Abstract
Differentiation of totipotent mouse embryonic stem (ES) cells to various lymphohematopoietic cells is an in vitro model of the hematopoietic cell development during embryogenesis. To understand this process at cellular levels, differentiation intermediates were investigated. ES cells generated progeny expressing CD34, which was significantly enhanced by vascular endothelial growth factor (VEGF). The isolated CD34+ cells were enriched for myeloid colony-forming cells but not significantly for erythroid colony-forming cells. When cultured on OP9 stroma cells in the presence of interleukin-2 and interleukin-7, the CD34+ cells developed two types of B220+ CD34−lymphocytes: CD3− cytotoxic lymphocytes and CD19+ pre-B cells, and such lymphoid potential was highly enriched in the CD34+ population. Interestingly, the cytotoxic cells expressed the natural killer (NK) cell markers, such as NKR-P1, perforin, and granzymes, classified into two types, one of which showed target specificity of NK cells. Thus, ES cells have potential to generate NK-type cytotoxic lymphocytes in vitro in addition to erythro-myeloid cells and pre-B cells, and both myeloid and lymphoid cells seem to be derived from the CD34+intermediate, on which VEGF may play an important role.
DURING DEVELOPMENT of the hematopoietic system, certain splanchnopleuric mesodermal cells, originated from pluripotent epiblasts, become committed to one of several developmental pathways, thereby giving rise to precursors of the hematopoietic as well as the endothelial lineage.1 However, the molecular and cellular events leading to the generation of multipotential hematopoietic stem cells (HSCs) from the mesoderm are still unresolved. Our approach to studying these issues is based on the capacity of totipotent embryonic stem (ES) cells, isolated from the inner cell mass of the mouse preimplantation embryo, to differentiate in vitro into cells of the hematopoietic lineage.2 Upon induction of differentiation, ES cells from a cell aggregate called the embryoid body (EB), which contains germ layer-like structures that mimic, to some extent, the early postimplantation embryo.3 Although a variety of cell types derived from three germ layers are generated, some EBs develop clustered, nucleated erythroid cells (primitive erythrocytes) resembling a blood island in the E7.5-day yolk sac, followed by the generation of macrophages, mast cells, and definitive erythrocytes.2,4-9 Myeloid progenitor cells are also detected in the yolk sac at about the same time of development, which are also followed by the appearance of macrophages and mast cells.10-12 In this manner, the transition from ES cells to hematopoietic cells in vitro parallels the in vivo events observed in the early yolk sac.5
The hematopoietic potential of ES cells is not restricted to the erythro-myeloid lineages. Using two different methods of in vitro differentiation (the EB formation method as described and the coculture method with hematopoietic stroma cell lines), B- as well as T-lymphocyte progenitors are generated from ES cells, although derivation of the third lymphoid lineage, natural killer (NK) cell, has not been reported.13-17 Marrow reconstituting activity (MRA) is generated with both methods as well.18,19 The lymphocyte progenitors and MRA derived from ES cells may represent the earliest lymphocyte progenitors and the earliest HSC developed during embryogenesis. Although the first site of lymphocyte development in the mammalian embryo remains to be definitively allocated,20-25recent experiments have provided strong evidence that this may be an intraembryonic site.26 Likewise, HSC for the entire definitive hematopoietic system seems to be generated spontaneously at an intraembryonic site designated as aorta–gonad–mesonephros (AGM).27-29 Thus, the in vitro differentiation of ES cells may represent not only the extraembryonic (yolk sac), primitive hematopoietic cell development, but also certain fundamental aspects of the intraembryonic, definitive hematopoietic cell development in vivo.
Epiblasts isolated from postimplantation mouse embryos also generate hematopoietic progenitor cells in vitro.30 31 Therefore, dissecting the differentiation pathway from ES cells to lymphohematopoietic cells by clarifying intermediate cell types using various cell-surface markers may shed light on the sequence of events that occurs in vivo during the differentiation of epiblasts to primitive and definitive hematopoietic progenitor cells. In this report, we show that CD34+ progeny are produced from ES cells in vitro and that vascular endothelial growth factor (VEGF) markedly enhances the generation of this cell population. More importantly, we report, for the first time, that (1) cytotoxic lymphocytes with characteristics of natural killer (NK) cells, the third lymphoid lineage, are generated in vitro from ES cells; and (2) the CD34+ cell population is highly enriched for both myeloid potential and NK as well as B-lymphoid potential.
MATERIALS AND METHODS
Cells and reagents.
The ES cell line, A3-1, was established as described before.32 The J7 ES cell was kindly provided by K. Stark (Amgen Inc, Thousand Oaks, CA), and 32Dcl3 myeloid progenitor cell line by C. Saris (Amgen). The OP9 stroma cell line was obtained from H. Kodama (Ohu University, Fukushima, Japan), and Yac-1 lymphoma and P815 mastocytoma cell lines were from G. Trail (Amgen). Iscove's modified Dulbecco's medium (IMDM), α-minimum essential medium (α-MEM) and Dulbecco's phosphate-buffered saline without Mg2+ and Ca2+ (PBSA) were purchased from GIBCO-BRL (Gaithersburg, MD). All tissue culture flasks and plates were from Falcon (Franklin Lakes, NJ).
Recombinant human erythropoietin (EPO), human leukemia inhibitory factor (LIF), human granulocyte colony-stimulating factor (G-CSF) human interleukin-2 (IL-2), mouse IL-3, human IL-6, rat stem cell factor (SCF), and human transforming growth factor (TGF)-β1 were prepared at Amgen (Thousand Oaks, CA). Recombinant mouse IL-4, mouse IL-7, human IL-11, mouse IL-12, human IL-15, mouse VEGF, mouse granulocyte macrophage colony-stimulating factor (GM-CSF) and mouse macrophage colony-stimulating factor (M-CSF) were purchased from R&D Systems (Minneapolis, MN). Monoclonal antibody (MoAb) for E-cadherin/uvomorulin (clone DECMA1) was from Sigma (St Louis, MO). MoAb for CD34 (clone RAM34) conjugated with biotin was purchased from Pharmingen (San Diego, CA). For fluorescence-activated cell sorter (FACS) analysis, MoAb for mouse CD16/CD32 (clone 2.4G2) from Pharmingen was added for blocking nonspecific staining. Fluorescein isothiocyanate (FITC)-conjugated streptavidin, and FITC-conjugated as well as phycoerythrin (PE)-conjugated MoAbs for mouse B220 (clone RA3-6B2), BP-1 (clone 6C3), CD3ε (clone 145-2C11), CD4 (clone RM4-4), CD8α (clone 53-6.7), CD19 (clone 1D3), CD43 (clone S7), CD44 (clone IM7), CD45 (clone 30F11), DX5, heat-stable antigen (HSA, clone M1/69), Mac-1 (clone M1/70), and Sca-1 (clone E13-161.7) were also purchased from Pharmingen.
Maintenance of ES cells and induction of differentiation by the EB formation method.
A3-1 as well as J7 ES cells were maintained as described,3,32 except that 10 ng/mL LIF was included in the medium. Differentiation of ES cells with the EB formation method was according to Keller et al,5 with some modifications according to Potocnik et al.16 Two days before initiation of differentiation, 3 × 105 ES cells were passaged onto a feeder-free, gelatinized 6-cm plate, and cultured in IMDM, 15% fetal calf serum (FCS) (GIBCO), 10 ng/mL LIF, 0.15 mmol/L monothioglycerol (MTG, Sigma). The day of differentiation, cells were harvested and resuspended in the differentiation medium, consisting of IMDM 15% FCS (Hyclone, Logan, UT), 100 μg/mL bovine transferrin (Sigma), 10 μg/mL bovine insulin (GIBCO), 0.45 mmol/L MTG, in the presence of various cytokines with or without 1% methylcellulose (Fluka, Ronkonkoma, NY), at 500 cells/mL for methylcellulose-containing culture, or at 5,000 cells/mL for liquid culture without methylcellulose. The cell suspension was plated at 15 mL per 10-cm bacterial grade dish and incubated in 5% CO2, 5% O2, at 37°C. The standard concentration of SCF was at 100 ng/mL, VEGF at 20 ng/mL, IL-3 at 10 ng/mL, EPO at 1 U/mL, and TGF-β1 at 1 ng/mL. For liquid culture, media was changed every day from day 5 to day 9, and then every other day. For methylcellulose culture, 10 mL 1% methylcellulose-containing differentiation medium was added on day 8 or day 9.
Harvesting and staining EB cells.
EBs were collected, washed, resuspended in 1 mL 1 mmol/L EDTA in PBSA, incubated at 37°C for 3 min, and dissociated into single-cell suspension by passage through a 27-gauge needle, once. The remaining small aggregates were removed by filtration through a 40-μm filter (Falcon). The EB cells were spun and resuspended in 0.5% (vol/vol) Path-O-Cyte (purified bovine serum albumin solution from Miles, Kankakee, IL) in PBSA at 0.5 to 1 × 107 cells/mL. As a control, undifferentiated ES cells were treated with the EDTA solution for 10 minutes, and single-cell suspension was made in 0.5% Path-O-Cyte solution at 0.5 × 107 cells/mL. The cell suspensions, 50 to 100 μL/well, were transferred to a V-bottom 96-well microtiter plate (Nunc, Naperville, IL) and stained with 10 to 20 ng/mL of antibodies. The stained samples were analyzed on FACScan (Becton Dickinson, San Jose, CA) or sorted for CD34 using FACStar plus (Becton Dickinson).
Colonogenic cell assay.
Single-cell suspensions made from EBs, FACS-sorted CD34+ EB cells, or CD34− EB cells were mixed with IMDM, 20% FCS (Stem Cell Technology, Vancouver, Canada) 10% BSA (Stem Cell Technology), 100 μg/mL bovine transferrin, 10 μg/mL bovine insulin, 0.1 mmol/L MTG, 1% methylcellulose, and distributed to four to six 35-mm bacterial-grade dishes at 104 to 105cells/plate. For erythroid progenitors, 100 ng/mL SCF, 10 ng/mL IL-3, and 1 U/mL EPO were added to the culture, and colony-forming unit erythroid (CFU-E), burst-forming unit-erythroid (BFU-E), and a combination of CFU-E Macrophage[M], CFU-n[neutrophil]E, CFU-mastE, CFU-nEM, and CFU-mastEM) were counted. For myeloid colonies, 100 ng/mL SCF, 10 ng/mL IL-3, 10 ng/mL GM-CSF, 50 ng/mL G-CSF, 50 ng/mL IL-6 were included, and CFU-M, CFU-n, CFU-mast, CFU-Mmix (combination of CFU-nM and CFU-mastM) were counted on day 8. By contrast, for pre-B cells, 100 ng/mL SCF and 25 ng/mL IL-7 with or without 50 ng/mL IL-11 were added, and CFU-pre-B and CFU-M were counted on day 11. All the plates were incubated under 5% O2 5% CO2 at 36°C. Average number of each colony type was adjusted per 105seeded cells.
Stroma coculture method for developing lymphokine-activated killer cells and pre-B cells.
The stroma cell line, OP9, was maintained as described.33Confluent culture of OP9 in a six-well plate was used for coculture. The coculture medium contained α-MEM, 5% FCS (Hyclone), 0.1 mmol/L MTG. For direct differentiation of ES cells on OP9 to lymphokine-activated killer (LAK) cells or to pre-B cells, we used a modified two-step culture method originally described by Nakano et al.17 Briefly, 4,000 ES cells/well were cultured on the confluent OP9 cells in the coculture medium containing an elevated level of FCS, 15%. On days 5 to 6, cells were harvested, and 5 × 104 live cells/well were transferred onto a new confluent OP9 cells. Culture medium was changed from day 6, to the standard coculture medium (containing 5% FCS), and 50 ng/mL IL-2 and 5 ng/mL IL-7 were added on day 7. On days 12 to 14, the culture was transferred, again, to a new OP9 well by scraping off the adherent cell layer with a rubber policeman (Falcon), followed by filtration through a 40-μm filter to remove large colonies of various adherent cell types. By day 21, LAK cells were detected as hallow-forming cells on the OP9 cell layer. Pre-B-like cell foci were also detected by this time in a lower frequency. Removal of IL-2 allowed them to expand in the culture.
For demonstration of lymphoid potential in the CD34+ EB cells, CD34+ cells sorted from day 6 to 7 EBs (formed in the presence of 100 ng/mL SCF and 20 ng/mL VEGF) were seeded on the OP9 stroma cells (103 to 105 cells/well of a 6-well plate) and cultured in the coculture medium containing 50 ng/mL IL-2 and 5 ng/mL IL-7. Medium was changed every 3 days, and small adherent LAK cells appeared in approximately 2 weeks. The LAK cells were always the dominant cell type under this condition, and removal of IL-7 on day 7 of culture had only a minimal effect on LAK cell generation. However, to select pre-B cells, IL-2 was removed on day 7 and maintained only in IL-7 for another 1 to 2 weeks.
Maintenance of LAK cells and pre-B cells.
CD34+ EB cell-derived as well as ES cell-derived LAK cells were initially cultured on OP9 cells in the coculture medium containing 50 ng/mL IL-2. Because these LAK cells do not need OP9 cells for proliferation, before performing biological assays or preparing DNA and RNA, they were transferred to a stroma-free culture. The type I LAK cells (Table 1) were cultured in the same medium, and type II LAK cells (Table 1) were kept in the presence of 500 ng/mL IL-2. Spleen LAK cells were generated as described.34 ES- and EB34-pre-B cells (Table 1) were maintained on a confluent layer of OP9 in the coculture medium with 5 ng/mL IL-7. For primary pre-B cells from bone marrow, 3 × 105 mouse bone marrow cells/well were cultured on OP9 cells under the same condition for 3 weeks. Resulting cells were CD43+ B220+ CD19+ (data not shown).
List of CD34+ EB Cell–Derived as Well as ES Cell–Derived Lymphocytes Generated on the OP9 Stroma Cells
| Name of the Generated Cells . | Sources . |
|---|---|
| Type I LAK cells | |
| LAKa | A3-1 ES cells |
| LAKj | J7 ES cells |
| LAK34a-1 | CD34+ A3-1 EB cells |
| LAK34a-3 | CD34+ A3-1 EB cells |
| Type II LAK cells | |
| LAK34a-2 | CD34+ A3-1 EB cells |
| Pre-B cells | |
| ES-pre-B | A3-1 ES cells |
| EB34-pre-B | CD34+ A3-1 EB cells |
| Control LAK and pre-B cells | |
| Spleen-LAK* | C57BL/6 mouse spleen |
| BM-pre-B | C57BL/6 mouse bone marrow |
| Name of the Generated Cells . | Sources . |
|---|---|
| Type I LAK cells | |
| LAKa | A3-1 ES cells |
| LAKj | J7 ES cells |
| LAK34a-1 | CD34+ A3-1 EB cells |
| LAK34a-3 | CD34+ A3-1 EB cells |
| Type II LAK cells | |
| LAK34a-2 | CD34+ A3-1 EB cells |
| Pre-B cells | |
| ES-pre-B | A3-1 ES cells |
| EB34-pre-B | CD34+ A3-1 EB cells |
| Control LAK and pre-B cells | |
| Spleen-LAK* | C57BL/6 mouse spleen |
| BM-pre-B | C57BL/6 mouse bone marrow |
*Spleen-LAK was established in suspension culture.
Proliferation assay and killer cell assay.
Sensitivity of the LAK cells and of pre-B cells to various cytokines was determined with alamar blue dye (Alamar Bioscience, Sacramento, CA). Briefly, cells were harvested, washed twice with the coculture medium and distributed at 5,000 to 20,000 cells/50 μL/well into a 96-well plate containing 50 μL/well of coculture medium with different concentrations of cytokines. Plates were incubated for 36 to 48 hours at 37°C in 5% CO2. Then, 10 μL/well of alamar dye was added and incubated further for 5 hours before fluorescence measurement (excitation wavelength 530 nm/emission wavelength 590 nm) by CytoFlor II fluorescence plate reader (PerSeptive Biosystems, Bedford, MA). For pre-B cells, the adherent population was harvested from the stroma layer with brief trypsin-EDTA treatment, followed by 30-minute incubation in a tissue culture flask in the corresponding FCS-containing medium to selectively remove the adherent stroma cells.
Cytotoxicity was quantified by the ability of effector cells to rupture the target cell membrane, which caused the release of 51Cr from labeled target cells to the medium. Two target cell lines, Yac-1 and P815, were labeled overnight with 2 μCi/mL Na251CrO4 (Amersham, Arlington Heights, IL), and a fixed number of the labeled target cells (8 × 104) were mixed with various numbers of effector cells in 0.6 mL coculture medium in a 48-well plate. After incubation for 4 to 18 hours at 37°C in 5% CO2, 0.3 mL each of the culture supernatant was removed for counting radioactivity with COBRA II gamma counter (Packard, Meriden, CT). For the 18-hour assay, the medium contained 50 ng/mL IL-2. Net release of radioactivity was divided by net total radioactivity in the target cells to obtain percentage specific killing. The former was determined by radioactivity in the medium subtracted with those caused by autonomous release from the target cells. The latter was obtained by radioactivity released from detergent-lysed target cells subtracted with the autonomous release.
Gene expression and chromosomal rearrangement in LAK cells and pre-B cells.
Total RNA was prepared from LAK cells, pre-B cells, 32Dcl3 cells, P815 cells, and OP9 cells according to Chirgwin et al.35 Oligo (dT)-primed reverse-transcription of the isolated RNAs was performed with Ready-To-Go T-prime reverse-transcription kit (Pharmacia, Piscataway, NJ) according to the manufacturer's recommendation. Balb/c mouse-derived spleen cDNA was purchased from Clontech (Palo Alto, CA). For LAK cells, expression of NKR-P1A (gene 2)36 NKR-P1B (gene 34),36 perforin-1,37 granzyme A,38 and granzyme B39,40 genes was measured by reverse transcriptase-polymerase chain reaction (RT-PCR). For EB34- and ES-pre-B cells, expression of the VpreB, Ig-β (B29), Rag-1 and Rag-2 genes, and the VhDJh-Cμ transcript was examined. The standard RT-PCR condition was 30-cycle amplification with incubation for 1 minute at 94°C, followed by 1-minute incubation at 60°C, and finally, by 1.75-minute incubation at 72°C per cycle. For the NKR-P1 family, perforin, the granzyme family, and VpreB, we used the nested PCR method to ensure the sequence specificity of the products. The sense-1 and antisense-1 primers were used for the initial amplification for 20 cycles, and the second amplification (20 cycles) was performed with the 1,000-fold diluted initial product, using the sense-2 and antisense-2 primers. Oligonucleotide primers for NK/CTL-specific genes synthesized are as follows (expected length of the individual PCR product is indicated in parentheses).
| NKR-P1A: (585 bp) | sense-1: 5′-CCC TGC TCA CCA GTT CAG-3′ antisense-1: 5′-CCA TAA CCC ACA TAG TTG CTC-3′ sense-2 and antisense-2 are according to Giorda and Trucco36 |
| NKR-P1B: (607 bp) | sense-1: 5′-CGG TGT CAA GTC CCT CCA TC-3′ antisense-1: 5′-TCT CTC CCC CAA ATC CCC TAA-3′ sense-2 and antisense-2 are according to Giorda and Trucco36 |
| Perforin-1: (609 bp) | sense-1 (= sense-2) and antisense-2 are as in Rolink et al41 antisense-1: 5′-CCT CTG TCG TGG GCA GCA GTC-3′ |
| Granzyme A: (438 bp) | sense-1: 5′-TTG ACT GCT GCC CAC TGT AAC-3′ sense-2: 5′-TTC ATT CTT GGG GCT CAC TCA-3′ antisense-1: 5′-ATC GGC GAT CTC CAC ACT TC-3′ antisense-2: 5′-GCC TCG CAA AAT ACC ATC ACA-3′ |
| Granzyme B: (344 bp) | sense-1: 5′-GAT ATG TGG GGG CTT CCT TA-3′ sense-2: 5′-GGC CCA CAA CAT CAA AGA A-3′ antisense-1: 5′-ACT TGG GTG CAA CTG TAT GAA-3′ antisense-2: 5′-CCG CAC ATA TCT GAT TGG TTT-3′ |
| β-Actin (336 bp) | sense-1 and antisense -2 are according to Rolink et al41 antisense-1 and sense-2 are as in Li et al.42 |
| NKR-P1A: (585 bp) | sense-1: 5′-CCC TGC TCA CCA GTT CAG-3′ antisense-1: 5′-CCA TAA CCC ACA TAG TTG CTC-3′ sense-2 and antisense-2 are according to Giorda and Trucco36 |
| NKR-P1B: (607 bp) | sense-1: 5′-CGG TGT CAA GTC CCT CCA TC-3′ antisense-1: 5′-TCT CTC CCC CAA ATC CCC TAA-3′ sense-2 and antisense-2 are according to Giorda and Trucco36 |
| Perforin-1: (609 bp) | sense-1 (= sense-2) and antisense-2 are as in Rolink et al41 antisense-1: 5′-CCT CTG TCG TGG GCA GCA GTC-3′ |
| Granzyme A: (438 bp) | sense-1: 5′-TTG ACT GCT GCC CAC TGT AAC-3′ sense-2: 5′-TTC ATT CTT GGG GCT CAC TCA-3′ antisense-1: 5′-ATC GGC GAT CTC CAC ACT TC-3′ antisense-2: 5′-GCC TCG CAA AAT ACC ATC ACA-3′ |
| Granzyme B: (344 bp) | sense-1: 5′-GAT ATG TGG GGG CTT CCT TA-3′ sense-2: 5′-GGC CCA CAA CAT CAA AGA A-3′ antisense-1: 5′-ACT TGG GTG CAA CTG TAT GAA-3′ antisense-2: 5′-CCG CAC ATA TCT GAT TGG TTT-3′ |
| β-Actin (336 bp) | sense-1 and antisense -2 are according to Rolink et al41 antisense-1 and sense-2 are as in Li et al.42 |
Primers for pre-B-cell–specific genes, VpreB (sense-1 and antisense-1), and Ig-β were according to Rolink et al,41and those for VpreB (sense-2 and antisense-2), Rag-1 and Rag-2 were based on Li et al.42 The VhDJh-Cμ transcript was detected by the nested RT-PCR method according to Nakano et al.17
Chromosomal DNAs from adherent as well as nonadherent pre-B cells, LAK cells, and OP9 cells were prepared by the standard SDS-protease K method.43 The DNA rearrangements at the immunoglobulin μ heavy chain locus (DJh and VhDJh) and at the κ light chain locus were, then, detected by PCR using the same primers (DSF, V7183 VQ52, Jh4, Vκ and Jκ2) described by Potocnik et al.16
RESULTS
Transient generation of CD34+ cell population is facilitated by exogenous VEGF.
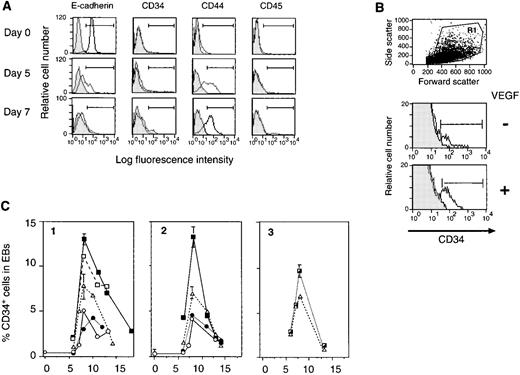
In an attempt to dissect the differentiation process of ES cells into lymphohematopoietic lineages, surface marker analysis was carried out by FACS phenotyping. Undifferentiated ES cells were CD34−CD44−, and expressed E-cadherin/uvomorulin.44,45 Upon differentiation, the E-cadherin expression diminished in five days, which was accompanied by a gain of CD44 expression (Fig 1A). The first sign of colony-forming cell (CFC) generation was observed at this time (data not shown). This finding is consistent with the fact that, during gastrulation, E-cadherin− mesodermal cells are differentiated from E-cadherin+ epiblasts.46 From day 7, some of the EB cells start expressing the hematopoietic stem/progenitor cell marker, CD34,47 followed by the appearance of leukocyte common antigen (CD45)+ cells (Fig1A). Because CD34+ cells from yolk sac contain erythromyeloid colony forming activity,48we further characterized these CD34+ progeny.
VEGF enhances transient appearance of CD34+cells during differentiation of ES cells in liquid culture. (A) Surface marker analysis of differentiating ES cells. Undifferentiated A3-1 ES cells (day 0), as well as cells from EBs cultured for 5 days and 7 days in the presence of 100 ng/mL SCF, 10 ng/mL IL-3, and 1 U/ml EPO were stained with anti-E-cadherin, CD34, CD44, and CD45 monoclonal antibodies. Results of the corresponding isotype control are shown in gray. Positive cell population (region indicated with ⊢⊣) on day 0, day 5, and day 7 for E-cadherin are 98.4%, 26.4%, and 0.3%, for CD34 are 0.2%, 1.4%, and 6.7%, for CD44 are 4.4%, 58.4%, and 68.7%, and for CD45 are 0.4%, 0.1%, and 1.0%, respectively. (B) Effect of VEGF on the CD34+ cell generation. The day 8 EBs derived from A3-1 ES cells cultured in the absence of cytokine (−), and in the presence of 20 ng/mL VEGF (+) were harvested and analyzed for CD34 expression. Staining results of the gate R1 (top dot plot) are shown in histograms. Background staining is indicated in gray. Positive cell population (⊢⊣) is 3.9% (−) and 8.5% (+). (C) Kinetics of CD34+ cell generation in EBs cultured in various growth factors. A3-1 ES cells (1) and J7 ES (2, 3) cells were induced to differentiate in liquid culture in the absence of cytokine (○), and in the presence of 100 ng/mL SCF (▵), 20 ng/mL VEGF (□), 100 ng/mL SCF, and 1 ng/mL TGF-β1 (•), 100 ng/mL SCF, 10 ng/mL IL-3, and 1 U/mL EPO (┌), and 100 ng/mL SCF and 20 ng/mL VEGF (▪). From day 6 to day 18, EBs were harvested and analyzed for CD34 expression. For A3-1, only day 6 and day 8 data were average values of duplicated samples, and for J7, all but “no cytokine” and “SCF + TGF-β1” were average values of duplicated samples. The vertical bar indicates standard deviation.
VEGF enhances transient appearance of CD34+cells during differentiation of ES cells in liquid culture. (A) Surface marker analysis of differentiating ES cells. Undifferentiated A3-1 ES cells (day 0), as well as cells from EBs cultured for 5 days and 7 days in the presence of 100 ng/mL SCF, 10 ng/mL IL-3, and 1 U/ml EPO were stained with anti-E-cadherin, CD34, CD44, and CD45 monoclonal antibodies. Results of the corresponding isotype control are shown in gray. Positive cell population (region indicated with ⊢⊣) on day 0, day 5, and day 7 for E-cadherin are 98.4%, 26.4%, and 0.3%, for CD34 are 0.2%, 1.4%, and 6.7%, for CD44 are 4.4%, 58.4%, and 68.7%, and for CD45 are 0.4%, 0.1%, and 1.0%, respectively. (B) Effect of VEGF on the CD34+ cell generation. The day 8 EBs derived from A3-1 ES cells cultured in the absence of cytokine (−), and in the presence of 20 ng/mL VEGF (+) were harvested and analyzed for CD34 expression. Staining results of the gate R1 (top dot plot) are shown in histograms. Background staining is indicated in gray. Positive cell population (⊢⊣) is 3.9% (−) and 8.5% (+). (C) Kinetics of CD34+ cell generation in EBs cultured in various growth factors. A3-1 ES cells (1) and J7 ES (2, 3) cells were induced to differentiate in liquid culture in the absence of cytokine (○), and in the presence of 100 ng/mL SCF (▵), 20 ng/mL VEGF (□), 100 ng/mL SCF, and 1 ng/mL TGF-β1 (•), 100 ng/mL SCF, 10 ng/mL IL-3, and 1 U/mL EPO (┌), and 100 ng/mL SCF and 20 ng/mL VEGF (▪). From day 6 to day 18, EBs were harvested and analyzed for CD34 expression. For A3-1, only day 6 and day 8 data were average values of duplicated samples, and for J7, all but “no cytokine” and “SCF + TGF-β1” were average values of duplicated samples. The vertical bar indicates standard deviation.
To optimize the level of CD34+ population, we searched extensively for protein factors to be included in the differentiation medium, focusing on those known to affect embryonic hematopoiesis in vivo such as SCF,49,50 VEGF,51-53 and TGF-β1.54 Although various hematopoietic cytokines used in the previous reports,4 5 including IL-3, IL-11, and EPO were also tested, VEGF gave the strongest stimulatory effect as a single factor on the enhancement of the CD34+ cell population in EBs (Fig 1B and C-1). SCF showed a weaker but significant effect, whereas TGF-β1 demonstrated only an antagonistic effect to SCF (Fig 1C). VEGF showed a strong additive effect to SCF (Fig 1C-1). The addition of IL-3 + EPO to SCF also exhibited some enhancement (Fig1C-3), albeit at lower levels than that achieved with VEGF (Fig 1C-2). Thus, SCF + VEGF was the optimal factor combination for generating CD34+ population in EBs. Neither SCF + VEGF nor SCF + IL-3 + EPO changed the transient nature of the appearance of CD34+ EB cells. Commonly, they peaked around day 8 (J7 ES cells) to day 9 (A3-1 ES cells) and disappeared by day 14, suggesting that these cytokines were unable to prolong the viability of, or block differentiation of the CD34+ EB cells.
Erythro-myeloid CFC potential in the CD34+ EB cell population.
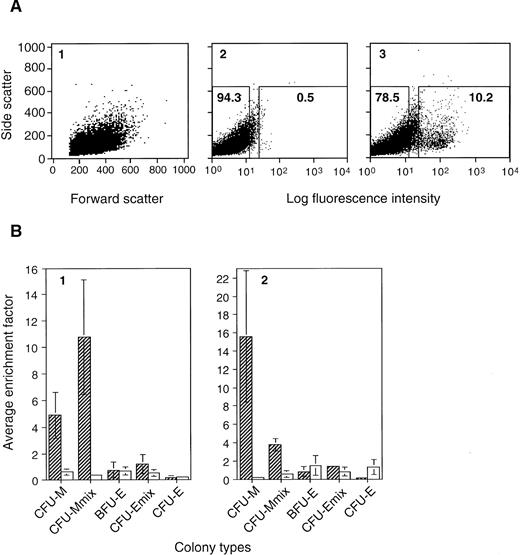
Hematopoietic progenitor cell activity within the CD34+ EB cell fraction was analyzed. We used two combinations of factors, SCF + IL-3 + EPO and SCF + VEGF, for differentiating ES cells, both of which allowed significant levels of CD34+ cells to be generated (Fig 1C). CD34+ EB cells were sorted from day 7 EBs (Fig2A), and subjected to CFC analysis. The isolated CD34+ cells contained erythro-myeloid CFCs but were not necessarily enriched for early (bipotential or multipotential) progenitor cell types (Fig 2B). Rather, they were preferentially enriched for myeloid CFCs (CFU-M, CFU-mast [data not shown], and bipotential CFU-Mmix), and not significantly for erythroid CFCs (CFU-E, BFU-E, and bipotential, as well as tripotential CFU-Emix). The total myeloid CFCs represented approximately 0.6% to 0.7% of total EB cells (740 ± 459 [standard deviation] CFCs/105SCF + IL-3 + EPO EB cells, and 653 ± 308 CFCs/105SCF + VEGF EB cells). Interestingly, the occurrence of myeloid CFCs in the CD34+ cell population was consistently higher when EBs were formed under SCF + VEGF (approximately 7%, 7,361 ± 1,563 CFCs/105 CD34+ cells) than when cultured in SCF + IL-3 + EPO (approximately 3%, 2,692 ± 648 CFCs/105CD34+ cells). Nevertheless, considering the population of CD34+ cells to be approximately 7% to 10% of total EB cells (Figs 1C and 2A), most of the myelomonocytic progenitors seemed to be CD34+, especially when developed in SCF + VEGF, and cells with erythroid potential were probably heterogeneous for the CD34 expression.
Types of colony-forming cells enriched in the CD34+ EB cell fraction. A3-1 ES cells were induced to differentiate in methylcellulose culture in the presence of 100 ng/mL SCF, and 20 ng/mL VEGF (A, B2), or 100 ng/mL SCF, 10 ng/mL IL-3, and 1 U/mL EPO (B1). EBs were obtained on day 7, stained with RAM34, and subjected to cell isolation by FACS. (A) One of the staining results of SCF + VEGF EB cells is shown: (1) the scatter pattern of the RAM34 stained sample, (2) staining pattern of isotype control, and (3) that of RAM34, in dot plots. Regions for sorting CD34+ (right) and CD34− (left) cell populations are indicated in boxes (2 and 3). Number in each box indicates percentage of total EB cells. Similar scatter pattern and staining pattern were obtained from EBs developed with SCF + IL-3 + EPO. (B) Erythro-myeloid CFC in the CD34+ and CD34− cell populations. Average numbers of CFU-M, CFU-Mmix, BFU-E, CFU-Emix, and CFU-E in 105 presorted EB cells, 105 sorted CD34+ EB cells, and 105 sorted CD34− EB cells were determined. The average colony numbers from CD34+ cells and CD34− cells were divided by corresponding numbers from presorted EBs to obtain enrichment factors for individual CFCs. Such enrichment factors obtained from three (SCF + VEGF) to four (SCF + IL-3 + EPO) independent experiments were then averaged to obtain average enrichment factor. Hatched bar indicates results for CD34+ EB cells, and open bar is for CD34− EB cells. Factor 1 means no enrichment. Vertical line indicates standard deviation.
Types of colony-forming cells enriched in the CD34+ EB cell fraction. A3-1 ES cells were induced to differentiate in methylcellulose culture in the presence of 100 ng/mL SCF, and 20 ng/mL VEGF (A, B2), or 100 ng/mL SCF, 10 ng/mL IL-3, and 1 U/mL EPO (B1). EBs were obtained on day 7, stained with RAM34, and subjected to cell isolation by FACS. (A) One of the staining results of SCF + VEGF EB cells is shown: (1) the scatter pattern of the RAM34 stained sample, (2) staining pattern of isotype control, and (3) that of RAM34, in dot plots. Regions for sorting CD34+ (right) and CD34− (left) cell populations are indicated in boxes (2 and 3). Number in each box indicates percentage of total EB cells. Similar scatter pattern and staining pattern were obtained from EBs developed with SCF + IL-3 + EPO. (B) Erythro-myeloid CFC in the CD34+ and CD34− cell populations. Average numbers of CFU-M, CFU-Mmix, BFU-E, CFU-Emix, and CFU-E in 105 presorted EB cells, 105 sorted CD34+ EB cells, and 105 sorted CD34− EB cells were determined. The average colony numbers from CD34+ cells and CD34− cells were divided by corresponding numbers from presorted EBs to obtain enrichment factors for individual CFCs. Such enrichment factors obtained from three (SCF + VEGF) to four (SCF + IL-3 + EPO) independent experiments were then averaged to obtain average enrichment factor. Hatched bar indicates results for CD34+ EB cells, and open bar is for CD34− EB cells. Factor 1 means no enrichment. Vertical line indicates standard deviation.
Generation of two different B220+ lymphocytes from CD34+ EB cells in vitro.
Since the ability of ES cells to differentiate into B- and T-lymphocyte progenitors in vitro had been demonstrated,14,16,17 we examined lymphoid potential of the CD34+ cell population. However, the sorted CD34+ cells obtained from day 6 and day 7 EBs in the same way as shown in Fig 2A did not give rise to any pre-B colonies in the presence of IL7 and SCF,55 nor in the presence of SCF, IL-7, and IL-11.56 Therefore, we examined a coculture method similar to what was previously described by Nakano et al,17 except that two cytokines that affect B lymphopoiesis in vivo, IL-257 and IL-7,58 were added. First, ES cells were cultured directly on OP9 in the presence of 50 ng/mL IL-2 and 5 ng/mL IL-7. In 2 to 3 weeks, B220+cells and CD43+ cells emerged, suggesting the successful generation of lymphoid cells. Removal of IL-7 leaving IL-2 in culture led to accumulation of B220+ CD43− large granular lymphocytes (LGLs) (LAKa and LAKj, Table 1), morphologically similar to LAK cells from spleen (spleen-LAK, Table 1). Interestingly, these LGL showed IL-2–dependent cytotoxicity to OP9 stroma cells. By contrast, removal of IL-2 enriched for B220+CD43+ BP-1− HSA+CD19+ small round pre-B–like cells (ES-pre B, Table 1). Cells of the same surface phenotype were also obtained when bone marrow cells were cultured for 2 to 3 weeks on OP9 with IL-7 (BM-pre-B, Table1).
Next, the sorted CD34+ cells were cultured on the OP9 stroma cells with IL-2 and IL-7. A3-1 ES cells were primarily used because the pre-B-cell potential was reproducibly higher than J7, and the SCF + VEGF condition was employed because IL-3 might be inhibitory for the pre–B-cell generation.59 Two types of B220+ cells emerged from the CD34+ cell fraction in approximately 2 weeks of coculture (Table2). When IL-7 was removed on day 7, B220+ CD43− cells with LGL morphology and cytotoxic characteristics to OP9 cells were expanded. We isolated such LAK cells from three independent experiments and designated them LAK34a-1, LAK34a-2, and LAK34a-3 (Table 1). The LAK34a-1 and LAK34a-3 were large LAK cells, morphologically similar to adherent LAK cells derived directly from ES cells and from spleen (Fig3A). However, LAK34a-2 was a relatively small cell with typical LGL morphology (Fig 3A). The former group of CD34+ EB cell–derived and ES cell–derived LAK cells was designated collectively as type I LAK cells, and the latter type II LAK cells (Table 1). Removal of IL-2 on day 7 of coculture generated B220+ CD43+ B lymphocyte like cells. These cells, designated EB34-pre-B, were morphologically similar to ES-pre B as well as BM–pre-B cells (Table 1; Fig 3A and C). Table 2 summarizes the results of these coculture experiments. From all the experiments, either or both LAK cells and pre-B cells were developed from 104 to 105 CD34+ EB cells. However, no such lymphocytes were generated from the same numbers of CD34− EB cells. Only in one case (experiment 4), 106 CD34− EB cells gave rise to type I-like LAK cells. Thus, progenitor cells for both pre-B cells and LAK cells were enriched in the CD34+ EB cell fraction.
LAK Cells and Pre-B Cells Were Generated Specifically From CD34+ EB Cells
| Plated Cell No./Well . | Experiment 1* . | Experiment 2† . | Experiment 3† . | Experiment 4† . | |||
|---|---|---|---|---|---|---|---|
| IL-2 + IL-7 . | IL-2 . | IL-7 . | IL-2 . | IL-7 . | IL-2 . | IL-7 . | |
| CD34+ | |||||||
| 105 | LAK‡ + 1-153/(pre-B+)1-155# | LAK++ | HPC¶ | HPC | pre-B1-160+++ | LAK1-164++ | HPC++ |
| 104 | ND | LAK1-161++ | HPC | —1-162 | (pre-B+) | (LAK+) | HPC |
| 103 | ND | — | — | — | — | — | — |
| CD34− | |||||||
| 106 | ND | —# | —# | —# | —# | (LAK+)# | HPC |
| 105 | — | — | — | HPC# | — | — | HPC |
| 104 | ND | — | — | — | — | — | — |
| Plated Cell No./Well . | Experiment 1* . | Experiment 2† . | Experiment 3† . | Experiment 4† . | |||
|---|---|---|---|---|---|---|---|
| IL-2 + IL-7 . | IL-2 . | IL-7 . | IL-2 . | IL-7 . | IL-2 . | IL-7 . | |
| CD34+ | |||||||
| 105 | LAK‡ + 1-153/(pre-B+)1-155# | LAK++ | HPC¶ | HPC | pre-B1-160+++ | LAK1-164++ | HPC++ |
| 104 | ND | LAK1-161++ | HPC | —1-162 | (pre-B+) | (LAK+) | HPC |
| 103 | ND | — | — | — | — | — | — |
| CD34− | |||||||
| 106 | ND | —# | —# | —# | —# | (LAK+)# | HPC |
| 105 | — | — | — | HPC# | — | — | HPC |
| 104 | ND | — | — | — | — | — | — |
Different numbers of sorted CD34+ EB cells and CD34− EB cells were cultured on a confluent layer of OP9 cells established in a six-well plate. Hematopoietic progenitor cells (HPCs) proliferating under the OP9 stroma cells were monitored. When the culture reached near-saturation, cells were harvested and surface expression of B220, CD19, and CD43 was analyzed by FACScan. B220+ CD19− CD43− cytotoxic cells were designated as LAK; B220+ CD19+CD43+ small noncytotoxic cells as pre-B; and noncytotoxic cells that ceased proliferating before analysis as HPC.
Abbreviation: ND, not determined.
Continuously cultured in IL-2 (50 ng/mL) and IL-7 (5 ng/mL).
As described under Experimental Procedures, on day 7, added cytokine was changed to either IL-2 or IL-7.
Type I LAK cells (LAK34a-1) were derived from this culture.
Relative frequency of LAK, pre-B, and HPC is shown based on culture period required for the cells to proliferate to a near-saturation level. +++: the culture was saturated, and transferred to a T25 flask within 2 weeks, ++: in 3 weeks, and + in 4 weeks or longer.
LAK or pre-B in parentheses indicates that either the culture was lost mainly as a result of ES-like cell out growth or the cell failed to expand after the first round of surface marker analysis.
#ES-like cells outgrew.
¶HPC that initially grew under the OP9 layer and ceased proliferating before reaching to a near saturation level are indicated as HPC without +, ++, or +++.
EB34-pre-B cells were derived from this culture.
Type I LAK cells (LAK34a-3) were derived from this culture.
Type II LAK cells (LAK34a-2) were derived from this culture.
No sign of hematopoietic cell proliferation.
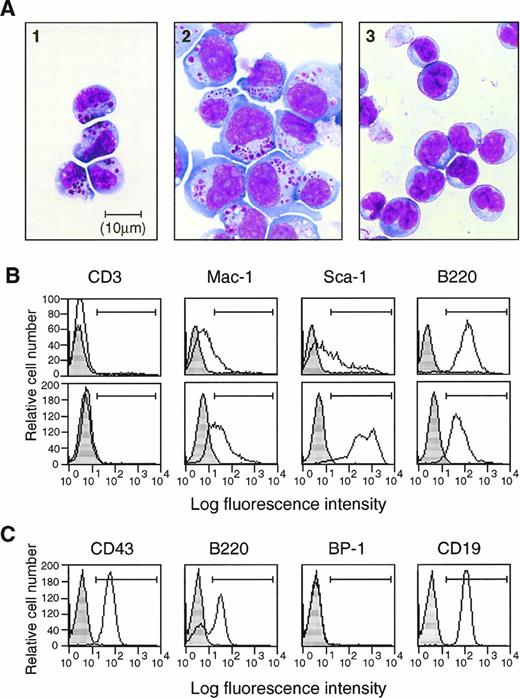
Two different B220+ lymphocytes derived from the CD34+ EB cell fraction. (A) Morphological characterization by Wright-Giemsa staining (Objective ×100 Oil). Type II LAK (LAK34a-2) cells (left), type I LAK (LAK34a-3) cells (center), and EB34-pre-B cells (right) were spun on slides and stained with Wright-Giemsa. Large granules are present in both small LAK34a-2 cells and larger LAK34a-3 cells, but not in EB34-pre-B cells. (B) Phenotypic analysis of CD34+ EB cell– derived LAK cells. Type II LAK cells (LAK34a-2, upper panels) and type I LAK cells (LAK34a-3, lower panels) were stained with PE-conjugated anti-CD3, Mac-1, Sca-1, and B220 monoclonal antibodies. Positive cells for CD3, Mac-1, Sca-1, and B220 (region ⊢⊣) are 0.1%, 23.0%, 40.5%, and 99.1%, respectively, in LAK34a-2, and 0.2%, 62.9%, 98.9%, and 98.5%, respectively, in LAK34a-3. Note the difference in the Sca-1 pattern between LAK34a-2 and LAK34a-3. Results of the corresponding isotype control are indicated in gray. (C) Phenotypic analysis of EB34-pre-B cells. Adherent EB34-pre-B cells were collected from the OP9 stroma layer, and stained with PE-conjugated anti-CD43, B220, BP-1, and CD19 monoclonal antibodies. Positive cells for CD43, B220, BP-1, and CD19 (region ⊢⊣) are 99.8%, 66.5%, 0%, and 99.5%, respectively. Background staining is indicated in gray.
Two different B220+ lymphocytes derived from the CD34+ EB cell fraction. (A) Morphological characterization by Wright-Giemsa staining (Objective ×100 Oil). Type II LAK (LAK34a-2) cells (left), type I LAK (LAK34a-3) cells (center), and EB34-pre-B cells (right) were spun on slides and stained with Wright-Giemsa. Large granules are present in both small LAK34a-2 cells and larger LAK34a-3 cells, but not in EB34-pre-B cells. (B) Phenotypic analysis of CD34+ EB cell– derived LAK cells. Type II LAK cells (LAK34a-2, upper panels) and type I LAK cells (LAK34a-3, lower panels) were stained with PE-conjugated anti-CD3, Mac-1, Sca-1, and B220 monoclonal antibodies. Positive cells for CD3, Mac-1, Sca-1, and B220 (region ⊢⊣) are 0.1%, 23.0%, 40.5%, and 99.1%, respectively, in LAK34a-2, and 0.2%, 62.9%, 98.9%, and 98.5%, respectively, in LAK34a-3. Note the difference in the Sca-1 pattern between LAK34a-2 and LAK34a-3. Results of the corresponding isotype control are indicated in gray. (C) Phenotypic analysis of EB34-pre-B cells. Adherent EB34-pre-B cells were collected from the OP9 stroma layer, and stained with PE-conjugated anti-CD43, B220, BP-1, and CD19 monoclonal antibodies. Positive cells for CD43, B220, BP-1, and CD19 (region ⊢⊣) are 99.8%, 66.5%, 0%, and 99.5%, respectively. Background staining is indicated in gray.
B220+ CD43− LGL from CD34+ EB cells contains LAK activity with target specificity of NK cells.
NK1.1+ spleen LAK cells and NK progenitor cells in bone marrow are known to be B220+.41,60 Therefore, we sought to determine whether the CD34+ EB cell-derived B220+ LAK cells could be categorized in the NK cell lineage. C57BL/6 mouse NK cells are surface CD3−NK1.1+ LGLs.61 NK1.1 has been molecularly defined as a member of the activating NK-receptor family, NKR-P1.62 Because the NK1.1 marker is not available for NK cells derived from the 129Sv mouse line,63 from which both ES cells are originated, we tested another pan-NK cell marker, DX5.64 All the CD34+ EB cell-derived and ES cell-derived LAK cells were B220+ CD3−CD8− CD19− CD34−CD43− cells, suggesting that they were neither typical B cells nor T cells (Fig 3B). However, expression of DX5 was not detected. This may be attributable to the long-term culture of the LAK cells in IL-2, because even spleen-LAK cells, initially containing DX5+ cells, lost expression after a few weeks of culture in IL-2 (data not shown). Weak expression of Mac-1 was found on type I LAK cells and, in even lower levels, on type II LAK cells (Fig 3B). Interestingly, Sca-1 differentiated the two types of LAK cells. It was highly expressed on type I cells, whereas most type II cells expressed at very low levels. Therefore, type I LAK cells are B220+CD3− CD8− CD19−CD34− CD43− Mac-1loSca-1+, and type II LAK cells are B220+CD3− CD8− CD19−CD34− CD43− Mac-1lo and Sca-1lo.
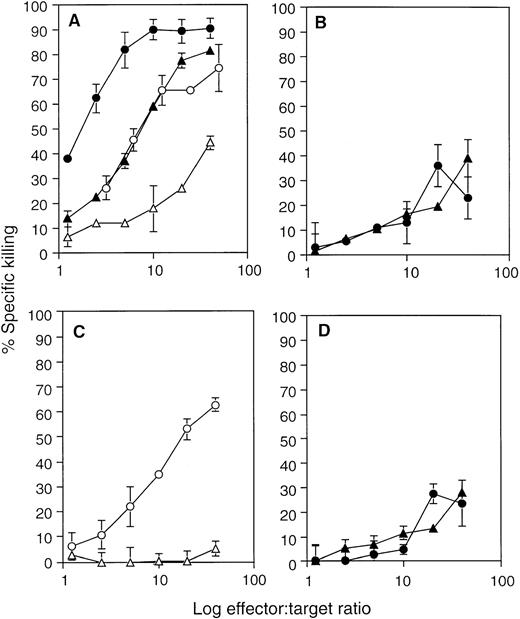
Cytotoxicity of the LAK cells was quantified by 51Cr release from two different target cells: NK-sensitive Yac-1 lymphoma and NK-resistant p815 mastocytoma cell lines. Primary LAK cells from spleen showed strong killing activity to both target cells in 4 hours (Fig 4A). However, type I LAK cells required a much longer period (18 hours) to show appreciable cytotoxicity, and they showed a similar degree of cytotoxicity to both Yac-1 and P815 cells (Fig 4B and D). By contrast, type II cells, LAK34a-2, exhibited strong cytotoxicity on Yac-1 cells, comparable to spleen-LAK cells (Fig 4A and C) and did not kill NK-resistant P815 cells in the standard 4-hour assay. Thus, CD34+ EB cell-derived as well as ES cell-derived LAK cells were cytotoxic, of which type II LAK cells showed target specificity equivalent to that of NK cells.
Cytotoxicity of the CD34+ EB cell–derived LAK cells to NK-sensitive Yac-1 cells and NK-resistant P815 cells. The cytotoxicity of spleen LAK (A), type I LAK (LAK34a-1) (B), LAK34a-3 (D), and type II LAK (LAK34a-2) (C) effector cells was compared by the51Cr release assay. A fixed number of51Cr-labeled target cells, Yac-1 (○, •) and P815 (▵, ▴), were mixed with different numbers of effector cells, and incubated for 4 hours (○, ▵), or 18 hours (•, ▴). Averaged values of percent specific killing are shown with the corresponding standard deviation (vertical line).
Cytotoxicity of the CD34+ EB cell–derived LAK cells to NK-sensitive Yac-1 cells and NK-resistant P815 cells. The cytotoxicity of spleen LAK (A), type I LAK (LAK34a-1) (B), LAK34a-3 (D), and type II LAK (LAK34a-2) (C) effector cells was compared by the51Cr release assay. A fixed number of51Cr-labeled target cells, Yac-1 (○, •) and P815 (▵, ▴), were mixed with different numbers of effector cells, and incubated for 4 hours (○, ▵), or 18 hours (•, ▴). Averaged values of percent specific killing are shown with the corresponding standard deviation (vertical line).
Effects of IL-2 and IL-12 on the CD34+ EB cell-derived LAK cells.
Both CD34+ EB cell-derived and ES cell-derived LAK cells were able to be maintained in a stroma-free condition in the presence of 50 ng/mL IL-2. However, quantitative survival/proliferation assay showed that type I LAK cells strongly responded to IL-2, whereas type II LAK cells showed only a weak response to IL-2 (Fig5A). IL-12 and IL-15 stimulate the generation of LAK cells and cytotoxic T lymphocytes (CTLs), as well as the proliferation of NK cells.65-69 IL-15, which shares its receptor subunits with the IL-2 receptor,69-71 showed a weak effect on both types of LAK cells in a high concentration range (50 to 250 ng/mL). In addition, IL-12, which acts as a co-stimulator with IL-2 and IL-15,65-68 synergized with a low concentration of IL-2 (3 ng/mL) to stimulate survival/proliferation of type II LAK cells (Fig 5B). To a lesser degree, SCF also showed a synergistic effect. However, IL-12 exhibited no synergy with IL-2 or IL-15 in type I LAK cells (data not shown). Therefore, type II LAK cells had cytokine dependency equivalent to the NK/CTL cell-types. Individually added IL-3, CSF-GM, CSF-M, IL-4, IL-7, IL-12, or SCF had no appreciable effects on both types of LAK cells.
Sensitivity of the CD34+ EB cell–derived LAK cells to various cytokines. (A) Difference in response to IL-2. Type I LAK cells (LAK34a-1; □, ▪, ✺; and LAK34-a-3: ▵, ▴, ✺), and type II LAK cells (LAK34a-2: ○, •, ✺) were cultured at 5,000 cell/well/0.1 mL coculture medium for 48 hours in the presence of different concentration of IL-2 (□, ○, ▵), IL-12 (✺), IL-15 (▪, •, ▴), and SCF (✺). The 1 (100) × dilution corresponds to 0.5 μg/mL of added cytokine except that, in the case of SCF, 1× dilution is 10 μg/mL. The cell viability was measured with alamar blue dye. Note that singly added IL-12 and SCF do not show any signal. (B) Effect of IL-12 and SCF on the IL-2–activated type II LAK cells. LAK34a-2 cells were cultured in a fixed concentration of IL-2 (3 ng/mL) (□), with various concentrations of IL-12 (0.5 μg/mL at 1× dilution) (○), and SCF (10 μg/mL at 1× dilution) (▵). The cell viability was measured with alamar blue dye.
Sensitivity of the CD34+ EB cell–derived LAK cells to various cytokines. (A) Difference in response to IL-2. Type I LAK cells (LAK34a-1; □, ▪, ✺; and LAK34-a-3: ▵, ▴, ✺), and type II LAK cells (LAK34a-2: ○, •, ✺) were cultured at 5,000 cell/well/0.1 mL coculture medium for 48 hours in the presence of different concentration of IL-2 (□, ○, ▵), IL-12 (✺), IL-15 (▪, •, ▴), and SCF (✺). The 1 (100) × dilution corresponds to 0.5 μg/mL of added cytokine except that, in the case of SCF, 1× dilution is 10 μg/mL. The cell viability was measured with alamar blue dye. Note that singly added IL-12 and SCF do not show any signal. (B) Effect of IL-12 and SCF on the IL-2–activated type II LAK cells. LAK34a-2 cells were cultured in a fixed concentration of IL-2 (3 ng/mL) (□), with various concentrations of IL-12 (0.5 μg/mL at 1× dilution) (○), and SCF (10 μg/mL at 1× dilution) (▵). The cell viability was measured with alamar blue dye.
Thus, type I LAK cells were IL-2–dependent B220+CD3− Sca-1+ cells with weak nonspecific cytotoxicity, and type II LAK cells were IL-2 + IL-12–dependent B220+ CD3− Sca-1lo cells with strong cytotoxicity to NK-sensitive Yac-1 cells. These observations provide additional support to the similarity of type II LAK cells to IL-2–activated NK cells.
The CD34+ EB cell-derived LAK cells express NK-specific genes.
The mouse NK cell can be molecularly defined as a CD3−NKR-P1+ lymphocyte.72 The activating NK receptor (NKR-P1) family is composed of NKR-P1A, NKR-P1B, and NKR-P1C (NK1.1) in mice and is the specific marker for NK cells.36,62,73 Furthermore, upon activation by IL-2, NK cells increase the expression level of a set of proteins essential for eliciting cytotoxicity. These include pore-forming protein, perforin, and a serine protease family, granzyme, which are also specific markers for CTLs and NK cells.74,75 We first determined if the NK receptor gene was expressed in the CD34+ EB cell-derived and ES cell–derived LAK cells by RT-PCR. Since the NK1.1 (NKR-P1C) gene is known to be highly polymorphic,63 and sequence information of 129Sv mouse–derived NK1.1 cDNA was unavailable, we examined expression of the NKR-P1A and NKR-P1B genes. As shown in Fig6, mRNA for NKR-P1A was detected specifically in all the LAK cells we tested, and NKR-P1B mRNA expression was found in spleen-LAK and type II LAK cells.
Expression of NK cell-specific mRNAs in the CD34+ EB cell–derived and ES cell–derived LAK cells. RT-PCR analysis was performed with total RNAs extracted from the C57BL/6 mouse spleen-LAK cells, type I LAK (LAKa, LAKj, LAK34a-1, and LAK34a-3) cells, type II LAK (LAK34a-2) cells, ES-pre-B cells, EB34-pre-B cells, P815 cells, 32Dcl3 cells, and OP9 cells, and analyzed on 1.2% agarose gels. The first lane is for size standard, and the rightmost lane is a negative control for cDNA. Results of the 2× 20-cycle amplification with different set of primers for NKR-P1A (top), NKR-P1B, granzyme A (gzm A), granzyme B (gzm B), perforin, and β-actin (bottom) genes are shown.
Expression of NK cell-specific mRNAs in the CD34+ EB cell–derived and ES cell–derived LAK cells. RT-PCR analysis was performed with total RNAs extracted from the C57BL/6 mouse spleen-LAK cells, type I LAK (LAKa, LAKj, LAK34a-1, and LAK34a-3) cells, type II LAK (LAK34a-2) cells, ES-pre-B cells, EB34-pre-B cells, P815 cells, 32Dcl3 cells, and OP9 cells, and analyzed on 1.2% agarose gels. The first lane is for size standard, and the rightmost lane is a negative control for cDNA. Results of the 2× 20-cycle amplification with different set of primers for NKR-P1A (top), NKR-P1B, granzyme A (gzm A), granzyme B (gzm B), perforin, and β-actin (bottom) genes are shown.
Next, expression of perforin, granzyme A, and granzyme B mRNAs was examined. These mRNAs were expressed in all the LAK cells we tested (Fig 6). Among non-LAK cells, 32Dcl3 myeloid cells expressed granzyme B mRNA, and P815 mastocytoma expressed both perforin and granzyme B mRNA as described.76 77 However, OP9 cells, and ES– as well as EB34–pre-B cells did not show any sign of expression of these genes. Thus, LAK cells generated from CD34+ EB cells and ES cells seemed to express the perforin/granzyme system for exerting cytotoxicity similar to CTL and NK cells.
In summary, the IL-2–dependent B220+ CD3−Sca-1+ type I LAK cells were NKR-P1A+perforin+ granzyme A/B+, and IL-2 + IL-12–dependent B220+ CD3−Sca-1lo type II LAK cells were NKR-P1A/B+perforin+ granzyme A/B+. These results suggest that CD34+ EB cell-derived as well as ES cell–derived LAK cells are likely to be IL-2–activated NK cells.
B220+ CD43+ cells derived from CD34+ EB cells show characteristics of pre-B cells.
Although EB34– as well as ES–pre-B cells responded to IL-7, maintenance of these cells was strictly dependent on both OP9 and IL-7 analogous to the PA6 + IL-7 pre-B cells described by Hayashi et al.78 Pre-B cells developed directly from ES cells have been described.14,16 17 To confirm that the CD34+ EB-derived pre-B-like cells were equivalent to BM-pre-B cells as well as the previously described ES cell-derived pre-B cells, we investigated expression of genes whose products are involved in pre-B-cell development (Table3). RT-PCR analysis clearly showed that mRNAs coding for the pre-B-cell receptor proteins such as VpreB, and Ig-β, and those encoding proteins for the immunogloblin gene rearrangement, Rag-1 and Rag-2, were expressed in these cells, but not in LAK cells nor in other non-B cells tested. Only spleen LAK cells showed some signal for Ig-β expression, probably because of contamination of spleen B cells. The adherent EB34-pre-B cells isolated from the OP9 stromal layer expressed somewhat lower levels of Rag-1 and Rag-2 than those expressed by the nonadherent population.
RT-PCR Analysis of Gene Expressions in the Pre-B Cells Derived from CD34+ EB Cells
| Primers . | cDNA . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Spleen* . | Pre-B Cells . | LAK Cells . | Controls . | ||||||||
| ES . | ad† EB34 . | Non-ad‡ EB34 . | Spleen2-153 . | 34a-1 . | 34a-2 . | 34a-3 . | p815 . | 32Dcl3 . | OP9 . | ||
| VpreB | −2-155 | + | + | + | − | − | − | − | − | − | − |
| Ig-β | + | + | + | + | +/− | − | − | − | − | +/− | − |
| VHDJH-Cμ | + | + | +/− | + | − | − | − | − | − | − | − |
| Rag-1 | − | + | +/− | + | − | − | − | − | − | − | − |
| Rag-2 | − | + | +/− | + | − | − | − | − | − | − | − |
| β-Actin | + | + | + | + | + | + | + | + | + | + | + |
| Primers . | cDNA . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Spleen* . | Pre-B Cells . | LAK Cells . | Controls . | ||||||||
| ES . | ad† EB34 . | Non-ad‡ EB34 . | Spleen2-153 . | 34a-1 . | 34a-2 . | 34a-3 . | p815 . | 32Dcl3 . | OP9 . | ||
| VpreB | −2-155 | + | + | + | − | − | − | − | − | − | − |
| Ig-β | + | + | + | + | +/− | − | − | − | − | +/− | − |
| VHDJH-Cμ | + | + | +/− | + | − | − | − | − | − | − | − |
| Rag-1 | − | + | +/− | + | − | − | − | − | − | − | − |
| Rag-2 | − | + | +/− | + | − | − | − | − | − | − | − |
| β-Actin | + | + | + | + | + | + | + | + | + | + | + |
*Balb/c mouse-derived spleen cells.
Adherent cells, cells grown under the OP9 stroma layer.
Nonadherent cells, cells grown in medium and cells loosely attached to the surface of OP9 cells.
C57BL/6 mouse-derived spleen LAK cells.
Expression levels are semiquantitatively displayed relative to the ES-pre-B signals as follows: +, high level; +/−, low level; −, no expression.
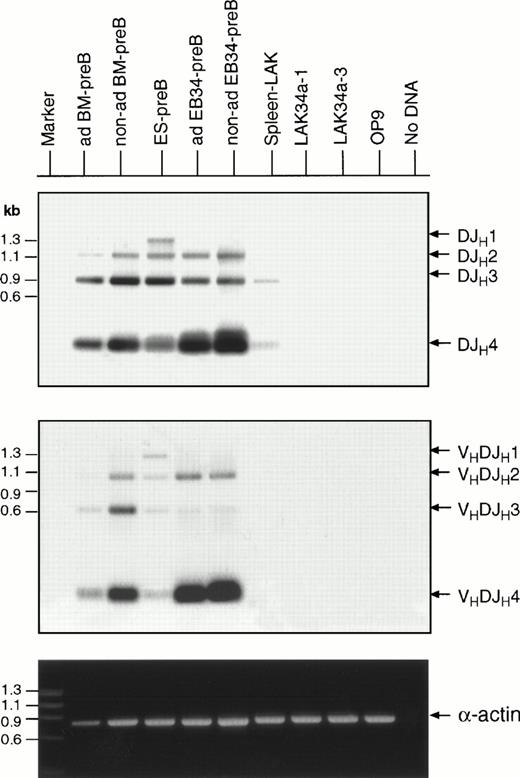
Next, chromosomal rearrangement at the immunoglobulin heavy chain locus was examined by PCR (Fig 7). Amplification with Dsf and Jh4 primers generated products indicating that, as in BM–pre-B cells, the four possible DJh recombinations, DJh1, DJh2, DJh3, and DJh4, took place in both EB34– and ES–pre-B cells (Fig 7 top). The spleen LAK cells showed again a sign of B cell contamination. The result with V7183 and Jh4 primers indicated that some of the EB34– as well as ES–pre-B cells underwent VhDJh recombination as well (Fig 7 middle). PCR with Vq52 and Jh4 primers gave a similar result (data not shown). Non-B cells such as type I LAK, type II LAK, and OP9 cells did not give any signal for the DJh and VhDJh recombination. The VhDJh-Cμ transcript was also detected specifically in ES– and EB34–pre-B cells, albeit at lower levels in the adherent population than in the nonadherent population (Table 3). By contrast, all LAK cells and OP9 stroma cells were negative. Chromosomal rearrangement at the light chain locus was also noted using Vκ and Jκ2 primers in BM–pre-B, ES–pre-B, and nonadherent EB34–pre-B cells (data not shown).
VhDJh as well as DJh rearrangements in the Hμ-locus in the EB34-pre-B and ES-pre-B cells. Large molecular weight DNAs from adherent (ad) BM-pre B cells, nonadherent (non-ad) BM-pre-B cells, ES-pre-B cells, adherent EB34-pre-B cells, nonadherent EB34-pre-B cells, LAK34a-1 cells, LAK34a-3 cells, and OP9 cells were subjected to PCR analysis using the DSF sense primer and the Jh4 antisense primer for DJh recombinations (top), and the V7183 sense primer and the Jh4 primer for detecting VhDJh recombinations (middle), and the α-actin primers (bottom), and separated on 1.2% agarose gels. Specificity of the DJh as well as VhDJh products were further ensured by Southern blotting, followed by visualization with the Jh probe.16
VhDJh as well as DJh rearrangements in the Hμ-locus in the EB34-pre-B and ES-pre-B cells. Large molecular weight DNAs from adherent (ad) BM-pre B cells, nonadherent (non-ad) BM-pre-B cells, ES-pre-B cells, adherent EB34-pre-B cells, nonadherent EB34-pre-B cells, LAK34a-1 cells, LAK34a-3 cells, and OP9 cells were subjected to PCR analysis using the DSF sense primer and the Jh4 antisense primer for DJh recombinations (top), and the V7183 sense primer and the Jh4 primer for detecting VhDJh recombinations (middle), and the α-actin primers (bottom), and separated on 1.2% agarose gels. Specificity of the DJh as well as VhDJh products were further ensured by Southern blotting, followed by visualization with the Jh probe.16
These observations suggest that the pre-B-like cells derived from CD34+ EB cells on OP9 correspond to pre-B cells from bone marrow, and thus, are similar to ES cell-derived pre-B cells described previously.16 17 We have therefore concluded that the CD34+ cells from EBs formed in the presence of VEGF contain B-lymphoid potential.
DISCUSSION
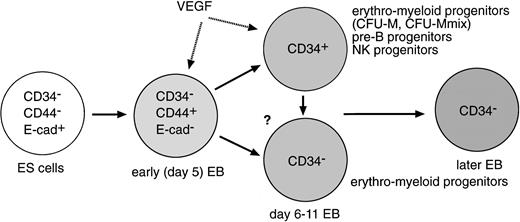
We have shown that the CD34+ cell population appears during the in vitro differentiation of ES cells, which becomes readily detectable in the presence of VEGF, and that ES cells have the potential to generate in vitro the third lymphoid lineage, NK cells, through the CD34+ progeny. Furthermore, the CD34+ cell fraction appears to represent the B-lymphoid potential and part of the erythro-myeloid cell potential of ES cells as well (Fig 8). Therefore, the CD34+ cell appears to be a critical developmental intermediate between ES cells and the lymphohematopoietic lineages.
Intermediate cell types from ES cells to hematopoietic progenitor cells.
The dramatic effect of VEGF on the elevation of the CD34+cell population in EBs was curious because VEGF is known to be a specific mitogen for vascular endothelial cells.79,80However, one of the VEGF receptors (VEGFR), flk-1, was originally isolated from fetal liver HSC fraction,81 and phenotypes of mutant mice that lack either the VEGF gene or the flk-1 gene indicate that VEGF signaling is essential for both embryonic hematopoiesis and vasculogenesis.51-53 Therefore, it is conceivable that the VEGF effect on the CD34+ EB cell generation may be a reflection of the in vivo role of VEGF on embryonic hematopoiesis.
Direct target(s) of VEGF in EBs remain(s) unclear. It is possible that proliferation of CD34+ lymphohematopoietic progenitor cells in EBs is stimulated by VEGF (Fig 8). It is equally possible that VEGF has a stimulatory effect on nonhematopoietic CD34+ cells such as VEGFR+ endothelial cells because endothelial cell development and vasculogenesis in EBs have already been demonstrated.82 In addition, VEGF may affect cells developed earlier than the CD34+ cells (Fig 8). Thus, detection and characterization of VEGFR+ cells in developing EBs are of great interest to us. In this regard, it is worth noting that two reports published during the preparation of this manuscript have described that proliferation of a common progenitor cell for both primitive and definitive erythropoiesis is stimulated by VEGF83 and that flk-1+ cells are actually generated during the differentiation of ES cells, which contain erythro-myeloid cell potential.84 Nevertheless, this result supports the possibility of the in vitro differentiation culture of ES cells to elucidate novel roles of known factors or to find novel factors for the early hematopoietic cell development.
The cytotoxic lymphocyte formation from ES cells in vitro was a novel observation although generation of B and T lymphocytes had already been reported by several groups.14,16,17 Therefore, the B220+ CD3− LAK cells from the CD34+ EB cell fraction were subjected to lineage analysis. Since NK cells in adult mice are CD3− NKR-P1+cytotoxic LGLs, we determined that the LAK cells were activated NK cells. All the LAK cells obtained expressed the activating NK receptor (NKR-P1) gene, as well as genes encoding NK/CTL specific cytolytic proteins, perforin, and granzymes. Furthermore, one type of LAK cells (type II) was sensitive to the NK cell stimulatory factor, IL-12,67 and showed cytotoxicity specific to NK-sensitive Yac-1 cells. Therefore, we concluded that these LAK cells were probably originated from NK progenitor cells present in the CD34+ EB fraction. Embryonic origin of NK cells in mice and humans has been traced back to fetal liver.34,85 86 Generation of NK-like killer cells from ES cells certainly suggests that at least NK potential would appear at a much earlier stage of embryogenesis in mice.
On the basis of the differences in morphology, target specificity, and cytokine responsiveness, the established LAK cells were classified into two types (Table 1). The type I LAK cells were large vacuolated LGLs with weak nonspecific cytotoxicity. Type II cells were smaller LGL, demonstrating sensitivity to IL-12 and target specificity to NK-sensitive Yac-1 cells (Figs 3-5). These differences may be attributed to difference in their maturity. At first, the LAK cells appeared to be smaller, and morphologically similar to type II LAK cells. These small cytotoxic cells did not necessarily proliferate rapidly in our culture condition. Occasionally, we failed to expand these cells (Table 2), or fast-growing larger cytotoxic cells, type I LAK cells, often took over the culture later. Therefore, the larger type I LAK cells may be derived from the smaller type II LAK cells. In support of this hypothesis, the occurrence of the LAK progenitor cells was much higher when type II LAK cells were established, so that they were not cultured as long before all the analyses were performed. Furthermore, long-term maintenance of type II LAK cells under the stroma-free condition was detrimental for their strong cytotoxicity to Yac-1 cells (data not shown). However, we cannot rule out the possibility that the two types of LAK cells simply represent different cytotoxic cell-types.
While erythro-myeloid potential of the CD34+ EB cell population could be predicted from characteristics of the yolk sac CD34+ cells,48 our observation that B cell as well as NK cell potential is also enriched (Table 2) has provided an additional support for the multipotentiality of the CD34+EB cell fraction. This prompted us to speculate that there might be a multipotential HSC in this cell fraction. We are currently investigating MRA in the CD34+ EB cell fraction using adult mice as well as newborn mice.
ACKNOWLEDGMENT
First, we acknowledge C.-Y. E. Han for excellent technical support in many aspects of this work, and T. Black and L. Chiu for help of FACS analysis as well as sorting. We also thank L. Souza for G-CSF, T. Boone for IL-3 and IL-6, K. Langley for SCF, W. Kenney for IL-2, B. Samal for LIF, and T. Arakawa and K. Westcott for TGF-β1. Special thanks to K. Stark for the initial set up for dissecting embryos, and also for providing us J7 ES cells, and to K. Klopchin for preparation of mice. Much appreciation to C. Saris for providing 32Dcl3 cells, to G. Trail for P815 and Yac-1 cells, and to H. Kodama (Ohu University) and K. Arai (University of Tokyo) for OP9 stroma cells. We are grateful to M. Hu, E. Medlock, C. Saris, and H.-M. Wang for sharing information, interesting discussion, and particularly, critical reading of this manuscript. Also, warm thanks to L. Souza for support and encouragement and to W. Boyle and R. Bosselman for critical reading of this manuscript.
Address reprint requests to Naoki Nakayama, PhD, Department of Cell Biology, Amgen Inc, 1840 Dehavilland Dr, Thousand Oaks, CA 91320.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal