Abstract
In this study, we show that both all-trans-retinoic acid (atRA) and 9-cis-retinoic acid (9-cis-RA) are potent inducers of tissue transglutaminase (TGase II), an enzyme involved in apoptosis, at the level of both enzyme activity and mRNA in the human myeloma cell line RPMI 8226. RPMI 8226 cells were shown to express mRNAs for all the retinoid receptors subtypes, ie, RARα, RARβ, RARγ, RXRα, RXRβ, and RXRγ. To identify which of these receptors are involved in regulating TGase II expression, several receptor-selective synthetic retinoids were used. Neither CD367, a very potent retinoid that selectively binds and activates receptors of the RAR family, nor CD2425, an RXR-selective agonist, induced TGase II when used alone. However, combination of CD367 and CD2425 resulted in nearly full induction of the enzyme. Moreover, when used in combination with atRA, CD367 partially inhibited the atRA-dependent induction of TGase II, whereas CD2425 enhanced it. The effects of Am 580, CD417, and CD437, three synthetic retinoids selective for the RARs subtypes RARα, RARβ, and RARγ, respectively, were also investigated. None of these compounds was able to induce TGase II when used alone; however, the combination of each of them with CD2425 resulted in strong induction of the enzyme activity, reaching 30% to 50% of the values obtained in the presence of retinoic acid and suggesting functional redundancy between the RAR subtypes. Finally, treatment with atRA or the combination of CD367 and CD2425, but not with CD367 or CD2425 alone, was also shown to trigger apoptosis in RPMI 8226 cells, with prominent accumulation of TGase II immunoreactivity in apoptotic cells. Taken together these data suggest that the induction of TGase II expression and apoptosis in the RPMI 8226 myeloma cell line required ligand-dependent activation of both the RAR and RXR receptors.
TRANSGLUTAMINASES (EC 2.3.2.13, TGase) are Ca++-dependent enzymes catalyzing the formation of ε(γ-glutamyl)lysine cross-links between polypeptide chains.1 Several members of this gene family have been identified including the type II (tissue) TGase.2,3 TGase II, the biological functions of which remain unclear, is an intracellular enzyme found in many cell types.4 TGase II is implicated in the activation of several cytokines5,6 and in signal transduction.7Expression of TGase II has been found associated with apoptosis in several cell types, and a role for TGase II in this process has been suggested.8-10
Apoptosis is a genetically controlled process of cell death and is important for the elimination of cells during morphogenesis, in embryonic development as well as in many adult tissues,10,11 and in cancer. Although the function of TGase II in apoptosis has yet to be elucidated, it has been proposed that TGase II may cross-link cellular proteins, thereby preventing the release of intracellular macromolecules.12
all-trans-retinoic acid (atRA) induces TGase II expression in various cell types.2,13-21 In mouse macrophages, regulation of TGase II expression by atRA occurs at the transcriptional level.2 Most of the effects ofatRA on gene expression are mediated by the activation of nuclear retinoid receptors, RARs and/or RXRs.22 The RAR and RXR gene family comprises three subtypes named α, β, and γ.22 These subtypes are expressed in a developmental stage– and cell type–specific manner,22 and each may regulate the expression of different genes. In this study, we examined the retinoid signaling pathways involved in the induction of TGase II (and apoptosis) in the human myeloma cell line RPMI 8226, using several retinoid receptor-selective agonists. This cellular model was chosen because atRA has been proposed for the treatment of multiple myeloma.
Our results provide evidence indicating that the induction of TGase II gene expression and apoptosis by atRA in RPMI 8226 cells is mediated through specific retinoid signaling pathways that involve both RAR(s) and RXR(s).
MATERIALS AND METHODS
Retinoids.
atRA and 9-cis-retinoic acid (9-cis-RA) were purchased from Sigma Chemical Co (St Louis, MO); Am580, CD367, CD417, CD437, and CD2425 were obtained from CIRD Galderma (Sophia Antipolis, France). Retinoids were dissolved in dimethyl sulfoxide at an initial 10−2 mol/L stock concentration and stored at −20°C in the dark. Dilutions were performed in ethanol. The final concentration of ethanol in culture was 0.1%.
Cell line and cell treatments.
RPMI 8226 cells (CCL-155 American Type Culture Collection) were cultured routinely at 37°C, 5% CO2 in complete medium (RPMI 1640 with Glutamax I, 10% fetal calf serum [FCS] inactivated by heating, penicillin [100 U/mL], and streptomycin [100 μg/mL]). Viability (higher than 95%) was determined by trypan blue exclusion. TGase induction by retinoids was performed in cells grown in standard medium (RPMI 1640 with Glutamax I, penicillin [100 U/mL], streptomycin [100 μg/mL], insulin [5 μg/mL], transferrin [5 μg/mL], and sodium selenite [5 ng/mL]) supplemented either with 3% FCS at 7 hours for TGase activity measurement, or with 10% FCS at 48 hours for microscopic studies.
TGase activity measurement.
TGase activity of cell lysates was determined by Ca++-dependent incorporation of [3H]putrescine into N,N' dimethylated casein.23 A total of 107 cells were induced by retinoid at 5 × 105 cells/mL for 48 hours and lysed in 150 μL 10 mmol/L Tris-HCl pH 7.5, 1 mmol/L EDTA, 0.5% Triton X-100. Aliquots of cell extracts (0.2 mg of proteins) were incubated at 30°C in a total volume of 100 μL containing 20 mmol/L Tris-HCl pH 7.5, 5 mmol/L CaCl2, 10 mmol/L β mercaptoethanol, 2 mg/mL N,N' dimethylated casein, 2 μCi [3H]putrescine (30 to 60 Ci/mmol, NEN Life Science, LeBlanc Mesnil, France), and 0.2 mmol/L putrescine. After 20 minutes, aliquots were spotted on Whatman 3MM filter paper, fixed and washed in 10% trichloroacetic acid, 5% trichloroacetic acid, ethanol/acetone (v/v), and acetone. Protein-bound [3H]putrescine was determined by liquid scintillation counting. Background value was obtained by substitution of 1 mmol/L EGTA for CaCl2 in the reaction mixture added to cell extracts and lysis buffer. Enzyme activity was expressed as picomoles of [3H]putrescine incorporated into N,N' dimethylated casein per minute and per milligram of cell lysate protein. Protein amount was assayed by the Bradford method24 using fraction V Albumin (Boehringer Mannheim, Mannheim, Germany) as the standard.
Reverse transcriptase-polymerase chain reaction (RT-PCR) and Northern blot analysis.
Total RNA was isolated from 107 cells using RNAble (Eurobio, Les Ullis, France) and was used for RT-PCR and Northern blot analysis.
For PCR amplification, pairs of primers were selected using the cDNA sequences of human TGase II from umbilical vein endothelial cells, human β-actin, human RARα, RARβ, RARγ, and human RXRα, RXRβ, RXRγ (Table 1). TGase II primers are located in the coding region of two contiguous exons.25The reverse transcriptase reaction was performed in a final volume of 20 μL containing 1 μg total RNA (denatured at 68°C for 5 minutes), 20 U RNAsin Ribonuclease inhibitor (Promega, Charbonnieres, France), 5 μmol/L pd(N)6(Pharmacia Biotech, Uppsala, Sweden), 0.5 mmol/L dNTPs, and 200 U Moloney Murine Leukemia Virus Reverse Transcriptase (Promega) in RT buffer. An aliquot (5 μL) of the RT reaction product was mixed with 20 μL PCR buffer containing 1 U Taq DNA polymerase (Promega), 0.1 mmol/L dNTPs, and 100 ng of each primer. PCR was performed in a minicycler (MJ Research, Watertown, MA) as follows: 95°C for 5 minutes, then n (15 to 40) cycles consisting of 1 minute at 95°C, 1 minute at 54°C, and 1 minute at 72°C. After the last cycle, incubation at 72°C was prolonged for 10 minutes. TGase II, RARs, RXRs, and β-actin PCR reactions were performed separately. PCR products (15 μL) were analyzed on a 2% agarose gel in TAE buffer (40 mmol/L Tris, 40 mmol/L sodium acetate, 1 mmol/L EDTA, pH 8.4) using a DNA ladder 100-bp marker and a PCR product from human TGase II cDNA26 (kindly provided by Dr V. Gentile, Naples, Italy) as controls. Samples were analyzed by Southern blot using as a probe internal primers for amplified sequence (Table 1). Primers were labeled with 20 μCi γ[32P] adenosine triphosphate (ATP; 3,000 Ci/mmol Dupont-NEN) using 5 to 10 U/μL T4 polynucleotide kinase (Promega).
5′ to 3′ Sequence of PCR Primers and Probes
| . | Primer . | Sequence 5′ to >3′ . | EMBL Bank Accession Number . |
|---|---|---|---|
| TGase II | Upper | CAGTTCCAGTTCGTGCCATCA | M55153 |
| Lower | ATGCCTGTCTCCTCCTTCTCG | ||
| Internal | ATCACCCACACCTACAAATACCCAGAG | ||
| β-actin | Upper | ATCATGTTTGAGACCTTCAA | X00351 |
| Lower | CATCTCTTGCTCGAAGTCCA | ||
| Internal | GACCTGGCTGGGCCGGACCTGACTGACTAC | ||
| RAR α | Upper | CATTGAGACCCAGAGCAGC | X006614 |
| Lower | CCGTCTCCGCATCATCCATC | ||
| Internal | GCTCTGAGAGCTACACGCT | ||
| RAR β | Upper | GAATTGAAACACAGAGCACC | Y00291 |
| Lower | GCAGGAGTGGTGACTGACTG | ||
| Internal | GACAGCTGAGTTGGACGATC | ||
| RAR γ | Upper | TGGAGACACAGAGCACCAGCTCA | M38258 |
| Lower | GTCAGTCTGCTGCCTGAAGCC | ||
| Internal | TGAGCCCTCAGTTAGAAGAGC | ||
| RXR α | Upper | CTCCTCAGGCAAGCACTATG | X52773 |
| Lower | AGAGCTTAGCGAACCTTCCC | ||
| Internal | AGGAGCGGCAGCGTGGCAAGGAC | ||
| RXR β | Upper | TCAGGCAAACACTACGGGGT | X63522 |
| Lower | GCATACACTTTCTCCCGCAG | ||
| Internal | AAAGGACAAGGATGGGGATGGGGAG | ||
| RXR γ | Upper | CTCAGGAAAGCACTACGGGG | U38480 |
| Lower | CAGGGTCATTTGTCGAGTTC | ||
| Internal | AGAAGAAAGACAGAGGAGCCGAGAG |
| . | Primer . | Sequence 5′ to >3′ . | EMBL Bank Accession Number . |
|---|---|---|---|
| TGase II | Upper | CAGTTCCAGTTCGTGCCATCA | M55153 |
| Lower | ATGCCTGTCTCCTCCTTCTCG | ||
| Internal | ATCACCCACACCTACAAATACCCAGAG | ||
| β-actin | Upper | ATCATGTTTGAGACCTTCAA | X00351 |
| Lower | CATCTCTTGCTCGAAGTCCA | ||
| Internal | GACCTGGCTGGGCCGGACCTGACTGACTAC | ||
| RAR α | Upper | CATTGAGACCCAGAGCAGC | X006614 |
| Lower | CCGTCTCCGCATCATCCATC | ||
| Internal | GCTCTGAGAGCTACACGCT | ||
| RAR β | Upper | GAATTGAAACACAGAGCACC | Y00291 |
| Lower | GCAGGAGTGGTGACTGACTG | ||
| Internal | GACAGCTGAGTTGGACGATC | ||
| RAR γ | Upper | TGGAGACACAGAGCACCAGCTCA | M38258 |
| Lower | GTCAGTCTGCTGCCTGAAGCC | ||
| Internal | TGAGCCCTCAGTTAGAAGAGC | ||
| RXR α | Upper | CTCCTCAGGCAAGCACTATG | X52773 |
| Lower | AGAGCTTAGCGAACCTTCCC | ||
| Internal | AGGAGCGGCAGCGTGGCAAGGAC | ||
| RXR β | Upper | TCAGGCAAACACTACGGGGT | X63522 |
| Lower | GCATACACTTTCTCCCGCAG | ||
| Internal | AAAGGACAAGGATGGGGATGGGGAG | ||
| RXR γ | Upper | CTCAGGAAAGCACTACGGGG | U38480 |
| Lower | CAGGGTCATTTGTCGAGTTC | ||
| Internal | AGAAGAAAGACAGAGGAGCCGAGAG |
TGase II primers were the same as reported.25
Northern blot analysis was performed using 8 μg total RNA and cDNA probes for human TGase II, RARα, RARβ, RARγ, and RXRα, and for mouse RXRβ, RXRγ kindly provided by Dr V. Gentile, Pr P. Chambon (LGME, Strasbourg, France), and Drs R.M. Evans and D.J. Mangelsdorf (The Salk Institute, La Jolla, CA), respectively. cDNAs were labeled with 20 μCi α[32P]ATP (800 Ci/mmol) using a multilabeling kit (Boehringer Mannheim).
Autoradiograms were quantified by densitometric analysis with a rapid electrophoresis apparatus (Helena Laboratory, Beaumont, TX). Values were normalized using β-actin as external standard.
Morphological studies.
At the indicated times, 15 × 104 cells were taken to make cytospin preparations. Slides were stored at −20°C. Cell morphology was observed after staining with May-Grünwald-Giemsa (MGG). DNA staining with Hoechst 33258 (Bis [benzidine]) was performed as described by Galli and Fratelli.27 In situ DNA fragmentation was measured by using the to TUNEL method based on 3'OH end labeling of DNA breaks with deoxyuridine terminal deoxynucleotidyl transferase.28 Immunofluorescence detection29of TGase II was performed by using a mouse monoclonal antibody (IgG) raised against purified guinea pig TGase II30 and kindly provided by Dr P.J. Birckbichler (Ardmore, OK). A fluorescein conjugated anti-mouse IgG (Biosys, Compiègne, France) was used for detection by fluorescence microscopy.
RESULTS
Time course and dose-response induction by atRA of TGase II activity.
TGase II activity was undetectable in untreated RPMI 8226 cells.atRA treatment resulted in a time and dose-dependent induction of the enzyme (Fig 1). This induction was easily detected after 2 days of incubation and increased linearly until 4 days (Fig 1A). Interestingly 9-cis-RA, which activates both RARs and RXRs, appeared more potent than atRA in dose-response experiments (Fig 1B).
Induction of TGase II activity by retinoic acid in RPMI 8226 cells. Cells were treated with atRA or 9-cis-RA, and TGase II activity was assayed by [3H]putrescine incorporation into N,N' dimethylated casein. (A)Time course of the induction of TGase II activity by 10−7 mol/L atRA. (B) Dose-response curves after 48 hours of induction with various concentration of atRA or 9-cis-RA.
Induction of TGase II activity by retinoic acid in RPMI 8226 cells. Cells were treated with atRA or 9-cis-RA, and TGase II activity was assayed by [3H]putrescine incorporation into N,N' dimethylated casein. (A)Time course of the induction of TGase II activity by 10−7 mol/L atRA. (B) Dose-response curves after 48 hours of induction with various concentration of atRA or 9-cis-RA.
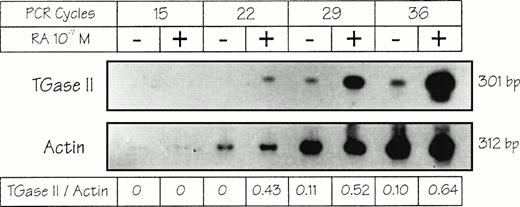
mRNA TGase II induction by atRA in RPMI 8226 cells.
TGase II induction was also detected at the mRNA level by using a RT-PCR assay (Fig 2). Whereas the expected 301-bp amplification product could not be detected before the 29th cycle of PCR in untreated cells, it was clearly observed after only 22 cycles in atRA-treated cells and increased dramatically after 29 and 36 cycles. The 301-bp band specificity was controlled using PCR performed with a bona fide cloned human TGase II cDNA26 andBamH1 cleavage, which yielded the expected digestion pattern (data not shown). Actin levels were not altered upon atRA treatment. Taken together these results showed that theatRA-dependent induction of TGase II in RPMI 8226 cells occurs at a pretranslational stage.
Induction of TGase II mRNA in RPMI 8226 cells. Cells were treated or not for 7 hours with 10−7 mol/L atRA in defined medium. Total RNA (1 μg) was submitted to RT and PCR (for 22, 28, 34, or 40 cycles), fractionated on 2% agarose gel, transferred to a Hybond N+ membrane, and hybridized to32P-labeled oligonucleotidic probes for human TGase II and actin. (For primers and probes see Materials and Methods).
Induction of TGase II mRNA in RPMI 8226 cells. Cells were treated or not for 7 hours with 10−7 mol/L atRA in defined medium. Total RNA (1 μg) was submitted to RT and PCR (for 22, 28, 34, or 40 cycles), fractionated on 2% agarose gel, transferred to a Hybond N+ membrane, and hybridized to32P-labeled oligonucleotidic probes for human TGase II and actin. (For primers and probes see Materials and Methods).
Expression of RARs and RXRs in RPMI 8226 cell line.
The induction of TGase II by atRA may be directly or indirectly mediated by nuclear retinoid receptors. To determine which retinoid receptors could be involved in this regulation, the expression of RARs and RXRs in RPMI 8226 cells was investigated. Untreated cells expressed RARα, RARβ, RARγ, RXRα, RXRβ, and RXRγ mRNAs at various levels (Fig 3). The size of the transcripts was checked by Northern Blot and found to be correct (data not shown). Treatment of RPMI 8226 cells for 7 hours with 10−7 MatRA resulted in an upregulation of RARβ and RARγ as shown by the sensitive RT-PCR assay used (Fig 3).
Retinoid-receptor profile of RPMI 8226 cell line. Cells were treated or not for 7 hours with 10−7 mol/LatRA in defined medium. Total RNA (1 μg) was submitted to RT and PCR (for 15, 20, 25, 30, or 35 cycles), fractionated on 2% agarose gel, transferred to a Hybond N+ membrane, and hybridized to 32P-labeled oligonucleotidic probes for human RARα, RARβ, RARγ, RXRα, RXRβ, RXRγ, or β-actin. (For primers and probes see Materials and Methods).
Retinoid-receptor profile of RPMI 8226 cell line. Cells were treated or not for 7 hours with 10−7 mol/LatRA in defined medium. Total RNA (1 μg) was submitted to RT and PCR (for 15, 20, 25, 30, or 35 cycles), fractionated on 2% agarose gel, transferred to a Hybond N+ membrane, and hybridized to 32P-labeled oligonucleotidic probes for human RARα, RARβ, RARγ, RXRα, RXRβ, RXRγ, or β-actin. (For primers and probes see Materials and Methods).
Effects of receptor-selective retinoids.
To evaluate the respective involvement of RARs and RXRs in the induction of TGase II, we used several retinoids that bind selectively to and activate specific RARs or RXRs (Fig4).
Structures of the natural and synthetic retinoids used. Structures of atRA, 9-cis-RA,4-[5,6,7,8-tetrahydro-5,5,8,8-tetramethyl-2-anthracenyl]-benzoic acid (CD367), (E)-2-[2-(5,6,7,8-tetrahydro-3,5,5,8,8-pentamethyl-2-naphthyl)propen-1-yl]-4-thiophenecarboxylic acid (CD2425 or AGN191701), [4-(5,6,7,8-tetrahydro-5,5,8,8-tetramethyl-2-naphthalenyl) carboxamido]benzoic acid (Am580), 6-(3-tert-butyl-4-methoxyphenyl)-2-naphthoic acid (CD417), and 6-[3-adamantyl-4-hydroxyphenyl]-2-naphtalene carboxylic acid (CD437). The selectivity of each compound is indicated according to previously published dissociation constant and EC50data.31,32 43 CD2425 does not transactivate RARs, but transactivates RXRs (EC50 = 54 nmol/L). It binds to RARs with a very weak affinity (kd = 10,000, 1470, and 712 nmol/L for RARα, RARβ, and RARγ, respectively; U. Reichert, personal communication, March 1997). No RXR binding data have been published for this compound.
Structures of the natural and synthetic retinoids used. Structures of atRA, 9-cis-RA,4-[5,6,7,8-tetrahydro-5,5,8,8-tetramethyl-2-anthracenyl]-benzoic acid (CD367), (E)-2-[2-(5,6,7,8-tetrahydro-3,5,5,8,8-pentamethyl-2-naphthyl)propen-1-yl]-4-thiophenecarboxylic acid (CD2425 or AGN191701), [4-(5,6,7,8-tetrahydro-5,5,8,8-tetramethyl-2-naphthalenyl) carboxamido]benzoic acid (Am580), 6-(3-tert-butyl-4-methoxyphenyl)-2-naphthoic acid (CD417), and 6-[3-adamantyl-4-hydroxyphenyl]-2-naphtalene carboxylic acid (CD437). The selectivity of each compound is indicated according to previously published dissociation constant and EC50data.31,32 43 CD2425 does not transactivate RARs, but transactivates RXRs (EC50 = 54 nmol/L). It binds to RARs with a very weak affinity (kd = 10,000, 1470, and 712 nmol/L for RARα, RARβ, and RARγ, respectively; U. Reichert, personal communication, March 1997). No RXR binding data have been published for this compound.
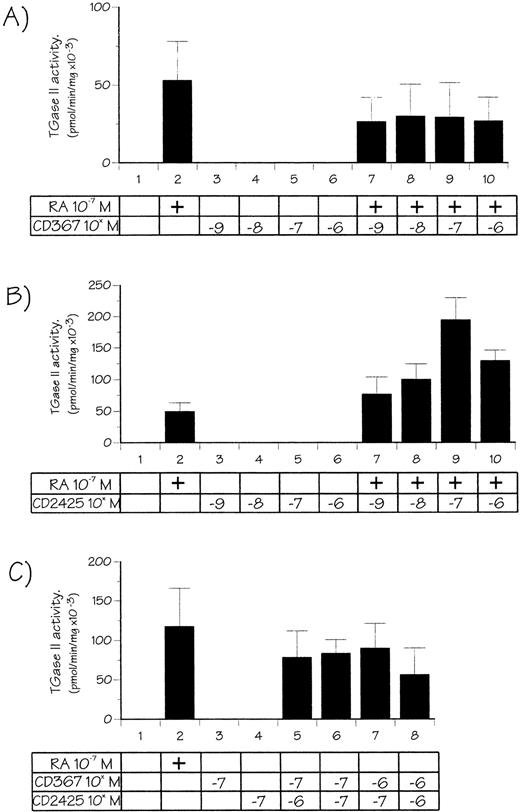
We first analyzed effects of CD367 and CD2425, two retinoids highly selective for the RAR and the RXR receptors families, respectively. CD367 binds to and activates RARα, RARβ, and RARγ at nanomolar concentrations,31 whereas CD2425 (or AGN 191701) is a RXR panagonist.32 RPMI 8226 cells were treated for 48 hours with these retinoids at concentrations ranging from 10−9 to 10−6 mol/L, either alone or in combination with 10−7 mol/L atRA. This concentration of atRA, yielding only suboptimal (about 30%) TGase II induction, was chosen to detect any positive or negative modulation of the response by the synthetic retinoid used. Results are presented in Fig 5. When used alone, and at every concentration tested, the RAR-selective retinoid CD367 did not cause any increase in the TGase II activity (Fig 5A). Moreover, it appeared able to partially inhibit the atRA-dependent TGase II induction. When used alone the RXR-selective compound, CD2425 appeared also unable to induce TGase II activity (Fig 5B). However, CD2425 in combination with atRA strikingly enhanced the atRA response in a dose-dependent manner, the maximum being observed at 10−7 mol/L CD2425 (threefold enhancement when compared with 10−7 mol/L atRA alone). At higher CD2425 concentrations (10−6 mol/L) combination with 10−7 mol/L atRA appeared less efficient. Finally, as shown in Fig 5C, combination of both RAR and RXR-selective agonists resulted in synergistic induction of TGase II, reaching 80% of the value obtained with 10−7 mol/L atRA. Taken together these observations suggested that activation of both RAR and RXR is required for induction of TGase II.
Effects of RAR and RXR-receptor–selective retinoids or both on TGase II activity in RPMI 8226 cells. Cells were treated for 48 hours with 10−7 mol/L atRA, varying doses of CD367 (A), CD2425 (B), and their combination (C), in defined medium for 7 hours, and then in 3% FCS-supplemented medium.
Effects of RAR and RXR-receptor–selective retinoids or both on TGase II activity in RPMI 8226 cells. Cells were treated for 48 hours with 10−7 mol/L atRA, varying doses of CD367 (A), CD2425 (B), and their combination (C), in defined medium for 7 hours, and then in 3% FCS-supplemented medium.
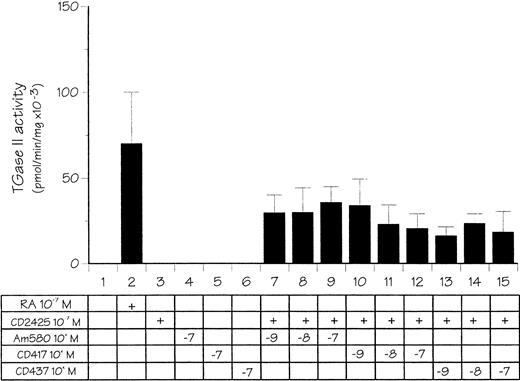
The next step in our studies was to determine if one of the RAR subtypes, ie, RARα, β, or γ, was selectively involved in the control of TGase II expression. RPMI 8226 cells were treated as before in the presence of either Am 580, CD417, or CD437, three synthetic retinoids that are RARα, β, or γ selective, respectively, when used at nanomolar concentrations.33 34 As shown in Fig 6, RAR subtype selective retinoids used alone were inactive. However, when used at 10−9 mol/L concentration in combination with CD2425, each of them induced TGase II activity to an extent varying from 25% to 45% of the level obtained after treatment with 10−7 mol/L atRA. Interestingly, no further induction was observed when the RAR subtype ligands were used at higher, nonselective concentrations. Thus, the various RAR subtypes that are all expressed in RPMI 8226 displayed functional redundancy with respect to the synergistic induction of TGase II in the presence of an RXR-selective ligand.
Effect of RARα-, β-, and γ-receptor–selective retinoids in combination with a RXR-receptor–selective retinoid on TGase II activity in RPMI 8226 cells. Cells were treated for 48 hours with either 10−7 mol/L atRA or 10−7mol/L CD2425 (RXR selective) and various doses of Am580 (RARα selective), CD417 (RARβ selective), or CD437 (RARγ selective) in defined medium for 7 hours, and then in 3% FCS-supplemented medium.
Effect of RARα-, β-, and γ-receptor–selective retinoids in combination with a RXR-receptor–selective retinoid on TGase II activity in RPMI 8226 cells. Cells were treated for 48 hours with either 10−7 mol/L atRA or 10−7mol/L CD2425 (RXR selective) and various doses of Am580 (RARα selective), CD417 (RARβ selective), or CD437 (RARγ selective) in defined medium for 7 hours, and then in 3% FCS-supplemented medium.
Correlation between the induction of TGase II and apoptosis in RPMI 8226 cells.
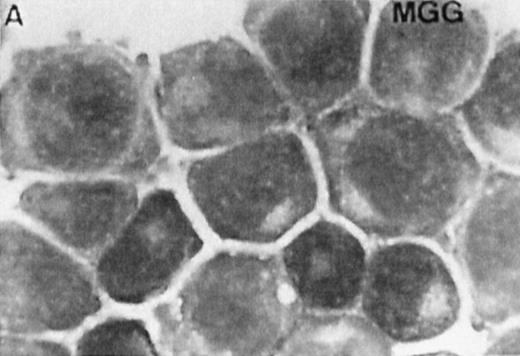
Several studies have suggested a role for TGase II in apoptosis.8-10 To examine whether the atRA-induced TGase II expression in RPMI 8226 cells was correlated with the induction of apoptosis, we performed both indirect immunofluorescence detection of TGase II protein (Fig 7, left panel) and MGG staining (Fig 7, right panel) of cells within 6 days of addition of different retinoids to the culture . Apoptosis was quantified by counting apoptotic cells after MGG and Hoechst 33258 staining. Count of TUNEL-positive cells gave similar results (Fig 8).
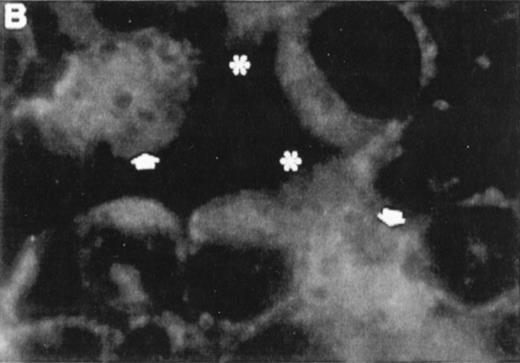
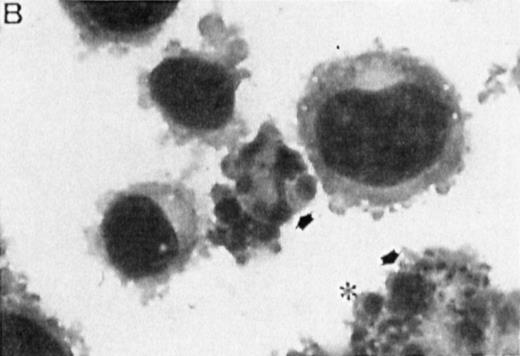
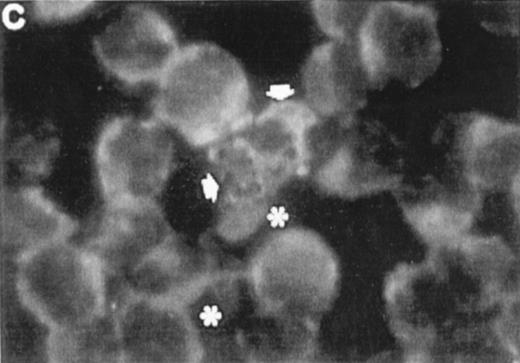
Correlation between the induction of TGase II protein and apoptosis in RPMI 8226 cells. Cells were incubated for 6 days in the absence of retinoids (A) or in the presence of 10−7 mol/LatRA (B) or the combination of both 10−7 mol/L CD367 and 10−7 mol/L CD2425 (C). TGase II protein expression was detected by indirect immunofluorescence using a monoclonal anti-TGase II antibody (left panel), and apoptotic cells were detected after MGG coloration on cytospin preparations (right panel).
Correlation between the induction of TGase II protein and apoptosis in RPMI 8226 cells. Cells were incubated for 6 days in the absence of retinoids (A) or in the presence of 10−7 mol/LatRA (B) or the combination of both 10−7 mol/L CD367 and 10−7 mol/L CD2425 (C). TGase II protein expression was detected by indirect immunofluorescence using a monoclonal anti-TGase II antibody (left panel), and apoptotic cells were detected after MGG coloration on cytospin preparations (right panel).
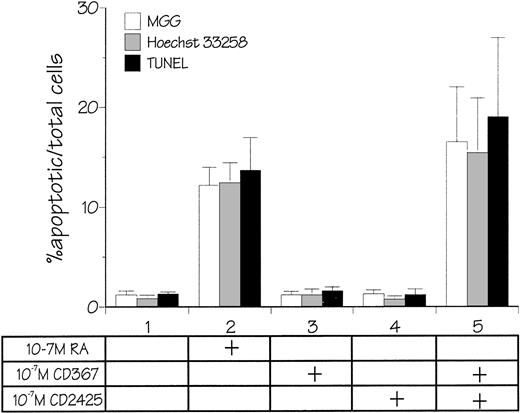
Quantification of apoptosis. Cells were incubated for 6 days in the absence of retinoids or in the presence of 10−7 mol/L atRA, 10−7 mol/L CD367, 10−7 mol/L CD2425, or the combination of both 10−7 mol/L CD367 and 10−7 mol/L CD2425. Count of apoptotic cells was performed on cytospin preparations after MGG coloration, Hoechst 33258 staining, or TUNEL assay. In each case the percentage of apoptotic cells was determined in duplicate counts of 400 cells. Mean values ±SD from six independent experiments are shown.
Quantification of apoptosis. Cells were incubated for 6 days in the absence of retinoids or in the presence of 10−7 mol/L atRA, 10−7 mol/L CD367, 10−7 mol/L CD2425, or the combination of both 10−7 mol/L CD367 and 10−7 mol/L CD2425. Count of apoptotic cells was performed on cytospin preparations after MGG coloration, Hoechst 33258 staining, or TUNEL assay. In each case the percentage of apoptotic cells was determined in duplicate counts of 400 cells. Mean values ±SD from six independent experiments are shown.
As shown in Fig 7A, after treatment of RPMI 8226 cells with 10−7 mol/L CD367 or 10−7 mol/L CD2425 alone, neither apoptotic cell death (right panel) nor TGase II (left panel) were induced. The apoptotic index in the CD367- or CD2425-treated cultures was not different from that in control (Fig 8). Thus, mere activation of endogenous RARs by a RAR-selective retinoid such as CD367 or of endogenous RXRs by a RXR-selective retinoid such as CD2425 is insufficient to induce apoptosis in RPMI 8226 cells.
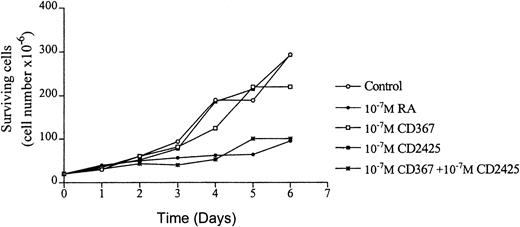
Interestingly, treatment with 10−7 mol/LatRA, the natural panagonist ligand, resulted in both TGase II induction (Fig 7B, left panel) and significant apoptosis. As shown in Fig 7B (right panel), numerous cells displayed the characteristics of a classical apoptotic phenotype with condensed nuclei, extensive cytoplasmic blebs, marked decrease in cellular volume, and cellular fragmentation. The apoptotic index of atRA-treated cells was approximately 12% (Fig 8). Finally, after 6 days of culture in the presence of both 10−7 mol/L CD367 and 10−7 mol/L CD2425, we observed the same morphological evidence of apoptosis as in atRA-treated cells (Fig 7C, right panel). The apoptotic index was in the 18% range (Fig 8). AfteratRA treatment TGase II appeared induced in cells undergoing apoptosis and accumulated in apoptotic bodies (Fig 7C, left panel). Taken together, our results showed that the induction of TGase II and apoptosis by retinoids in RPMI 8226 cells is linked and that the activation of both RARs and RXRs is required for the induction of apoptosis. Interestingly, the same requirement was observed for the inhibition of cell proliferation elicited by retinoids in RPMI 8226 cells (Fig 9).
RPMI 8226 cells growth in the presence of various natural and synthetic retinoids. Cell proliferation was evaluated by direct counting (Trypan blue assay).
RPMI 8226 cells growth in the presence of various natural and synthetic retinoids. Cell proliferation was evaluated by direct counting (Trypan blue assay).
DISCUSSION
In this study we showed that retinoic acid induces TGase II expression in a dose-dependent manner at both the mRNA and the enzyme activity levels in RPMI 8226 cells. A retinoic acid–dependent expression of the protein was also shown in intact cells by indirect immunofluorescence assay. In RPMI 8226 cells, there is no detectable basal expression of TGase II, a situation also observed in other cellular models, including HL-60 human leukemia cells, a model extensively used for the study of the mechanisms controlling TGase II expression.15Conversely, in some other cell types and tissues, significant basal expression of TGase II has also been reported.35-38 In most cellular models, however and whatever the basal level, retinoic acid induced TGase II, with only few exceptions,35 39 probably related to some poorly understood postreceptor resistance.
RPMI 8226 cells express all the retinoic acid receptors mRNA, suggesting that all the RARs and RXRs proteins are also expressed in these cells, which could therefore constitute a very attractive model for the elucidation of their respective role in the control of various target genes by retinoic acid. A more limited pattern of expression of the retinoic acid receptors has been reported in HL-60 and SPOC-1 cells, two models recently used for the study of the control of expression of TGase II by retinoids. HL-60 expressed only RARα, RXRα, and RXRβ, whereas SPOC-1 cells expressed only RARα, RARγ, and RXRβ.32 40
RAR subtype (α, β, or γ) and RXR-specific retinoids are extremely useful ligands that, when used at appropriate selective concentrations, allow the investigation of the respective contribution of the various RARs and RXRs in the molecular control of a given retinoic acid–induced response.34,41 Our results showed that in RPMI 8226 cells, TGase II induction required the activation of both the RAR and the RXR pathways. Our report is the first trying to elucidate the molecular mechanism of the control of TGase II by retinoic acid and its correlation to apoptosis in human myeloma cells. However, this regulation has been already investigated in other human cells by using the same kind of approach. In the HL-60 promyelocytic leukemia cell line, TGase II induction was observed in the presence of both RAR/RXR panagonists and RXR-selective ligands,42-45 implying that the activation of RXR alone is able to elicit the response. The implication of the RAR pathway is less clear, because conflicting results have been reported using TTNPB, an RAR-selective ligand, in two different HL-60 subclones. In the HL-60 cdm-1 clone, TTNPB was inactive,42 whereas in the HL-60 CCL240 clone it induced TGase II activity at the same level as retinoic acid– and RXR-specific ligands.45 These discrepancies are probably related to differences in the cellular context of the two subclones. Interestingly, the HL-60 cdm-1 line is a differentiation-resistant line.32 In NB4 cells, a human promyelocytic leukemia cell line expressing both the wild-type RARα and the PML-RARα fusion protein (resulting from the reciprocal chromosomal translocation t(15:17) that is the molecular hallmark of acute promyelocytic leukemia, RAR- and RARα-selective retinoids were able to induce TGase II, whereas RXR-selective retinoids were inactive.39Moreover, an RARα antagonist completely inhibited the expression of TGase II. Thus, in these cells expression of TGase II appeared to be controlled by RARs rather than by RXR, and a peculiar role for PML-RARα in the regulation has also been reported. Interestingly, the specific involvement of RARα in the control of TGase II expression has been also documented in rat and mouse fibroblasts and rat tracheobronchial cells by using both RAR- and RXR-selective retinoids40,46 and modulation of RARα level by transfecting sense and antisense RARα-expression vectors.47 RXR-selective ligands displayed neither activity when used alone nor synergy with RAR-selective ligands.40
Taken together these data indicate that, whereas retinoic acid is able to induce TGase II in a large variety of tissues and species, the molecular mechanism by which it exerts this control differs strikingly and involves different retinoid-receptor subtypes according to the cell model used. The pattern of expression of the various RARs and RXRs in a given cell, the promoter context of the TGase II gene, and more generally the cellular context are probably responsible for the diversity observed. Recently, the mouse TGase II promoter has been cloned and its inducibility by retinoids has been studied in CV-1 cells transfected with various RARs and RXRs.48 In the CV-1 cells context this promoter was shown to be activated by both RAR and RXR pathways, a result different from the RAR preference observed in mouse and rat cells.
We observed that activation of both the RAR and RXR pathways is required to obtain a synergistic induction of TGase II. Such a synergy has been already reported for other retinoic acid–induced responses.34 A potentiation of the effect of a RAR-selective ligand on HL-60 cell differentiation by RXR-selective ligands has been also reported.49 Such synergic and potentiating effects probably involve the activation of RAR-RXR heterodimers and a cooperation between the ligand-dependent activation functions of both partners.34 No retinoid response element has been identified on the partial sequence of the human TGase II promoter published so far,50 and the existence of such elements in this promoter remains to be documented. Our results suggest that a subtle equilibrium between the respective activation levels of RARs and RXRs has to be reached to obtain maximum synergy (see the opposite effects of the combination of CD2425 v CD367 withatRA in Fig 5A and B). The observed enhancing effect of CD2425, when added to 10−7 mol/L atRA, could be explained by a better activation of RXR, because atRA activates mainly RARs; its ability to activate RXRs also in living cells probably results from partial conversion into 9-cis-RA. Interestingly, 9-cis-RA, which activates both RARs and RXRs, appeared more efficient than atRA for the induction of TGase II (Fig 1B). Thus, in the presence of CD2425, the response to atRA was shifted to a 9-cis-RA–like response with better TGase II induction. Conversely, in the presence of CD367, the RAR-selective ligand, the balance was shifted toward lower induction (Fig 5A). This inhibitory effect could result from a disequilibrium between RAR and RXR activation. The relative level of activation of the two receptor types need probably to be finely tuned to obtain maximal activation, and in the presence of CD367 this equilibrium could be disturbed (with a too-high stimulation of the RAR partner, when compared with RXR). We previously showed that CD367 induces the formation of RARs homodimers in vitro51; however, the occurrence of such homodimers is not documented in vivo. Moreover, recent data obtained in our laboratory showed that CD367 and atRA do not interact in the same way with RARα and RARα mutants (B. Lefebvre et al, submitted; S. Mailfait, in preparation). In fact, CD367, although considered as a RAR panagonist, displays a variable efficiency according to the response tested (U. Reichert and S. Michel, CIRD Galderma, personal communication, March 1997). It is therefore possible that for the specific control of TGase II induction, CD367 behaves as a partial agonist, displaying also weak antagonist activity. Another possibility is an interference of CD367 with the metabolism of atRA and its isomerization into 9-cis-RA.
Whereas the combination of a RAR panagonist (CD367) with a RXR agonist (CD2425) resulted in a synergic induction of TGase II and apoptosis at a level close to the one obtained with atRA (but slightly lower, further supporting the hypothesis of a partial agonist activity in our model), the combination of the various RAR-subtypes–selective ligands tested with CD2425 resulted in similar but only partial synergic induction of TGase II.
These results suggest that full TGase II induction by atRA probably involved the activation of all the RAR subtypes, ie, RARα, β, and γ present in RPMI 8226 cells, and that these subtypes are partially redundant for this induction. Such a functional redundancy has been already observed in other models.34
Finally, our observation that the expression of TGase II and the induction of apoptosis are regulated by retinoids in a very similar way in our cells is in good agreement with previous reports suggesting that TGase II plays a role in apoptosis.8-10,52 53 With their complete equipment in RARs and RXRs receptor subtypes, RPMI 8226 cells afford a very suitable model for further studies of the molecular control of apoptosis by retinoids in human myeloma.
ACKNOWLEDGMENT
We thank Dr Birckbichler for the gift of the monoclonal antibody to TGase II, Dr V. Gentile for the gift of the TGase II plasmid, and Drs D.J. Mangelsdorf, P. Chambon, and R.M. Evans for the gift of the RARs and RXRs plasmids. We are also indebted to B. Masselot, P. Segard, P. Plouvier, and S. Tournay for technical and secretarial assistance. We thank Drs Uwe Reichert and Serge Michel (CIRD GALDERMA) for providing RAR- and RXR-selective ligands and for helpful discussion.
Supported by grants from the Institut National de la Santé et de la Recherche Médicale (INSERM), the Ligue du Nord Contre le Cancer, the Association de la Recherche contre le Cancer (ARC), the CIRD GALDERMA, and Université de Lille II. INSERM U459 belongs to IFR 22 supported by INSERM, CH et U de Lille, Université de Lille II, and Centre Oscar Lambret.
Address reprint requests to Pierre Formstecher, INSERM U459 ≪Signaux, Récepteurs et Différenciation Cellulaire», Facultéde médecine, 1 place de Verdun 59045 Lille cedex, France.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

![Fig. 1. Induction of TGase II activity by retinoic acid in RPMI 8226 cells. Cells were treated with atRA or 9-cis-RA, and TGase II activity was assayed by [3H]putrescine incorporation into N,N' dimethylated casein. (A)Time course of the induction of TGase II activity by 10−7 mol/L atRA. (B) Dose-response curves after 48 hours of induction with various concentration of atRA or 9-cis-RA.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/7/10.1182_blood.v91.7.2423/3/m_blod4072801.jpeg?Expires=1765102256&Signature=lEym~rhseeM3FWbon3-e3zT5uspk8W2b033h9GG02Al7vYMcrYSPuCj~Yfamh5Cy-kdO6Tk~ba9Hta0QZcMden48YKu3mhVpnlZ~SpqqMICrJTzph2tjHXS-j~x0xb0E1Ym0hU1bjfLkQd7hQ0hMGXkLhgW5QLoVAuFiALNxlOAAvU84fGs1V3UzN5OOwSHJC2zMD7abc37vUuMwUBhatzAChQ-YSisOxevKTQ8RrI9cMfH3JUHQTOvmoB75FkUE7cLMZPZRv~NS~INdwHROTcvZPFw1uq2tk0drqyBnF3xwOLI0h2U7nbWIsu~~fe1IatU5hnBpR9xHX4HBE5yzvg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Fig. 4. Structures of the natural and synthetic retinoids used. Structures of atRA, 9-cis-RA,4-[5,6,7,8-tetrahydro-5,5,8,8-tetramethyl-2-anthracenyl]-benzoic acid (CD367), (E)-2-[2-(5,6,7,8-tetrahydro-3,5,5,8,8-pentamethyl-2-naphthyl)propen-1-yl]-4-thiophenecarboxylic acid (CD2425 or AGN191701), [4-(5,6,7,8-tetrahydro-5,5,8,8-tetramethyl-2-naphthalenyl) carboxamido]benzoic acid (Am580), 6-(3-tert-butyl-4-methoxyphenyl)-2-naphthoic acid (CD417), and 6-[3-adamantyl-4-hydroxyphenyl]-2-naphtalene carboxylic acid (CD437). The selectivity of each compound is indicated according to previously published dissociation constant and EC50data.313243 CD2425 does not transactivate RARs, but transactivates RXRs (EC50 = 54 nmol/L). It binds to RARs with a very weak affinity (kd = 10,000, 1470, and 712 nmol/L for RARα, RARβ, and RARγ, respectively; U. Reichert, personal communication, March 1997). No RXR binding data have been published for this compound.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/7/10.1182_blood.v91.7.2423/3/m_blod4072804.jpeg?Expires=1765102256&Signature=TCyCdZjUxhlifDTbAUzrVgCYdAtqcrKy860yYwa7Lkrx0zC6znWpzyTIFbRrwNgJpVa5Hq4fn2isPSDY3Q3BZG9TEYuiRpDsxlH2rU-ZBIgbW~4~niMUBsEYIBt1u4bTYhi5Ynoyj-lZzQCriackQN5qxnTllP68FyCq~fnwkjZxpkC25nCp1km4Dy-ST3IXcPSrl8pg50lXhNKqdoCHSwoK3Q4LtzfZud0HxHN7B5kMDGtWG3zkChMFWECrfmf-AIN8O2dHdImyzwjJvs9JIWJJpha3PC50HTrpyEu92nHB-U0FSQKMobwzoWPzsVbs1yhEw98-SiSsJSihOMnPRg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal