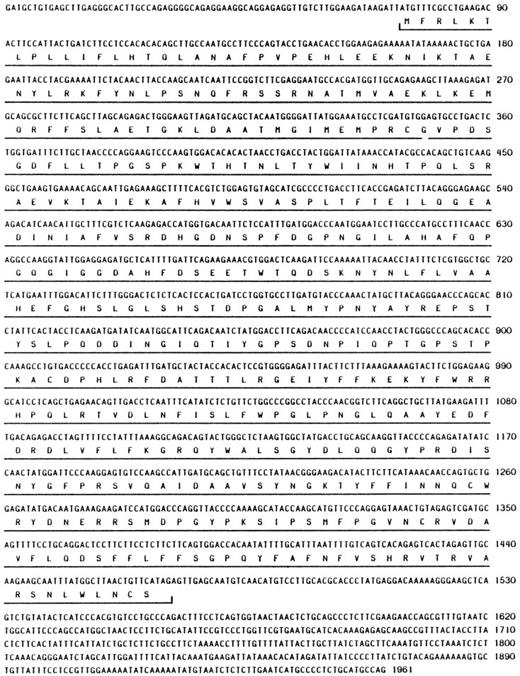
Sequence analysis of the longest clone (mNC 4-1) revealed the presence of a complete open reading frame encoding 465 amino acid residues (Fig1; Genbank Accession No. U96696). There appears to be a relatively short 5′ untranslated region (UTR) of 73 nucleotides (nt) and a 3′ UTR of about 500 nt. We have not determined if there are additional 5′ untranslated sequences; however, the hNC 5′ UTR is approximately 95 nt, so it is likely that the clone we isolated represents the full-length mRNA.
At the amino acid level, the human and mouse proteins share 73% identity, although several functional domains exhibit much higher similarity (Fig 2). The hNC zymogen is normally latent and can be activated autolytically in response to various stimuli.25,26 An important region required for latency in all MMPs is the cysteine switch region found within the propeptide domain that is thought to interact with the zinc atom bound to the active site.27 Box 1 in Fig 2 highlights the residues that comprise the cysteine switch region; the sequence within this region is identical in hNC and mNC. Box 2 shows that the sequence of the Zn binding domain/active site is also identical when comparing the two proteins.
Other sites within hNC that are important for regulation of enzyme activity include the autolytic activation and degradation sites, as well as the signal peptidase.25 Each of these residues is indicated in Fig 2. The residues corresponding to the signal peptidase (arrow 3) and autolytic activation (arrow 4) cleavage sites are conserved between the two proteins, whereas the second residue at the degradation site (arrow 5) is proline rather than leucine.
Expression of mNC during neutrophil maturation.
The MPRO cell line can be induced to undergo neutrophil maturation in response to superphysiologic concentrations of ATRA.16 By 72 hours in the presence of 10 μmol/L ATRA, greater than 90% of the cells exhibit neutrophil morphology and express SGP transcripts such as LF and NG.
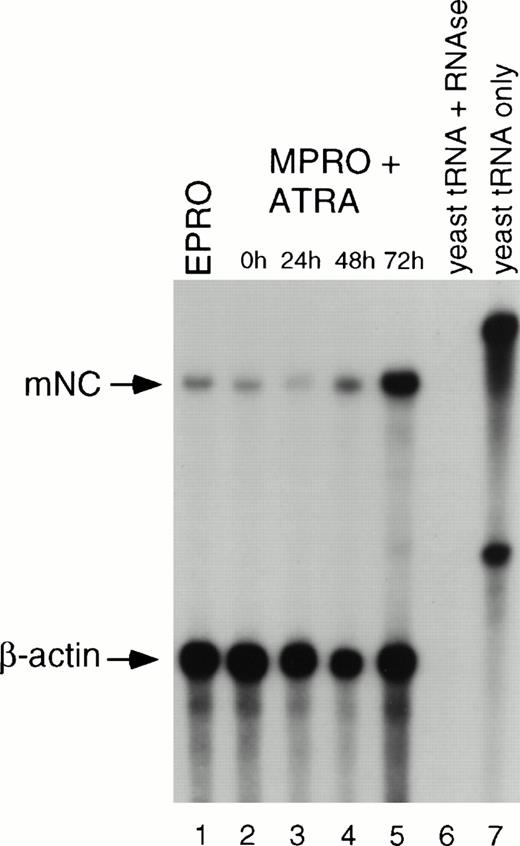
To determine if mNC is induced upon neutrophil maturation, we analyzed mNC levels after exposure of MPRO cells to 10 μmol/L ATRA. By RPA analysis, uninduced MPRO cells express mNC at a low level (Fig 3, lane 2). Upon initial exposure to ATRA, levels of mNC transcript increase slightly (lanes 3 and 4) and peak at 72 hours (lane 5), a time at which more than 90% of the cells are neutrophils (data not shown). As a positive control, uninduced EPRO mRNA (500 ng) was included in the assay (lane 1). Lanes 6 and 7 show the probes hybridized to yeast RNA with and without RNAse digestion to confirm the digestion of single-stranded probe while confirming the absence of hybridization to yeast RNA and to confirm the size and integrity of undigested probe, respectively. As a control for the target RNA integrity and amount, a β-actin riboprobe was simultaneously hybridized to all samples (lanes 1 to 7).
RPA of mNC expression in MPRO cells. RNA was isolated from uninduced and ATRA-induced cells at the indicated time points and subjected to RPA using riboprobes for mNC and β-actin. The mNC riboprobe is approximately 500 nt and protects a fragment of 424 nt. The β-actin probe is 330 nt, and the protected fragment is 250 nt. Lane 1, EPRO polyA RNA (500 ng); lane 2, uninduced MPRO; lane 3, MPRO induced with ATRA for 24 hours, lane 4, 48 hours, and lane 5, 72 hours; lane 6, yeast RNA digested with RNase; lane 7, yeast RNA alone, indicating size of undigested riboprobes.
RPA of mNC expression in MPRO cells. RNA was isolated from uninduced and ATRA-induced cells at the indicated time points and subjected to RPA using riboprobes for mNC and β-actin. The mNC riboprobe is approximately 500 nt and protects a fragment of 424 nt. The β-actin probe is 330 nt, and the protected fragment is 250 nt. Lane 1, EPRO polyA RNA (500 ng); lane 2, uninduced MPRO; lane 3, MPRO induced with ATRA for 24 hours, lane 4, 48 hours, and lane 5, 72 hours; lane 6, yeast RNA digested with RNase; lane 7, yeast RNA alone, indicating size of undigested riboprobes.
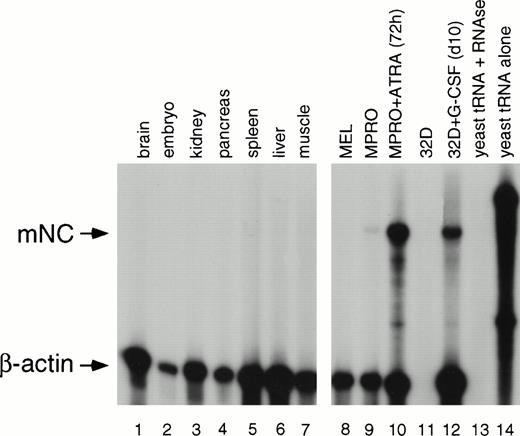
To further show that the cDNA we isolated was a neutrophil-specific gene, we assayed for its expression in a variety of mouse tissues and cell lines. A number of tissues and murine hematopoietic cell lines were analyzed for expression of mNC. All mouse tissues assayed do not express a transcript corresponding to mNC (Fig 4, lanes 1 to 7). Furthermore, uninduced MEL and 32D C13 cells fail to express mNC; the β-actin control is absent for the uninduced 32D C13 sample, indicating probable degradation of this sample, although we have confirmed nonexpression in uninduced 32D C13 cells by Northern blot analysis (Fig 5).Uninduced MPRO cells (Fig 3, lane 9) express a low level of mNC which is upregulated upon induction with ATRA (lane 10). After induction with G-CSF, 32D C13 cells undergo terminal maturation to neutrophils, with corresponding upregulation of SGP transcripts.4 14 As expected, mNC is also induced after treatment of 32D C13 cells with G-CSF (lane 12). As a control for the RNA amount and integrity, β-actin was included in the hybridization (lanes 1 to 14). As previously described, both riboprobes were hybridized with yeast RNA in the presence (lane 13) and absence (lane 14) of RNase.
RPA of mNC expression in mouse tissues and cell lines. RNA from a variety of tissues was subjected to RPA. Lane 1, brain; lane 2, embryo; lane 3, kidney; lane 4, pancreas; lane 5, spleen; lane 6, liver; lane 7, muscle; lane 8, mouse erythroleukemia cells; lane 9, uninduced MPRO cells; lane 10, MPRO cells induced with ATRA for 72 hours; lane 11, uninduced 32D C13 cells; lane 12, 32D C13 induced for 10 days with G-CSF; lane 13, yeast RNA digested with RNase; lane 14, yeast RNA alone.
RPA of mNC expression in mouse tissues and cell lines. RNA from a variety of tissues was subjected to RPA. Lane 1, brain; lane 2, embryo; lane 3, kidney; lane 4, pancreas; lane 5, spleen; lane 6, liver; lane 7, muscle; lane 8, mouse erythroleukemia cells; lane 9, uninduced MPRO cells; lane 10, MPRO cells induced with ATRA for 72 hours; lane 11, uninduced 32D C13 cells; lane 12, 32D C13 induced for 10 days with G-CSF; lane 13, yeast RNA digested with RNase; lane 14, yeast RNA alone.
Northern analysis of 32D cells overexpressing CDP. 32D/CDP and 32D/neo cells were induced with G-CSF, and RNA was harvested at the indicated time points. Northern analysis was performed, and the blot was sequentially hybridized with probes for mLF, mNG, mNC, human p47, and β-actin. Lane 1, uninduced 32D/neo cells; lane 2, 32D/neo induced for 4 days with G-CSF; lane 3, uninduced 32D/CDP cells; lane 4, 32D/CDP induced with G-CSF for 2 days and 4 days (lane 5); lane 6, 32D C13 induced with G-CSF for 10 days.
Northern analysis of 32D cells overexpressing CDP. 32D/CDP and 32D/neo cells were induced with G-CSF, and RNA was harvested at the indicated time points. Northern analysis was performed, and the blot was sequentially hybridized with probes for mLF, mNG, mNC, human p47, and β-actin. Lane 1, uninduced 32D/neo cells; lane 2, 32D/neo induced for 4 days with G-CSF; lane 3, uninduced 32D/CDP cells; lane 4, 32D/CDP induced with G-CSF for 2 days and 4 days (lane 5); lane 6, 32D C13 induced with G-CSF for 10 days.
Inhibition of mNC expression by CDP.
CDP has been found to silence the expression of a number of genes, including the myeloid-specific oxidase component gp91phox.20,22,28 Recently, we have found that CDP binds to an element within the LF promoter and serves to inhibit transcription in uninduced 32D C13 cells.21 Upon G-CSF–induced neutrophil differentiation, CDP no longer binds to this site and LF expression increases. We have established a 32D C13 cell line that overexpresses CDP and fails to induce expression of LF upon induction with G-CSF while undergoing normal morphologic differentiation.21 In an effort to provide further evidence for common regulation of SGP transcript expression, we performed Northern analysis using 32D/CDP cells and probes for the known mouse SGP homologs. 32D/CDP and 32D/neo cells were induced with G-CSF, and RNA was isolated for Northern blot analysis on days 0, 2, and 4. Figure5 shows a representative Northern blot of bulk cell populations sequentially probed with mLF, mNG, mNC, and human p47 cDNA fragments. In 32D/neo cells, mLF, mNG, and mNC are upregulated by day 4 in response to G-CSF (lane 2), whereas in cells overexpressing CDP, both of these transcripts fail to be upregulated (Fig 5, lane 2 v5). In contrast, the oxidase component p47, which is known to be unaffected by CDP,22 does not exhibit repression in the presence of excess CDP (lane 5). The blot was probed with β-actin to confirm equal loading of sample RNA. This pattern of SGP gene expression has been noted in clonal 32D C13 cell lines overexpressing CDP (data not shown).
DISCUSSION
Neutrophils are crucial both for the host responses to bacterial infections and as critical mediators of inflammation. Secondary granules are secretory granules that fuse with the plasma membrane and release their contents upon neutrophil activation.3Although the functions of SGPs are not fully understood, they are thought to have both bactericidal and proinflammatory functions. LF is hypothesized to sequester iron away from bacteria, contributing to bacterial killing,29 30 and metalloproteases such as NG and NC are postulated to contribute to matrix modification at the site of inflammation.
The development of secondary granules is a late event during neutrophil maturation, and the detection of SGP expression correlates with the transition from the promyelocyte stage to later stages of neutrophil differentiation.3 Stage-specific expression of SGP mRNA can be detected in primary bone marrow from healthy donors and during in vitro differentiation of G-CSF–induced CD34+ progenitor cells.31 In contrast, leukemic cell culture models of myeloid differentiation, such as HL60 and NB4, show a defect in SGP gene expression2,12,13,32 and fail to express SGP transcripts even after induction of neutrophil maturation by chemical agents such as DMSO or ATRA.10 12 Therefore, to study the control of SGP gene transcription, it is necessary to use murine factor-dependent cell lines that have a more normal pattern of differentiation; this requires isolation of the corresponding murine SGP genes.
In this study, we have described the cloning and characterization of expression of mNC. While performing a RDA screen for differentially expressed genes, we isolated a fragment with significant homology to hNC. Sequence analysis of the corresponding cDNA showed a high degree of homology at the nucleotide level to hNC (data not shown). The mNC clone contains a 1,395-bp open reading frame (Fig 1), and the hypothetical translation of this sequence reveals 73% identity with hNC (Fig 2). Not surprisingly, the functional domains common to MMPs, ie, the zinc binding and cysteine switch regions, are identical between the mouse and human proteins, as are the signal peptidase and autolytic activation sites. Interestingly, the autolytic degradation site is not conserved between the two proteins (Fig 2, arrow 3): degradation occurs in hNC at the G262-L263 bond, but the L residue is a P residue in the mouse protein. Although the hydrophobic character of this residue is maintained, the potential structural ramifications and their likely impact on the rate of degradation remain unclear.
We have characterized mNC expression in a variety of tissues and hematopoietic cell lines. The RPA studies presented here show no evidence for expression of mNC in nonhematopoietic tissues. Unlike interstitial collagenase, which is widely expressed, NC expression has been thought to be restricted to neutrophils. Recent studies have suggested that hNC may also be expressed by chondrocytes,33especially following stimulation with IL-1β,34 and that it may play a role in articular destruction in arthritis. If this is also a characteristic of mNC, then isolation of the cDNA described here may have potential value in the investigation of mouse models of arthritis.
Our primary interest in mNC is focused on the pattern of coordinate tissue- and stage-specific expression that it may share with other SGPs. The studies presented here provide support for the hypothesis that the positive and negative regulation of these genes is linked. Within hematopoietic cell lines, our studies confirm that mNC follows the predicted pattern of expression shared by other SGP genes. The MPRO cell line is arrested at the promyelocyte stage and undergoes neutrophil maturation16 with expression of SGP transcripts upon induction with ATRA. The transcript for mNC is expressed at low levels in uninduced MPRO cells, in contrast to both mLF and mNG, while all three SGP genes are upregulated in response to ATRA. Furthermore, mNC is also expressed in G-CSF–induced 32D C13 cells in concert with the other SGP genes.
As already noted, expression of SGP genes is impaired in leukemic cell lines induced to undergo differentiation toward neutrophils, such as NB4 and HL60. A similar defect in SGP expression has been described in myelodysplasia and in the residual neutrophils of patients with newly diagnosed leukemia.35 36 We therefore hypothesize that the pathogenesis of these malignancies involves an event that disrupts the differentiation program even in cells that retain a capacity for limited maturation, as reflected in the coordinate absence of expression of SGP mRNA.
In this regard, the pattern of SGP gene repression in cells overexpressing CDP is striking. CDP was initially identified as a repressor of sperm H2B transcription in sea urchin embryos.28 It has since been shown to repress a wide variety of genes in many species, including gp91-phox, the γ-globin genes, and mouse NCAM and myosin heavy-chain.20,22,37,38Studies from our laboratory have shown that CDP binds to a region within the LF promoter that possesses silencing activity.21Furthermore, overexpression of CDP in the 32D C13 cell line results in inhibition of LF induction without blocking morphologic differentiation. We show here that expression of the mNC gene, characterized in this study, as well as mNG, is similarly repressed at the mRNA level by CDP overexpression. The cloning and characterization of mNC has thus allowed us to add further credence to the hypothesis that shared trans-regulatory elements may be responsible for both positive and negative regulation of SGP genes. Furthermore, we have identified CDP as the first transcriptional regulator that is a candidate for mediating coordinate repression of these genes, although potential shared activating factors remain to be found.
Supported by National Institutes of Health (NIH) Grants No. DK48053 and HD33184 (N.B.), a Scholar Grant from the Leukemia Society of America (N.B.), and NIH Training Grant No. GM07499 (N.D.L.).
Address reprint requests to Nancy Berliner, MD, Department of Internal Medicine, Section of Hematology WWW428a, Yale University School of Medicine, 333 Cedar St, New Haven, CT 06510.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal