Abstract
Eosinophil-derived neurotoxin (EDN) found in the granules of human eosinophils is a cationic ribonuclease toxin. Expression of the EDN gene (RNS2) in eosinophils is dependent on proximal promoter sequences in combination with an enhancer located in the first intron. We further define here the active region of the intron using transfections in differentiated eosinophilic HL60 cells. We show that a region containing a tandem PU.I binding site is important for intronic enhancer activity. This region binds multiple forms of transcription factor PU.I as judged by gel-shift analysis and DNA affinity precipitation. Importantly, introducing point mutations in the PU.I site drastically reduces the intronic enhancer activity, showing the importance of PU.I for expression of EDN in cells of the eosinophilic lineage.
EOSINOPHILS PLAY an important role in protection against parasitic diseases and are involved in the pathogenesis of allergic diseases.1,2 Parasite killing and tissue damage seen in allergic lesions are both linked to the release of eosinophil granule proteins by degranulation or exocytosis and production of toxic oxygen metabolites.1,3,4Eosinophil-derived cytotoxic proteins include major basic protein (MBP), eosinophil peroxidase (EPO), eosinophil cationic protein (ECP), and eosinophil derived neurotoxin (EDN).2 5
EDN is localized in the granule matrix in the human eosinophil and possesses neurotoxic, helminthotoxic, and ribonucleolytic activities.6 EDN is synthesized as a preprotein of 161 amino acids including a 27 amino acid signal peptide, which is processed to form the mature protein.7-10
The EDN gene consists of two exons separated by a single intron, of which the second exon contains the complete coding region and 3′-untranslated sequences.11 These structural features are shared with other genes of the RNAse gene superfamily, such as ECP.11 EDN mRNA can be detected in the RNA from hypodense eosinophils9 and during the promyelocytic stage of eosinophil development as judged by in vitro differentiation of cord blood mononuclear cells.12 Similarly, EDN mRNA expression can be detected in the promyelocytic HL60 cell line when it is induced to differentiate with interleukin-5 (IL-5)9 or butyric acid (BA).13
Previously, it was shown that optimal expression of EDN in HL60 cells is dependent on the interaction of the proximal promoter sequences and the intron.13 The EDN intron contains consensus binding sites for transcription factors AP-1, NF-AT, and PU.I.13Recently, it was shown that the NF-AT site plays an important role in the activity of the intronic enhancer in undifferentiated HL60 cells, whereas its contribution in the more eosinophil-differentiated HL60 clone 15 cell line was limited.14 By contrast, the contributions of the AP-1 and PU.I sites to the activity of the intron remain obscure. Because AP-1 was not previously shown to play a specific role in myeloid cells, we focused our attention on the role of PU.I.
PU.I was originally identified as the product of the Spi-I oncogene in Friend virus induced erythroleukemias (reviewed in Janknecht and Nordheim15). Transcription factor PU.I was then characterized as a member of the ets multigene family, which bind to a purine-rich sequence containing the GGAA core sequence motif.15,16 Members of the ets-family of transcription factors share a conserved 85 amino acid domain, the ets domain, which is responsible for sequence-specific DNA binding and has some homology with the winged helix-turn-helix family of proteins.15,17PU.I is expressed in hematopoietic cells, predominantly in the B-cell and myeloid lineages.18-21 The PU.I cis-acting DNA element is found to be essential for the activity of numerous myeloid-specific promoters, including the macrophage scavenger receptor gene,22 CD11b,23 CD18,24CD64,25 myeloperoxidase,26 neutrophil elastase,27 FcγRIIIA,28proteinase-3,29 the granulocyte colony-stimulating factor (G-CSF) receptor,30 the macrophage colony-stimulating factor (M-CSF) receptor,31 and the granulocyte-macrophage colony-stimulating factor (GM-CSF) receptor.32 It was also shown that PU.I can functionally interact with other classes of transcription factors, such as NF-EM5/PIP.33,34 Gene targeting experiments have shown that PU.I is required for the development of lymphoid and myeloid lineages during fetal liver hematopoiesis.35,36 Similarly, blocking PU.I function in vitro in ES cells37 or CD34+cells38 further demonstrates the essential role of PU.I during myelopoiesis.
We show here that a PU.I site in the intron of the EDN gene is important in optimal expression in eosinophilic HL60 cells. Multiple forms of PU.I that are expressed in differentiating HL60 cells bind to the intronic PU.I site. Mutation of the PU.I site strongly decreases the activity of the EDN promoter/intron combination. This is the first report showing the importance of PU.I for the expression of genes in the eosinophil lineage.
MATERIALS AND METHODS
Cells, plasmids, oligonucleotides, and antibodies.
HL60 cells were cultured in RPMI medium (Life Technologies, Breda, The Netherlands) containing 8% fetal calf serum (FCS). HL60 7.7 cells were generated by culturing HL60 cells for 2 months at pH 7.7 in RPMI containing 25 mmol/L N-[2-Hydroxyethyl]piperazine-N′-3-propane-sulfonic acid (EPPS; Sigma, St Louis, MO). To further differentiate HL60 7.7 cells, butyric acid (0.5 mmol/L; Sigma) was added for 2 to 5 days. HeLa cells were grown in Dulbecco's modified Eagles medium (DMEM; Life Technologies) containing 8% FCS.
The EDN promoter/intron (−300 to +298) was cloned by polymerase chain reaction (PCR) using human genomic DNA. The amplified fragment was then sequenced and cloned into the promoterless CAT reporter plasmid pBLCAT3. Intron deletion constructs (+193, +193δPU.I, +123, and +67) were generated by PCR and cloned into pBLCAT3. Site-directed mutagenesis was performed as described previously.39 The β actin reporter and the GAPDH probe are described elsewhere.40
The following oligonucleotides were used in this study: for PCR cloning of the EDN promoter/intron, EDN-F (CCTGTAAGAAAAGAAGAG) and EDN-R (CACCGCTCCTGTCAGCCA); for the generation of intron deletions, EDN +193 (CGGGATCCATTTCCTTTACTTCCTGTC), EDN+193 d PU.I (CGGGATCC ATTGCCTTTACTGCCTGTC), EDN +123 (CGGGATCCAGTTGCTGCCCCATTGC), and EDN +67 (CGGGATCCAGTCTCCGCGCTGTAG); for site-directed mutagenesis of the PU.I site, PU.I mut1 (CTTTGCAG ACAGGCCTTAAAGGAAATGGG), PU.I mut2 (CAGGAAGTAAAG GCCTTGGGACCCAGAGT), and PU.I mut3 (GCAGACAGGCAGTAA AGGCAATGGGACC); for band-shift analysis (only upper strand is shown), EDN PU.I wt (AGCTTGACAGGAAGTAAAGGAAATG) and EDN-PU.I mut (AGCTTGACAGGCAGTAAAGGCAATG); and for the PCR cloning of an EDN probe for Northern analysis, EDN-RNA-F (CTTCTGTTGGGGCTTCTG) and EDN-RNA-R (TTGGAGTTGTGAGGTTAC).
The following antibodies were used in this study: anti-PU.I (rabbit polyclonal IgG T-21, SC-352; Santa Cruz, Santa Cruz, CA) and anti-STAT3 (rabbit polyclonal IgG C-20, SC-482; Santa Cruz).
RNA isolation, reverse transcription, PCR, and Northern blotting.
Total cellular RNA was isolated by the guanidine isothiocyanate-acid phenol method.41 For the isolation of an EDN-specific probe for Northern blotting, RNA was reverse transcribed using murine Maloney leukemia virus (M-MLV) reverse transcriptase according to the manufacturer's protocols (Life Technologies). PCR was performed in a total volume of 20 μL with 5 μL cDNA synthesis mixture for 36 cycles of 94°C denaturation (1 minute), 52°C annealing (1.5 minutes), and 72°C extension (2 minutes). For Northern blotting, RNA (20 μg) was electrophoretically separated on 0.8% agarose gels and transferred to Hybond (Amersham, Arlington Heights, IL). Blots were hybridized with randomly 32P-labeled EDN or GAPDH fragments overnight at 42°C in hybridization buffer, washed. and exposed to film as described previously.40
Transient transfections.
For transfection experiments, HeLa cells were subcultured in 6-well dishes and transfected with 10 to 20 μg supercoiled plasmid DNA 16 hours later, as described previously.42 43 HL60 cells (10 × 106 cells per sample) were transfected with 20 μg plasmid DNA by electroporation (280 V, 960 μF). The CMV-LacZ plasmid (2 μg) was included to correct for differences in transfection efficiency between different samples. Transfected cells were harvested 48 hours later for CAT assays. CAT assays were performed as follows. Cells were lyzed by repeated freeze-thawing in 250 mmol/L Tris, pH7.4, 25 mmol/L EDTA. Twenty-five micrograms of cellular extract was then incubated in a total volume of 100 μL containing 250 mmol/L Tris, pH 7.4, 2% glycerol, 0.3 mmol/L Butyryl Coenzyme A (Sigma), and 0.05 μCi 14C Chloroamphenicol (Amersham) for 2 hours at 37°C. Reaction products were then extracted using 400 μL xylene/pristane (1:2), and the percentage of acetylated products was then determined using liquid scintillation counting. All experiments were performed at least four times.
In vitro translation, gel retardation assay, and DNA affinity precipitation.
PU.I cDNA was translated in vitro using the TnT-coupled reticulocyte lysate system (Promega, Madison, WI) according to the manufacturer's protocol. Nuclear extracts were prepared as described previously.42 Oligonucleotides were labeled by filling in the cohesive ends with [α-32P]dCTP using Klenow fragment of DNA polymerase I. Gel retardation assays were performed according to published procedures with slight modifications. Briefly, nuclear extracts (10 μg) were incubated in a final volume of 20 μL containing 20 mmol/L N-[2-hydroxyethyl]piperazine-N′-[2-ethanesulfonic acid] (HEPES), pH 7.9, 50 mmol/L KCL, 0.1 mmol/L ethylenediaminetetraacetic acid (EDTA), 2 mmol/L MgCl2, 10% (vol/vol) glycerol, 1 mmol/L dithiothreitol, 1 μg poly(dI-dC) (Pharmacia, Uppsala, Sweden), and 1.0 ng of 32P-labeled EDN-PU.I oligonucleotide for 20 minutes at room temperature. Complexes were then separated though nondenaturing 5% polyacrylamide gels and visualized by autoradiography. For supershift analysis, nuclear extracts were preincubated with 1 μg anti-PU.I antibody (SC352X; Santa Cruz) for 30 minutes on ice before the addition of the labeled probe.
For DNA affinity precipitation, nuclear extract was prepared from 20 × 106 cells. The extract was diluted to 400 μL with dilution buffer (10 mmol/L HEPES, pH 7.9, 50 mmol/L KCl, 1 mmol/L EDTA, 5 mmol/L MgCl2, 10% glycerol) and precleared with 25 μL streptavidin-agarose beads (Sigma) for 15 minutes at 4°C. The extract was then incubated with 25 μL of biotinylated EDN oligonucleotide coupled to streptavidin-agarose beads (Sigma) in the presence of 2 μg poly(dI-dC) and 20 μg bovine serum albumin. After incubation for 1 hour at 4°C, protein-DNA complexes were washed three times with dilution buffer. After boiling for 3 minutes in Laemmli sample buffer, samples were separated on 12% sodium dodecyl sulphate-polyacrylamide gels and electro-transferred to Immobilon-P membranes (Millipore, Bedford, MA). Membranes were blocked in TBST-buffer (150 mmol/L NaCl, 10 mmol/L Tris, pH 8.0, 0.3% Tween 20) containing 5% nonfat milk for 30 minutes and probed with anti-PU.I antibody (1 ng/mL; SC352X; Santa Cruz) for 1 hour. After three washes with TBST, the membranes were incubated for 1 hour with peroxidase-conjugated goat-antirabbit antibodies (DAKO, Glostrup, Denmark), followed by five washes with TBST. Proteins were visualized with enhanced chemiluminescence (Amersham).
Direct immunoprecipitation was performed as described previously.44
RESULTS
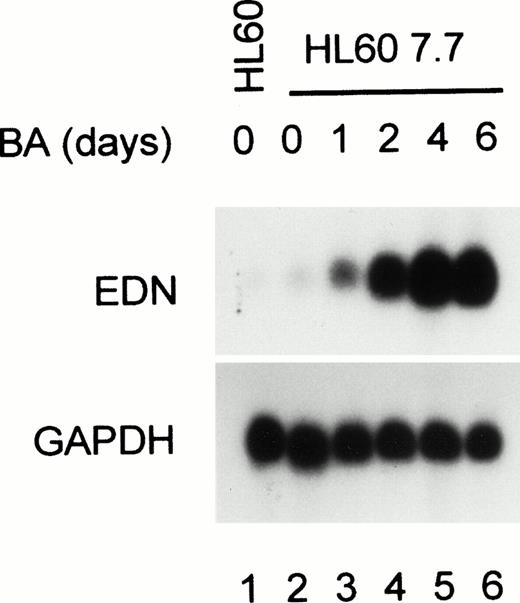
It was previously shown that EDN mRNA expression is strongly enhanced in BA-treated HL60 clone 15 cells.13 To investigate whether in HL60 cells differentiated at pH 7.7 EDN expression is also responsive to BA, RNA was isolated from HL60 7.7 cells treated for various periods with BA. Figure 1 shows that EDN mRNA is weakly expressed in parental HL60 and HL60 7.7 cells. However, EDN expression is strongly upregulated by BA treatment within 1 to 2 days, whereas EDN mRNA is maximal 4 days after BA addition. Reprobing the blot with a GAPDH probe shows that equal amounts of RNA were loaded in each lane. These results demonstrate that HL60 7.7 cells are suitable for studying the regulation of the EDN promoter.
Upregulation of EDN expression during eosinophilic differentiation of HL60 cells. HL60 7.7 cells were treated with BA (0.5 mmol/L) for 1, 2, 4, or 6 days, after which RNA was isolated and analyzed by Northern blotting. Hybridization of the blot with a probe specific for EDN shows that EDN mRNA expression is strongly induced by BA. Reprobing of the blot with a GAPDH probe shows that equal amounts of RNA were loaded in each lane. Scanning of the autoradiograph shows the following increase in EDN expression (relative to GAPDH) compared with wild-type HL60 cells: 2-, 5-, 45-, 160-, and 130-fold increase for 7.7 cells treated for 0, 1, 2, 4, and 6 days, respectively.
Upregulation of EDN expression during eosinophilic differentiation of HL60 cells. HL60 7.7 cells were treated with BA (0.5 mmol/L) for 1, 2, 4, or 6 days, after which RNA was isolated and analyzed by Northern blotting. Hybridization of the blot with a probe specific for EDN shows that EDN mRNA expression is strongly induced by BA. Reprobing of the blot with a GAPDH probe shows that equal amounts of RNA were loaded in each lane. Scanning of the autoradiograph shows the following increase in EDN expression (relative to GAPDH) compared with wild-type HL60 cells: 2-, 5-, 45-, 160-, and 130-fold increase for 7.7 cells treated for 0, 1, 2, 4, and 6 days, respectively.
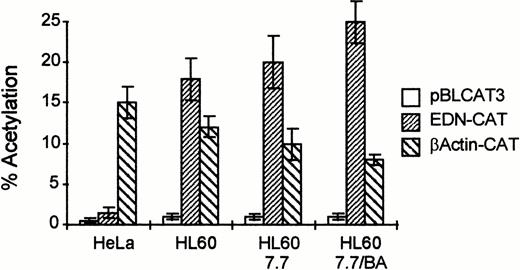
Next, we cloned the EDN promoter/intron into the pBLCAT3 reporter plasmid and tested its activity in HL60 cells. Figure 2 shows that this reporter is active in HL60 cells compared with a promoterless CAT construct. Moreover, its activity is upregulated by BA during HL60 differentiation when compared with a reporter containing the β actin promoter (β actin-CAT). Transfection of these constructs into HeLa cells shows that the EDN promoter is not active in all cell types, but has some hematopoietic specificity, as was also suggested by Tiffany et al.13
The EDN promoter/intron combination is active in HL60 cells and their differentiated derivatives. HL60, HL60 7.7, and HeLa cells (10 × 106 per transfection) were transfected with a CAT reporter construct containing the EDN promoter and the first intron (EDN-CAT; 20 μg), the empty vector without EDN sequences (pBLCAT3; 20 μg), or a positive control containing the promoter of the β-actin gene (β actin-CAT; 20 μg) by electroporation (HL60 cells, 280 V, 960 μF) or calcium phosphate precipitation (HeLa cells). Some cells received BA (0.5 mmol/L) 1 hour after transfection (HL60 7.7/BA). Two days posttransfection, cells were harvested and assayed for CAT activity. Bars indicate the mean percentage of acetylation of at least four independent experiments. The standard deviation is indicated by error bars. The EDN reporter construct is clearly active in HL60 cells and their differentiated derivatives, but not in nonhematopoietic HeLa cells.
The EDN promoter/intron combination is active in HL60 cells and their differentiated derivatives. HL60, HL60 7.7, and HeLa cells (10 × 106 per transfection) were transfected with a CAT reporter construct containing the EDN promoter and the first intron (EDN-CAT; 20 μg), the empty vector without EDN sequences (pBLCAT3; 20 μg), or a positive control containing the promoter of the β-actin gene (β actin-CAT; 20 μg) by electroporation (HL60 cells, 280 V, 960 μF) or calcium phosphate precipitation (HeLa cells). Some cells received BA (0.5 mmol/L) 1 hour after transfection (HL60 7.7/BA). Two days posttransfection, cells were harvested and assayed for CAT activity. Bars indicate the mean percentage of acetylation of at least four independent experiments. The standard deviation is indicated by error bars. The EDN reporter construct is clearly active in HL60 cells and their differentiated derivatives, but not in nonhematopoietic HeLa cells.
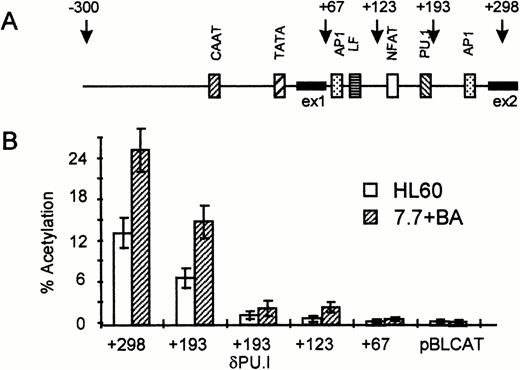
It was previously shown that the intron of the EDN gene is of major importance for the activity of the EDN promoter in HL60 c15 cells.13 To identify the sequences important for the activity of the intron, we constructed a number of intron-deletion constructs that are schematically shown in Fig 3A. The activity of these constructs was then tested in HL60 cells and HL60 7.7 cells treated with BA. As shown in Fig 3B, deletion of 105 nucleotides from the intron including the AP1 binding site (+193) reduces CAT activity by about 40%. Further deletion to +123 including a tandem PU.I site and an NFAT site strongly reduces CAT activity, whereas deletion of the complete intron (+67) completely abolishes CAT activity. To investigate the role of the PU.I site further, we constructed a +193 construct with a mutated PU.I site (+193 δ PU.I). Surprisingly, this reduces CAT activity down to the level of the +123 construct, suggesting that the PU.I site is of major importance for the activity of the EDN intron.
The PU.I site in the intron is important for the enhancer function of the EDN intron. (A) Schematic representation of the EDN promoter and intron. Putative transcription factor binding sites (CAAT box, AP1 sites, NFAT site, and PU.I site) are indicated as boxes. The arrows indicate the 3′ ends of the reporter constructs used in (B). ex1, exon 1. (B) The EDN reporter constructs described in (A) and the control vector pBLCAT3 were transfected into HL60 or HL60 7.7 cells as described in Fig 2. Construct +193 d PU.I contains point mutations in the PU.I site. The HL60 7.7 cells received BA 1 hour after transfection. CAT activity was determined 48 hours posttransfection. Bars indicate the mean percentage of acetylation of at least four independent experiments. The standard deviation is indicated by error bars. It is clear that the intron is essential for the activity of the EDN reporter construct, whereas the PU.I site seems to play a major role in the activity of the intron.
The PU.I site in the intron is important for the enhancer function of the EDN intron. (A) Schematic representation of the EDN promoter and intron. Putative transcription factor binding sites (CAAT box, AP1 sites, NFAT site, and PU.I site) are indicated as boxes. The arrows indicate the 3′ ends of the reporter constructs used in (B). ex1, exon 1. (B) The EDN reporter constructs described in (A) and the control vector pBLCAT3 were transfected into HL60 or HL60 7.7 cells as described in Fig 2. Construct +193 d PU.I contains point mutations in the PU.I site. The HL60 7.7 cells received BA 1 hour after transfection. CAT activity was determined 48 hours posttransfection. Bars indicate the mean percentage of acetylation of at least four independent experiments. The standard deviation is indicated by error bars. It is clear that the intron is essential for the activity of the EDN reporter construct, whereas the PU.I site seems to play a major role in the activity of the intron.
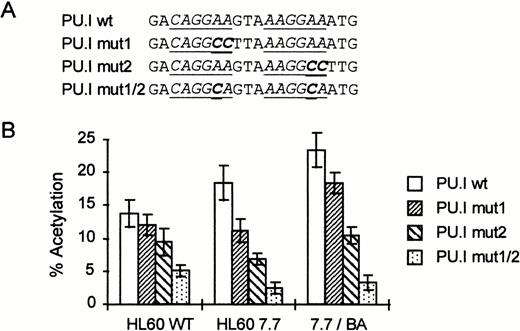
To further study the importance of the PU.I site for the intronic enhancer activity, we prepared different point mutations in the background of the complete promoter/intron sequence. We mutated either the first PU.I core element (PU.I mut1, Fig4A) or the second (PU.I mut2) or both (PU.I mut1/2). The activity of the different mutants was then tested in HL60, HL60 7.7, and HL60 7.7/BA cells (Fig 4B). Mutation of the first PU.I core sequence slightly reduces CAT activity, whereas mutation of the second site has a somewhat stronger effect. Importantly, mutation of both sites leads to a strong reduction in CAT activity, especially in HL60 7.7 and HL60 7.7/BA cells. Taken together, these results suggest that the PU.I site in the EDN intron plays a major role in EDN expression in eosinophilic cell lines.
The PU.I site is essential for the activity of the intronic enhancer. (A) Schematic representation of the mutant reporter constructs. Only the sequence around the tandem PU.I site is shown. The PU.I core binding sites are underlined. Mutations made are indicated in bold type. (B) EDN-CAT constructs containing the full promoter and intron with different mutations in the tandem PU.I site were transfected into HL60 and HL60 7.7 cells and treated with BA as described in Fig 2. Two days posttransfection, cells were harvested and assayed for CAT activity. Bars indicate the mean percentage of acetylation of at least four independent experiments. The standard deviation is indicated by error bars. The double mutant in the tandem PU.I site strongly reduces the activity of the EDN-CAT reporter construct.
The PU.I site is essential for the activity of the intronic enhancer. (A) Schematic representation of the mutant reporter constructs. Only the sequence around the tandem PU.I site is shown. The PU.I core binding sites are underlined. Mutations made are indicated in bold type. (B) EDN-CAT constructs containing the full promoter and intron with different mutations in the tandem PU.I site were transfected into HL60 and HL60 7.7 cells and treated with BA as described in Fig 2. Two days posttransfection, cells were harvested and assayed for CAT activity. Bars indicate the mean percentage of acetylation of at least four independent experiments. The standard deviation is indicated by error bars. The double mutant in the tandem PU.I site strongly reduces the activity of the EDN-CAT reporter construct.
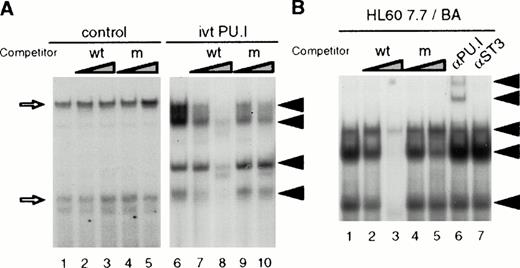
PU.I is a member of the multigene Ets transcription factor family (for a review, see Janknecht and Nordheim15). To determine the nature of the proteins binding to the PU.I site in the EDN intron, we performed band shift analysis using the PU.I site as a probe. We synthesized PU.I protein using a rabbit reticulocyte system and tested the binding of this protein to the EDN PU.I site. Figure 5A shows that unprogrammed lysate forms two complexes with the PU.I probe, which cannot be competed with either wild-type or mutant PU.I oligonucleotide (lanes 1 through 5). By contrast, in vitro translated PU.I binds as four different complexes to the EDN PU.I site (lane 6). The four complexes can be competed with a 100-fold molar excess of unlabeled wild-type PU.I site (lanes 7 and 8), but not with the mutant PU.I site (lanes 9 and 10). To determine whether PU.I also binds this site in HL60 7.7/BA cells, nuclear extracts were analyzed. Figure 5B shows that three different protein/DNA complexes can be observed, which can be competed with a 100-fold molar excess of unlabeled wild-type PU.I site (lanes 2 and 3), but not with the mutant PU.I site (lanes 4 and 5). Moreover, preincubation of the nuclear extract with an anti-PU.I antibody results in the formation of a supershift (lane 6), whereas an anti-STAT3 antibody did not alter binding. Similar results were obtained with HL60 and HL60 7.7 cells (not shown). These results indicate that the intronic PU.I site is capable of binding PU.I in nuclear extracts of HL60-derived cells.
PU.I binds to the PU.I site in the EDN intron. (A) PU.I protein was generated in vitro using a rabbit reticulocyte lysate. PU.I (lanes 6 through 10) or unprogrammed control reticulocyte lysate (lanes 1 through 5) was then tested in a bandshift assay for binding to the PU.I site from the EDN intron. Arrows indicate four PU.I/DNA complexes that are competed by excess PU.I oligo (lanes 7 and 8, 10- and 100-fold), but not by excess mutant PU.I oligo (lanes 9 and 10, 10- and 100-fold). Nonspecific complexes formed with control lysate are indicated by open arrows. The free DNA probe is not shown. (B) Nuclear extracts were prepared from HL60 7.7 cells treated for 4 days with BA. These extracts were then tested for PU.I binding activity in a band shift assay. Three protein/DNA complexes are observed (lane 1) that are competed by excess PU.I oligo (lanes 2 and 3, 10- and 100-fold), but not by excess mutant PU.I oligo (lanes 4 and 5, 10- and 100-fold). Preincubation with an anti-PU.I antibody (lane 6), but not with an anti-STAT3 control antibody (lane 7) results in the appearance of two supershifted complexes indicated by arrows. The free DNA probe is not shown.
PU.I binds to the PU.I site in the EDN intron. (A) PU.I protein was generated in vitro using a rabbit reticulocyte lysate. PU.I (lanes 6 through 10) or unprogrammed control reticulocyte lysate (lanes 1 through 5) was then tested in a bandshift assay for binding to the PU.I site from the EDN intron. Arrows indicate four PU.I/DNA complexes that are competed by excess PU.I oligo (lanes 7 and 8, 10- and 100-fold), but not by excess mutant PU.I oligo (lanes 9 and 10, 10- and 100-fold). Nonspecific complexes formed with control lysate are indicated by open arrows. The free DNA probe is not shown. (B) Nuclear extracts were prepared from HL60 7.7 cells treated for 4 days with BA. These extracts were then tested for PU.I binding activity in a band shift assay. Three protein/DNA complexes are observed (lane 1) that are competed by excess PU.I oligo (lanes 2 and 3, 10- and 100-fold), but not by excess mutant PU.I oligo (lanes 4 and 5, 10- and 100-fold). Preincubation with an anti-PU.I antibody (lane 6), but not with an anti-STAT3 control antibody (lane 7) results in the appearance of two supershifted complexes indicated by arrows. The free DNA probe is not shown.
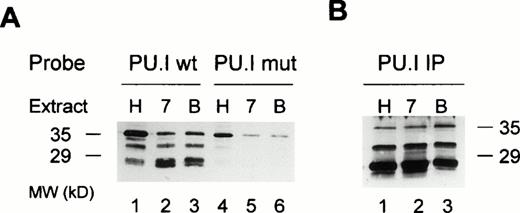
To more precisely determine the nature of the different protein/DNA complexes observed with the EDN PU.I site, we performed DNA affinity precipitation experiments. The PU.I site was labeled with biotinylated dCTP, coupled to streptavidin agarose, and used as a probe to fish out binding proteins from nuclear extracts. Interacting proteins were then resolved on a polyacrylamide gel, blotted onto nylon filters, and probed with an anti-PU.I antibody that is not cross-reactive with other Ets family members. Figure 6A shows that three different PU.I forms bind to the EDN PU.I site in HL60, HL60 7.7 and HL60 7.7/BA cells (lanes 1 through 3). Reprobing the blot with a control antibody (anti STAT3) showed that all three bands indeed are likely to represent different forms of PU.I (data not shown). When the mutant PU.I site was used as a probe, binding of the two faster migrating PU.I forms is strongly reduced, whereas the largest protein binds significantly less (75% decrease) compared with the wild-type probe (lanes 4 through 6). When we performed direct immuno-precipitations of PU.I from the cells, the same three complexes were observed, although in a slightly different ratio that might be caused by different affinity of the PU.I antibody for the different complexes in solution (Fig 6B) that are not evident when the antibody is used on denatured protein (Fig 6A). Taken together, these results strongly suggest that multiple forms of transcription factor PU.I are expressed in HL60-derived cells and that all these forms are capable of interacting with the intronic PU.I binding site.
Multiple forms of PU.I bind to the intron of the EDN gene. (A) DNA affinity precipitation of nuclear extracts from HL60 (H), HL60 7.7 (7), or BA-treated HL60 7.7 cells (B). Either wild-type (lanes 1 through 3) or mutant PU.I oligonucleotide (lanes 4 through 6) was used as a probe. Proteins bound to these probes were precipitated, separated through a 12% polyacrylamide gel, and blotted onto nylon membranes. The filter was then probed with an anti-PU.I antibody. Three different forms of PU.I bind to the wild-type PU.I probe, of which only the largest form binds (with lower affinity) to the mutant probe. (B) Immunoprecipitation of PU.I from HL60 (H), HL60 7.7 (7), or BA-treated HL60 7.7 cells (B) using an anti-PU.I antibody. The precipitates were visualized as in (A). The same three PU.I forms are detected as in (A).
Multiple forms of PU.I bind to the intron of the EDN gene. (A) DNA affinity precipitation of nuclear extracts from HL60 (H), HL60 7.7 (7), or BA-treated HL60 7.7 cells (B). Either wild-type (lanes 1 through 3) or mutant PU.I oligonucleotide (lanes 4 through 6) was used as a probe. Proteins bound to these probes were precipitated, separated through a 12% polyacrylamide gel, and blotted onto nylon membranes. The filter was then probed with an anti-PU.I antibody. Three different forms of PU.I bind to the wild-type PU.I probe, of which only the largest form binds (with lower affinity) to the mutant probe. (B) Immunoprecipitation of PU.I from HL60 (H), HL60 7.7 (7), or BA-treated HL60 7.7 cells (B) using an anti-PU.I antibody. The precipitates were visualized as in (A). The same three PU.I forms are detected as in (A).
DISCUSSION
Expression of EDN mRNA is observed in cells of the eosinophilic lineage as well as in human neutrophils9 and in human tissue containing phagocytes.45 In this report, we show that transcription factor PU.I plays an important role in the activity of the EDN promoter/intron, suggesting that PU.I is involved in expression of EDN in eosinophilic cells.
It was previously shown that PU.I is essential for the development of lymphoid and myeloid lineages during fetal liver hematopoiesis.35,36 Moreover, using PU.I −/− ES cells, it was shown that PU.I is essential for terminal differentiation of myeloid precursors to neutrophils and macrophages.37 In these cells, expression of early myeloid markers, such as GM-CSF-R, G-CSF-R, and myeloperoxidase was unaltered. By contrast, genes associated with terminal myeloid differentiation, such as CD11b, CD64, and M-CSFR, were not expressed in PU.I −/− ES cells.37 However, the role of PU.I for eosinophilopoiesis was not determined. Our results might suggest that PU.I might well play a role in gene regulation during terminal differentiation of eosinophils comparable to its role in neutrophils.
Tiffany et al13 have shown that the EDN intron is essential for optimal activity of the EDN promoter. The activity of the intron is only obvious in hematopoietic cells, such as U937, K562, Jurkat, HL60,13 and HL60 7.7 +/-BA (this study), but not in other cell lines such as 29313 or HeLa (this study). It is therefore likely that a transcription factor expressed mainly in hematopoietic cells is involved in the activity of the intron. Of the transcription factor binding sites present in the intron, both PU.I and NFAT, but not AP-1, fit this criterion. Using our intron deletion constructs, we have mapped the major determinant of the intron activity to the tandem PU.I site (Figs 3 and 4). However, the first 60 bp of the intron, containing the NFAT site, also contains some activity compared with an intronless promoter (Fig 3B). Very recently, Handen and Rosenberg14 have shown that mutation of the NFAT site strongly represses the activity of the EDN intron in wild-type undifferentiated HL60 cells. However, this effect is much weaker in the more differentiated eosinophilic subclone HL60 c15, which was subcloned from HL60 cells grown at pH 7.7,46 and is therefore likely to be comparable with our HL60 7.7 cells.14 By contrast, the effect of the PU.I mutation is stronger in the more differentiated HL60 7.7 and HL60 7.7/BA cells as compared with the undifferentiated parental HL60 cells (Fig 4B). These results suggest that the NFAT site might play a role in EDN regulation early during myelopoiesis, whereas PU.I is more important later during terminal eosinophilic differentiation. Interestingly, a similar role for PU.I was recently demonstrated by Olson et al,37 who have shown that PU.I is not essential for early myeloid gene expression but is required for terminal myeloid differentiation.
We have shown that multiple forms of PU.I interact with the EDN PU.I site (Figs 5 and 6). However, we cannot rule out the possibility that only one form of PU.I interacts directly with the PU.I site, whereas the other complexes represent PU.I forms interacting with the DNA bound PU.I form. These different forms might represent different serine or threonine phosphorylations of PU.I. Although the difference in size between the different forms of PU.I is rather large, it is not unprecedented, because different phosphorylated forms differing 9 kD were previously shown during monocytic differentiation of U937 cells with 12-O-tetradecanoylphorbol-13-acetate.47 Similarly, stimulation of macrophages with lipopolysaccharide also results in serine phosphorylation of PU.I.48 It was also suggested that these different forms might have distinct trans-activation potentials.48 Further experiments are necessary to study the role of the different PU.I forms in the regulation of the activity of the EDN intron.
EDN mRNA is strongly upregulated during eosinophilic differentiation of HL60 cells with BA (Tiffany et al13 and Fig 1). However, the activity of the EDN intron/promoter combination is only modestly increased during this process. This might indicate that other enhancers located more upstream or downstream from the sequences that we have studied are involved in EDN upregulation by BA. Alternatively, this facet of EDN regulation might involve mechanisms that cannot be studied in transient transfections, such as chromosome accessibility involving nucleosome displacement. On the other hand, the observed induction of EDN mRNA might well be caused by posttranscriptional mechanisms, such as changes in mRNA stability or RNA processing. Further study is necessary to shed light on the mechanism by which BA induces EDN mRNA expression in HL60 7.7 cells.
Taken together, we have shown that PU.I is likely to play an important role in the regulation of EDN expression in eosinophils. Importantly, in the highly homologous ECP gene, the tandem PU.I site is 100% conserved.14 Moreover, other genes that are expressed in eosinophils, such as CLC49 and the β chain of the IL-5R (van Dijk et al, manuscript in preparation), also contain PU.I sites in their regulatory regions. These data suggest that, besides its role in neutrophils and macrophages, PU.I is likely to play an important role in gene regulation during terminal differentiation of eosinophils.
ACKNOWLEDGMENT
The authors thank Dr Marc van Dijk for providing reagents.
Supported by a research grant from Glaxo-Wellcome bv.
Address reprint requests to Rolf P. de Groot, PhD, Department of Pulmonary Diseases, G03.550, University Hospital Utrecht, Heidelberglaan 100, 3584 CX Utrecht, The Netherlands.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal