Abstract
Abnormalities of chromosome 1q21 are common in B-cell malignancies and have been associated with a poor response to therapy. The nature of the involved gene(s) on chromosome 1q21 remains unknown. A cell line (CEMO-1) has recently been established from a patient with precursor-B–cell acute lymphoblastic leukemia (ALL), which exhibited a t(1;14)(q21;q32). To identify the gene involved in this translocation, we have cloned both rearranged IGHJ alleles using long-distance inverse polymerase chain reaction (LDI-PCR). TwoIGHJ fragments were amplified from CEMO-1 DNA and sequenced. One allele showed novel sequences upstream of JH5 with no homology to either IGH or any other sequences on the databases. Using a single-copy Xho I fragment immediately 5′ ofJH5, PAC clones were isolated and mapped to chromosome 1q21 on normal metaphases by fluorescence in situ hybridization (FISH), confirming that this allele represented the t(1;14)(q21;q32) breakpoint. Sequence analysis of the 1q21 XhoI fragment showed identity with an expressed sequence tag (EST), and this probe was therefore used to probe Northern blots. Two transcripts of 6.3 kb and 4.2 kb expressed at low level in mRNA from all tissues were detected: a third transcript of 1.6 kb was expressed only in thymus, spleen, and small intestine. Full-lengthBCL9 cDNA clones were obtained from a normal human fetal brain cDNA library supplemented by 5′ and 3′ RACE. Sequence analysis predicted a protein of 1394 amino acids containing 18% proline, 11% glycine, 11% serine, and 6% methionine, but no recognizable protein motifs or significant homologies to any other known proteins. The CEMO-1 1q21 breakpoint fell within the 3′ UTR of the BCL9 gene. Low-level expression of BCL9 was detected in Epstein-Barr virus-transformed normal B cells by Northern blot; in contrast, abundant BCL9 expression was observed in CEMO-1, indicating that deregulated expression of this gene was one pathological consequence of the translocation. Screening of a panel of 39 B-cell malignancies with 1q abnormalities by Southern blot showed one additional case with a breakpoint in the 3′ UTR ofBCL9, indicating that this was a recurrent breakpoint. FISH analysis using an 850-kb YAC spanning BCL9 identified a further case with t(1;22)(q21;q11) causing juxtaposition of BCL9 to theIGλ locus. Other breakpoints were heterogeneous, falling both centromeric (10 cases) and telomeric (10 cases) of the BCL9gene. These data suggest that BCL9 may be the target of translocation in some B-cell malignancies with abnormalities of 1q21 and that deregulated BCL9 expression may be important in their pathogenesis.
B-CELL MALIGNANCIES and, in particular, the B-cell non-Hodgkin's lymphomas (B-NHL) are often associated with chromosomal translocation in which segments of other chromosomes become juxtaposed to the IG loci.1 Most of these involve the IG heavy chain locus (IGH) adjacent to the telomere of the long arm of chromosome 14 at 14q32.3, but variant forms may involve the light chain loci at 2p12 (IGκ) and 22q11 (IGλ). Specific subtypes of B-cell disease are associated with specific translocations juxtaposing the IG loci with various oncogenes.2,3 For instance, the t(11;14)(q13;q32) involving the Cyclin D1 gene at 11q13 is seen in greater than 95% of mantle-cell B-NHL, the t(14;18)(q32;q21) involving the BCL2 gene in 80% of follicular B-NHL, the t(8;14)(q24;q32) involving the MYC gene in most cases of Burkitt's lymphoma (BL), and the t(3;14)(q27;q32) involving the BCL6 gene in diffuse large-cell lymphoma (DLCL).4-9 Translocation of the oncogene results in its deregulated expression, presumably in part due to the potent enhancers within the IG loci.10
The approach of molecular cloning of IGH translocation breakpoints continues to allow the identification of novel oncogenes and novel pathogenic mechanisms in B-NHL.11-13 To facilitate the cloning of IGH translocations, we have recently developed the method of long-distance inverse polymerase chain reaction (LDI-PCR) for the rapid molecular cloning of such translocation breakpoints.14
Abnormalities of chromosome 1q21 are a common feature of B-cell malignancies of all stages of differentiation. No association with a particular disease subtype has been reported, although abnormalities of this region have been found in 39% of cases of marginal zone B-cell lymphoma.15 In B-NHL, 1q21 abnormalities are generally secondary, occurring in some 10% to 15% of cases, and have been found by multivariate analysis to be associated with a poor prognosis, particularly in patients with DLCL.16,17 Abnormalities include the recurrent translocations t(1;6)(q21;q23-q26), t(1;11)(q21;p15), t(1;12)(q21;q24), t(1;14)(q21;q32), and t(1;22)(q21;q11)16-18 (I.W., unpublished observations), together with the duplication dup(1)(q21q32) which, after the MYC translocations, is the second most common cytogenetic abnormality found in BLs, occurring in greater than 30% of cases.19 However, no oncogene involved in lymphoid malignancy has yet been identified within this region.
A new pre-B acute lymphoblastic leukemia (ALL) cell line, CEMO-1 (cIgm+/sIg−/CD19+/CD20+/−/CD22+/CD34−), was recently established.20 Both the original patient material and cell line initially exhibited t(1;14)(q21;q32) as the sole cytogenetic abnormality, although with continued growth in vitro the cell line developed the additional translocation, t(9;9)(p24;q32).
We report here the molecular cloning using LDI-PCR of the t(1;14)(q21;q32) in the CEMO-1 cell line, in which IGHJ had become juxtaposed to novel sequences on 1q21, and the isolation of a new gene (BCL9) of unknown functions. We have also shown that translocation breakpoints in some B-NHL cases with 1q21 breakpoints were located in close proximity to BCL9. In addition, we have found evidence of deregulated overexpression of BCL9 in the CEMO-1 cell line, suggesting that overexpression of BCL9 may be of pathogenic significance in a proportion of B-cell malignancies with breakpoints at 1q21.
MATERIALS AND METHODS
Cell lines, cell culture, and cytogenetics.
The CEMO-1 cell line was derived from the peripheral blood of a previously healthy 30-year-old man with precursor B-cell ALL. He achieved complete remission after intensive induction therapy, but sustained a bone marrow relapse 3 months later and died of progressive disease. Cytogenetic analysis of the original patient material and the cell line initially showed the presence of the translocation t(1;14)(q21;q32), but with subsequent growth in vitro the cell line developed a further t(9;9)(p24;q32). Immunophenotype, cytogenetics and Southern blot analysis of IGHJ rearrangements have since remained stable during further culture.20 BL cell lines were kindly provided by Prof G. Lenoir (IARC, Lyon, France). Details of these and of other cell lines and their derivation can be obtained on request. Cell lines were grown in RPMI supplemented with 10% fetal calf serum under standard conditions.
Cytogenetic analysis on fresh patient material was performed using conventional methods as previously described.15 21 Case no. 2 (Table 1) was a 57-year-old woman with stage IIIA follicular lymphoma (CD10+, bcl2+, Igλ+); karyotype, obtained from LN biopsy at diagnosis, was as follows: 49, XX, t(1;22)(q21;q11), del(4)(q21q27), del(6)(q12q25), +7, +8, +9, t(14;18)(q32;q21). She was initially diagnosed in 1994 and underwent autologous bone marrow transplantation in 1996. Case no. 6 (Table 1) was an 87-year-old woman with mantle cell lymphoma (CD5−, CD19+, CD22+, IgM+, CD25−, FMC7+, CD23−); karyotype, obtained from peripheral blood at diagnosis, was as follows: 45, XX, del(1)(p11p31), der(1)(q), add(2)(p21), add(5)(p14), add(9)(p21), add(10)(p15), t(11;14)(q13;q32), add(17)(p13), −18. She was admitted in 1995 with gastritis, leukocytosis, lymphadenopathy, and splenomegaly and died 2 months later of progressive disease despite multiagent chemotherapy.
Analysis of BCL9 in Cases With 1q21 Breakpoints
| Case No. . | Cytogenetic 1q21 Abnormality . | Histology . | Southern Blot (X665) . | BCL9 Overexpression . | Location of YAC 882B3 Signals . |
|---|---|---|---|---|---|
| CEMO-1 | t(1;14)(q21;q32) | BCP-ALL | R | ++ | |
| Patient material | |||||
| 1 | der(14)t(1;14)(q21;q32) | B-NHL | NT | NT | 1, 1 |
| 2 | t(1;22)(q21;q11) | B-NHL/FL | G | NT | 1, der(1), der(22) − split |
| 3 | t(1;22)(q21;q11) | B-NHL/DLCL | G | NT | NT |
| 4 | t(1;22)(q21;q11) | B-NHL/FL | NT | NT | 1, der(1) |
| 5 | der(22)t(1;22)(q21;q11) | B-NHL | NT | NT | 1, 1, der(22) |
| 6 | der(1q) | B-NHL/MCL | R | NT | 1, der(1) |
| 7 | add(1), der(19)t(1;19)(q21;p13) | B-NHL | NT | NT | add(1) amplified + der(19) |
| 8 | t(1;3)(q21;q27) | B-NHL/FL | NT | NT | 1, der(1) |
| 9 | der(3)t(1;3)(q21;q27), der(17)t(1;17) (q21;13) | B-NHL | NT | NT | NT |
| 10 | der(3)t(1;3)(q21;p22) | B-NHL | G | NT | NT |
| 11 | t(1;5)(q21;p14) | B-NHL/MCL | G | NT | 1, der(5) |
| 12 | der(6)t(1;6)(q21;q22) | B-NHL | NT | NT | 1, 1, der(6) |
| 13 | der(10)t(1;10)(q21;q24) | B-NHL | NT | NT | 1, 1, der(10) |
| 14 | der(12)t(1;12)(q21;q13) | B-NHL/DLCL | G | NT | NT |
| 15 | der(15)t(1;15)(q21;pter) | B-NHL | NT | NT | 1, 1, der(15) |
| 16 | der(16)t(1;16)(q21;q24) | B-NHL/FL | G | NT | 1, 1, der(16) |
| 17 | der(16)t(1;16)(q21;q21) | ALL | G | NT | NT |
| 18 | der(17)t(1;17)(q21;q25) | B-NHL | NT | NT | 1, 1 |
| 19 | der(20)t(1;20)(q21;p12) | B-NHL | G | NT | 1, 1 |
| 20 | der(21)t(1;21)(q21;q22) | B-NHL | NT | NT | 1, 1 |
| 21 | add(1)(q12) | B-NHL | NT | NT | 1, der(1) |
| 22 | dup(1)(q21q24) | ALL | G | NT | 1, duplicated on dup(1) |
| Cell lines | |||||
| UOCB-1 | der(12)(1;12)(q21;q13) | BCP-ALL | NT | − | NT |
| AS283A | der(4)t(1;4)(q21;q35) | Burkitt's | NT | − | NT |
| BL 32 | dup(1)(q21q32) | Burkitt's | G | − | 1, duplicated on dup(1) |
| BL 92 | dup(1)(q21q25) | Burkitt's | NT | − | NT |
| BL 103 | invdup(1)(q21q25) | Burkitt's | NT | − | NT |
| BL 104 | t(1;3)(q21;p25) | Burkitt's | NT | NT | 1, der(1) |
| BL 115 | dup(1)(q21q31) | Burkitt's | G | − | 1, duplicated on dup(1) |
| BL 136 | der(1)(1pter-q21∷1q21-q36∷1q21q36∷1q21-qter) | Burkitt's | NT | − | NT |
| Namalwa E6 | dup(1)(q21q32) | Burkitt's | NT | − | NT |
| Case No. . | Cytogenetic 1q21 Abnormality . | Histology . | Southern Blot (X665) . | BCL9 Overexpression . | Location of YAC 882B3 Signals . |
|---|---|---|---|---|---|
| CEMO-1 | t(1;14)(q21;q32) | BCP-ALL | R | ++ | |
| Patient material | |||||
| 1 | der(14)t(1;14)(q21;q32) | B-NHL | NT | NT | 1, 1 |
| 2 | t(1;22)(q21;q11) | B-NHL/FL | G | NT | 1, der(1), der(22) − split |
| 3 | t(1;22)(q21;q11) | B-NHL/DLCL | G | NT | NT |
| 4 | t(1;22)(q21;q11) | B-NHL/FL | NT | NT | 1, der(1) |
| 5 | der(22)t(1;22)(q21;q11) | B-NHL | NT | NT | 1, 1, der(22) |
| 6 | der(1q) | B-NHL/MCL | R | NT | 1, der(1) |
| 7 | add(1), der(19)t(1;19)(q21;p13) | B-NHL | NT | NT | add(1) amplified + der(19) |
| 8 | t(1;3)(q21;q27) | B-NHL/FL | NT | NT | 1, der(1) |
| 9 | der(3)t(1;3)(q21;q27), der(17)t(1;17) (q21;13) | B-NHL | NT | NT | NT |
| 10 | der(3)t(1;3)(q21;p22) | B-NHL | G | NT | NT |
| 11 | t(1;5)(q21;p14) | B-NHL/MCL | G | NT | 1, der(5) |
| 12 | der(6)t(1;6)(q21;q22) | B-NHL | NT | NT | 1, 1, der(6) |
| 13 | der(10)t(1;10)(q21;q24) | B-NHL | NT | NT | 1, 1, der(10) |
| 14 | der(12)t(1;12)(q21;q13) | B-NHL/DLCL | G | NT | NT |
| 15 | der(15)t(1;15)(q21;pter) | B-NHL | NT | NT | 1, 1, der(15) |
| 16 | der(16)t(1;16)(q21;q24) | B-NHL/FL | G | NT | 1, 1, der(16) |
| 17 | der(16)t(1;16)(q21;q21) | ALL | G | NT | NT |
| 18 | der(17)t(1;17)(q21;q25) | B-NHL | NT | NT | 1, 1 |
| 19 | der(20)t(1;20)(q21;p12) | B-NHL | G | NT | 1, 1 |
| 20 | der(21)t(1;21)(q21;q22) | B-NHL | NT | NT | 1, 1 |
| 21 | add(1)(q12) | B-NHL | NT | NT | 1, der(1) |
| 22 | dup(1)(q21q24) | ALL | G | NT | 1, duplicated on dup(1) |
| Cell lines | |||||
| UOCB-1 | der(12)(1;12)(q21;q13) | BCP-ALL | NT | − | NT |
| AS283A | der(4)t(1;4)(q21;q35) | Burkitt's | NT | − | NT |
| BL 32 | dup(1)(q21q32) | Burkitt's | G | − | 1, duplicated on dup(1) |
| BL 92 | dup(1)(q21q25) | Burkitt's | NT | − | NT |
| BL 103 | invdup(1)(q21q25) | Burkitt's | NT | − | NT |
| BL 104 | t(1;3)(q21;p25) | Burkitt's | NT | NT | 1, der(1) |
| BL 115 | dup(1)(q21q31) | Burkitt's | G | − | 1, duplicated on dup(1) |
| BL 136 | der(1)(1pter-q21∷1q21-q36∷1q21q36∷1q21-qter) | Burkitt's | NT | − | NT |
| Namalwa E6 | dup(1)(q21q32) | Burkitt's | NT | − | NT |
Twenty-two cases of fresh patient material and 3 malignant B-cell lines derived from various disease subtypes were studied by either Southern blot using the 3′ BCL9 probe X665 and/or the nonchimeric BCL9 YAC 882B3: cell lines were additionally studied by Northern blot. One case showed a der(14)t(1;14)(q21;q32) similar to that seen in CEMO-1: FISH with the BCL9 YAC showed a telomeric 1q21 breakpoint. Four cases showed an apparent variant translocation t(1;22)(q21;q11): direct involvement ofIGλ was shown in 2 cases by FISH using cosmids spanning the IGλ locus (I.W., unpublished observations). One of these cases exhibited a breakpoint within theBCL9 YAC, whereas the breakpoint was telomeric in 1 case and centromeric in another: the final case was untested due to insufficient material. One case with no cytogenetic abnormality of 1q21 and no abnormality of YAC 882B3 showed rearrangement with the probe X665 on southern blot (Fig 6): this was a case of mantle cell lymphoma. Also, 1 case with t(1;19)(q21;p13) showed amplification of the BCL9YAC. Otherwise, none of the other cases appeared to involve theBCL9 locus. These cases included 2 with t(1;3)(q21;q27) in which both breakpoints were telomeric of BCL9, 2 with t(1;12)(q21;q13) and 6 cases with dup(1)(q21qter) with a variable distal breakpoint cytogenetically: other partner chromosomes were unique. 1q21 breakpoints in these cases were heterogeneous, falling both centromeric and telomeric of the BCL9 gene.
Abbreviations: BCP-ALL, B-cell precursor ALL; FL, follicular lymphoma; MCL, mantle cell lymphoma; R, rearranged; G, germline; NT, not tested; ++, overexpression; −, no overexpression.
Southern blot analysis.
DNA blotting was performed as previously described.22Briefly, high molecular weight DNA was digested with Bgl II,HindIII, Kpn I, and Xba I (GIBCO-BRL, Gaithersburg, MD) for 3 to 4 hours at 37°C, electrophoresed in 0.8% agarose and transferred to positively charged nylon membranes (Hybond N+; Amersham, Amersham, UK) by overnight capillary transfer. Hybridization to a radiolabeled IGHJ probe (clone C76R51A) and to BCL9 probes was performed as previously described.22 Filters were autoradiographed at −80°C with intensifying screens for up to 96 hours.
LDI-PCR cloning of IGHJ rearrangements.
High molecular weight DNA from CEMO-1 was digested to completion withHindIII and ligated at low concentration to promote the formation of monomeric circles. The DNA was then purified using a Wizard column (Promega, Madison, WI) and 10 ng was amplified in a nested PCR reaction using primers designed to anneal to the JHand IGH enhancer regions. The sequences of these primers were as follows: External J6 (J6E), 5′-CCCACAGGCAGTAGCAGAAAACAA-3′; and External Hind (JHE), 5′-TGGGATGCGTGGCTTCTGCT-3′; and the nested primers Internal J6 (J6I), 5′-TCTGGGCTCGAGTCGACGCAGAAAACAAAGGCCCTAGAGGG-3′; and Internal Hind (JHI), 5′-GCCCTTGTTAATGGACTTGGAGGA-3′. After PCR, aliquots of the reaction were run in a 0.8% agarose gel. The bands were then excised from the gel, purified using Qiaquick spin columns (Qiagen, Hilden, Germany), and ligated into the TA cloning vector pCR2.1 (Invitrogen, San Diego, CA). Aliquots of the ligation reaction were transformed by electroporation and grown in selective media. Plasmid DNA was prepared using standard methods.
DNA sequencing.
Automated sequencing was performed using an ABI PRISM automated sequencer and AmpliTaq polymerase, FS (Perkin Elmer, Foster City, CA). Sequences were checked against the Genbank and EMBL databases using BLAST and FASTA programs made available through the Human Genome Mapping Project Resource Centre (HGMP-RC; Hinxton Hall, Cambridge, UK). Larger clones were sequenced by primer walking or by subcloning into pBluescript (Stratagene, La Jolla, CA).
Isolation of PAC and YAC clones and fluorescence in situ hybridization (FISH).
PAC and YAC clones containing BCL9 sequences were isolated by radioactive screening using BCL9 cDNA probes from gridded PAC and YAC libraries provided by the HGMP-RC. DNA was extracted from PAC and YAC clones according to standard techniques.
FISH was performed according to standard protocols as previously described.23 Briefly, the probes were labeled by nick-translation with biotin-16 deoxyuridine triphosphate (Boehringer Mannheim, Mannheim, Germany). Competition hybridization by preannealing with a 100- to 200-fold excess of Cot-1 DNA (GIBCO-BRL) was performed before overnight hybridization on slides with previously denatured DNA. Probes were visualized by subsequent incubations with streptavidin-fluorescein isothiocyanate (FITC), biotinated goat antistreptavidin, and streptavidin-FITC (Vector Laboratories, Burlingame, CA). Cells and chromosomes were counterstained with diamidino-phenylindole (DAPI) and embedded in Vectashield (Vector Laboratories). Digital images were captured using a Photometrics cooled-coupled device (CCD) camera mounted on a Zeiss Axioplan fluorescence microscope equipped with selective filters for fluorescein and DAPI and controlled by a Quadra 950 MacIntosh computer using SmartCapture software (Digital Scientific, Cambridge, UK).
Northern blot analysis.
A commercial multiple tissue Northern blot with 2 μg poly(A+) RNA from a variety of tissues (Clontech, Palo Alto, CA) was screened with a 665-bp Xho I fragment single-copy probe (probe X665) derived from the 1q21 breakpoint region. For cell lines and patient samples, total RNA was isolated by the guanidium thiocyanate/cesium chloride method and was electrophoresed on a 1.0% agarose gel containing 2.2 mol/L formaldehyde and transferred to nylon membranes (Hybond N; Amersham). Hybridization with radiolabeled GAPDH and BCL9cDNA probes (X665) was performed at 68°C for 90 minutes using Rapid-Hyb buffer (Amersham). The filters were washed with 2× SSC, 0.05% sodium dodecyl sulfate (SDS) at room temperature for 30 minutes and with 0.1× SSC, 0.1% SDS at 65°C for 30 minutes. Filters were autoradiographed at −80°C for up to 7 days.
Isolation of cDNA clones.
cDNA clones were isolated by screening an oligo (dT)-primed normal human fetal brain cDNA λZAP library (Stratagene) with probe X665. Positive phage clones were excised from the λZap phage as pBluescript SK− plasmids, subcloned, and sequenced. Rapid amplification of cDNA ends (RACE) was performed using Marathon Ready skeletal muscle cDNA (Clontech) and Advantage cDNA PCR kit (Clontech). PCR primers were sense 5′-GCTCTCCAGGCATGATGATGT-3′ (nucleotide 4866) and 5′-CATGATGGGACCCAACAGAACATC-3′ (nucleotide 4897) for 3′ RACE and antisense 5′-TCTGGGGGGACTTGAGGTTCC-3′ (nucleotide 3381) and 5′-CATTGGTGACTGGACTTGGTGCAT-3′ (nucleotide 3341) followed by antisense 5′-GCTGTGCTTCTCTCTGTCTTGTTGTTAG-3′ (nucleotide 1365) and 5′-GTTCTGGATGTGGA-AAGAGACGATAGTT-3′ (nucleotide 1333) for 5′ RACE. RACE products were cloned into the TA cloning vector pCR2.1 (Invitrogen) and sequenced.
RESULTS
Molecular cloning of a t(1;14) breakpoint in CEMO-1.
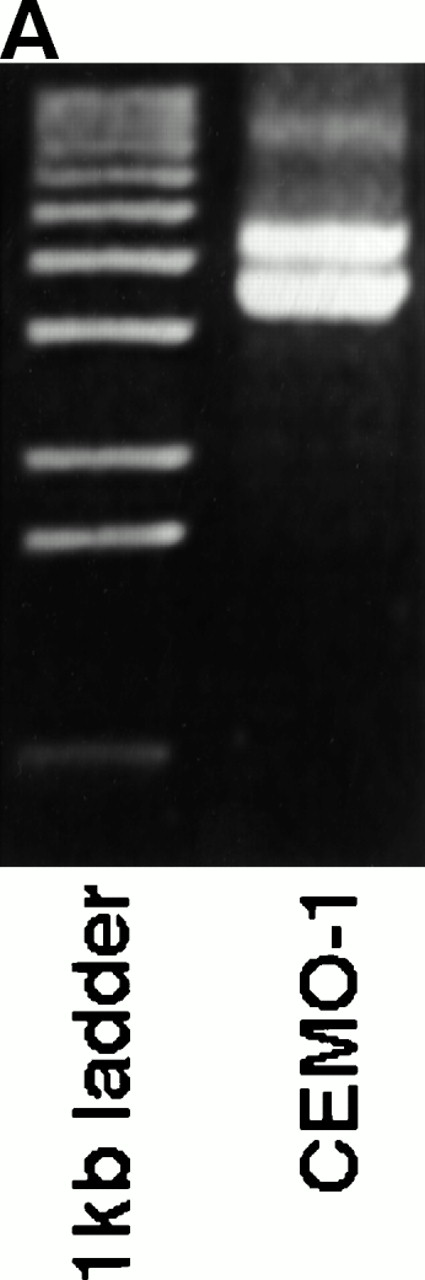
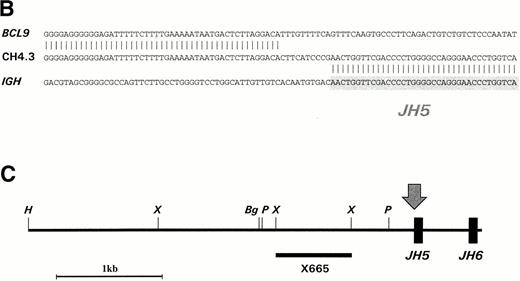
Southern blot analysis of HindIII-digested DNA from CEMO-1 using the JH probe showed two rearranged bands of 5.2 and 5.8 kb. These rearrangements were amplified by long-distance inverse PCR, resulting in two products of 3.7 and 4.3 kb (Fig 1A), which were cloned and sequenced. Sequence analysis of the 3.7-kb rearrangement showed a productive and unmutated VH6DIR1DN1JH6 rearrangement (data not shown). In the other allele, there was loss of homology with IGH sequences upstream of JH5 (Fig 1B). A restriction map of this 4.3-kb clone (CH4.3) is shown in Fig 1C. Further sequencing of this clone showed a region of identity with an EST (accession no.W23092) cloned from a human retinal cDNA library, but no homology with any other sequences on the database. A single-copy 665-bp Xho I fragment (probe X665) containing this region of identity was used to screen a gridded PAC library, resulting in the isolation of two PAC clones that were mapped back to chromosome 1q21 on normal metaphases by FISH (Fig 2A), confirming that this rearrangement represented the t(1;14) translocation breakpoint. In addition, the probe X665 gave a rearranged signal on Southern blot that comigrated with the 5.8-kb rearranged JHHindIII fragment (data not shown).
(A) LDI-PCR products from HindIII-digested CEMO-1 DNA. Bands of 3.7 and 4.3 kb were amplified, corresponding to the rearrangements seen on Southern blot. Five microliters of PCR product was analyzed on 0.8% agarose stained with ethidium bromide. (B) DNA sequence analysis of translocation breakpoint from 4.3-kb LDI-PCR product (clone CH4.3). DNA sequence is compared with germline IGHJsequence and BCL9 cDNA sequence. Shaded region denotesJH5 sequence. (C) Restriction map of LDI-PCR clone CH4.3. The breakpoint in CEMO-1 is indicated by an arrow. Probe X665 is indicated by heavy horizontal bar. This probe was used to screen PAC library and for Northern and Southern analysis. Bg, Bgl II;H, HindIII; P, Pst I; X,Xho I.
(A) LDI-PCR products from HindIII-digested CEMO-1 DNA. Bands of 3.7 and 4.3 kb were amplified, corresponding to the rearrangements seen on Southern blot. Five microliters of PCR product was analyzed on 0.8% agarose stained with ethidium bromide. (B) DNA sequence analysis of translocation breakpoint from 4.3-kb LDI-PCR product (clone CH4.3). DNA sequence is compared with germline IGHJsequence and BCL9 cDNA sequence. Shaded region denotesJH5 sequence. (C) Restriction map of LDI-PCR clone CH4.3. The breakpoint in CEMO-1 is indicated by an arrow. Probe X665 is indicated by heavy horizontal bar. This probe was used to screen PAC library and for Northern and Southern analysis. Bg, Bgl II;H, HindIII; P, Pst I; X,Xho I.
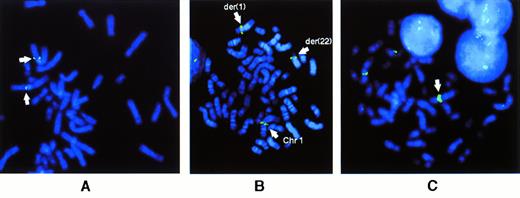
FISH results for BCL9 probes. (A) Localization of PAC containing BCL9 sequences to chromosome 1q21 on normal metaphases. (B) Rearrangement of CEPH YAC 882B3 in case no. 2 with t(1;22)(q21;q11), showing hybridization signals at the breakpoints of both der(1) and der(22) chromosomes. (C) Metaphase of case no. 7 showing YAC 882B3 hybridization signals on normal 1 and der(19) chromosomes. Amplification of hybridization signal on the add(1) is shown by a solid arrow. Note the extra unidentified material between centromere and YAC signal.
FISH results for BCL9 probes. (A) Localization of PAC containing BCL9 sequences to chromosome 1q21 on normal metaphases. (B) Rearrangement of CEPH YAC 882B3 in case no. 2 with t(1;22)(q21;q11), showing hybridization signals at the breakpoints of both der(1) and der(22) chromosomes. (C) Metaphase of case no. 7 showing YAC 882B3 hybridization signals on normal 1 and der(19) chromosomes. Amplification of hybridization signal on the add(1) is shown by a solid arrow. Note the extra unidentified material between centromere and YAC signal.
Isolation of BCL9 cDNA clones.
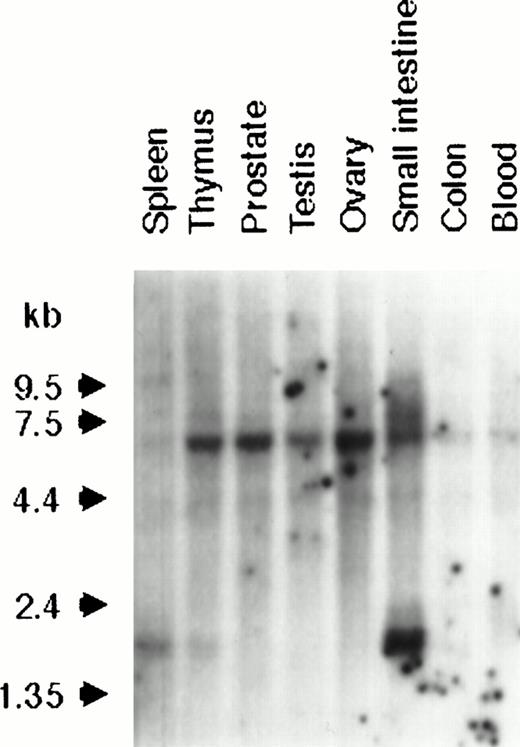
Because of its identity with an EST, probe X665 was hybridized to multiple tissue Northern blots containing 2 μg poly(A+) RNA from a variety of different tissues. Two low-level transcripts were detected in all tissues, a major transcript of 6.3 kb and a less prominent 4.2-kb species, whereas a third transcript of 1.6 kb was detected only in spleen, thymus, and, most strongly, in small intestine (Fig 3).
Northern analysis of BCL9 expression in a variety of normal human tissues. A multiple tissue Northern blot was hybridized with the BCL9 probe X665. Reprobing with a GAPDH probe showed equal RNA loading in all samples (data not shown).
Northern analysis of BCL9 expression in a variety of normal human tissues. A multiple tissue Northern blot was hybridized with the BCL9 probe X665. Reprobing with a GAPDH probe showed equal RNA loading in all samples (data not shown).
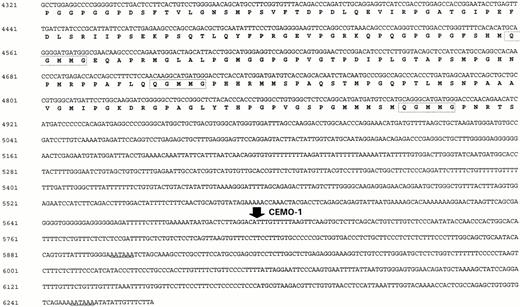
Probe X665 was therefore used to screen a normal fetal brain cDNA library and two overlapping clones covering a region of 1.5 kb were cloned and sequenced. In addition, primers were designed within the region of identity with the EST for 3′ RACE. Amplification in the 3′ direction yielded a major product of 1.6 kb that was cloned and sequenced. To clone the 5′ end of the gene, 5′ RACE was performed. In the first stage, a major product of 2.1 kb was amplified, cloned, and sequenced. Primers were designed from this sequence and a further round of 5′ RACE was performed, resulting in a product of 1.3 kb. The full-length BCL9 sequence was found to be 6,268 bp, with no further regions of nucleic acid homology on the databases other than identity with some ESTs. This sequence (accession no. Y13620) is shown together with the predicted amino acid translation in Fig 4. Sequence analysis showed a long open reading frame of 4.1 kb with termination codons in all three reading frames 5′ of it. The first ATG codon of this ORF was at nucleotide 740; it was unclear whether this represented the site of translation initiation because it did not adhere to the Kozak consensus sequence.24 The second ATG codon, at nucleotides 818, showed a closer resemblance to a potential translation initiation site.
BCL9 cDNA sequence (Y13620) together with predicted amino acid translation. (i) Breakpoint in CEMO-1 in theBCL9 3' UTR is indicated by a solid arrow. (ii) Polyadenylation sites are underlined. (iii) Potential nuclear localization signal is indicated by shaded box. (IV) Region with strong (90%) amino acid homology to Drosophila embryo EST (accession no.AA202331) is underlined. Outside this region, the EST lacked homology with BCL9. (v) Pentapeptide repeats are indicated by open boxes (sequence with residue numbers in parentheses): SPSGN (11-15, 35-39, 878-882)(; SSTPL (294-298, 1020-1024); SSTP (988-992, 1019-1023); and QGMMG (1274-1278, 1324-1328, 1385-1389).
BCL9 cDNA sequence (Y13620) together with predicted amino acid translation. (i) Breakpoint in CEMO-1 in theBCL9 3' UTR is indicated by a solid arrow. (ii) Polyadenylation sites are underlined. (iii) Potential nuclear localization signal is indicated by shaded box. (IV) Region with strong (90%) amino acid homology to Drosophila embryo EST (accession no.AA202331) is underlined. Outside this region, the EST lacked homology with BCL9. (v) Pentapeptide repeats are indicated by open boxes (sequence with residue numbers in parentheses): SPSGN (11-15, 35-39, 878-882)(; SSTPL (294-298, 1020-1024); SSTP (988-992, 1019-1023); and QGMMG (1274-1278, 1324-1328, 1385-1389).
Analysis of the BCL9 predicted protein of 1,394 amino acids showed that it was rich in proline (18%), glycine (11%), serine (11%), and methionine (6%) residues. Several pentapeptide repeats (SPSGN, SSTPL, PSSTP, QGMMG, and PGPVG) were identified. A potential nuclear localization signal (KRRK) was identified at residues 243-246, but there were no other recognizable protein domains. There was also no homology with other proteins on the databases, although one region showed amino acid homology (90% homology >30 amino acids) to an EST cloned from a Drosophila embryo cDNA library. The sequences of the shorter transcripts of 4.2 and 1.6 kb have not yet been determined.
The translocation t(1;14) in CEMO-1 results in overexpression of BCL9.
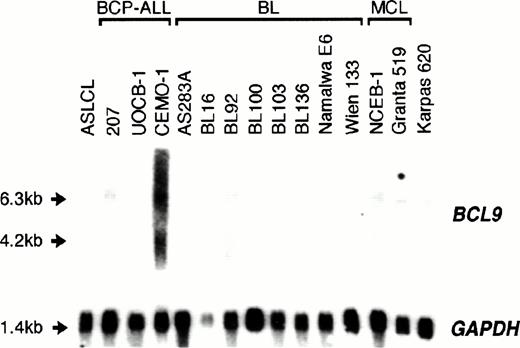
Sequence analysis showed that the t(1;14)(q21;q32) breakpoint in CEMO-1 fell within the 3′ UTR of the BCL9 gene: in this the translocation was comparable to those involving IGJH and the major breakpoint region of BCL2, where there is no disruption of the ORF. To assess whether the juxtaposition of BCL9 toIGH resulted in its deregulation, expression of BCL9was determined in CEMO-1 and also in other malignant and nonmalignant B-cell lines. At least 50-fold overexpression ofBCL9, as calculated by densitometric analysis of hybridization bands, was detected in CEMO-1 as compared with ASLCL, an Epstein-Barr virus (EBV)-transformed normal B-lymphoblastoid cell line (Fig 5). Both a truncated BCL9transcript of 6.3 kb and the 4.2-kb transcript were overexpressed, but not the 1.6-kb transcript. However, no such overexpression was detected in other cell lines with and without abnormalities of 1q21, including 2 precursor B-cell ALL cell lines, 10 BL cell lines, 2 mantle cell lymphoma cell lines, and 1 plasma cell leukemia cell line (Karpas 620). These data are shown in Table 1.
Northern blot analysis of 10 μg total RNA from various cell lines of B-cell lineage. ASLCL, an EBV-transformed normal B-lymphoblastoid cell line acted as negative control. Both a truncated 6.3-kb and the 4.2-kb BCL9 transcript were overexpressed in the CEMO-1 cell line but not in any other B-cell lines with 1q21 abnormalities (Table 1).
Northern blot analysis of 10 μg total RNA from various cell lines of B-cell lineage. ASLCL, an EBV-transformed normal B-lymphoblastoid cell line acted as negative control. Both a truncated 6.3-kb and the 4.2-kb BCL9 transcript were overexpressed in the CEMO-1 cell line but not in any other B-cell lines with 1q21 abnormalities (Table 1).
Rearrangements of BCL9 in other cases with abnormalities of 1q21.
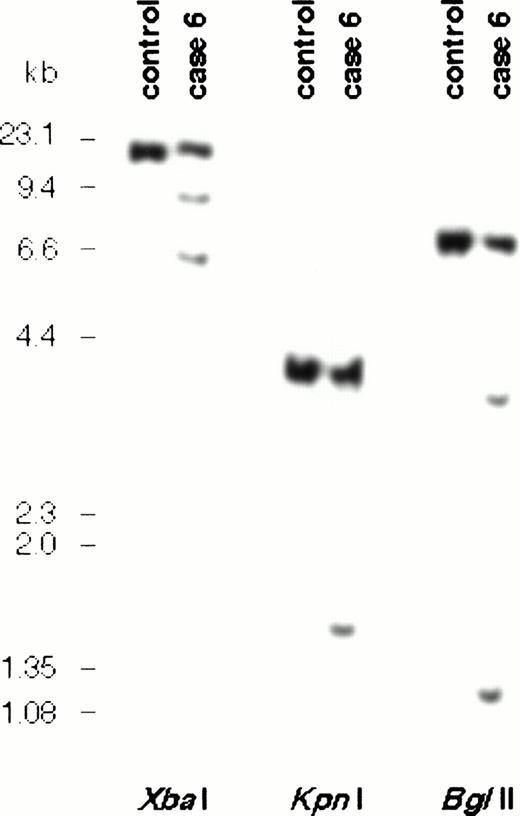
To assess the extent of involvement of BCL9 in other cases of B-cell malignancy with abnormalities of 1q21 but in which no RNA was available, rearrangements of BCL9 were sought in a panel of 39 cases with 1q21 abnormalities. Conventional DNA blot using the 3′ probe X665 detected additional rearrangements in multiple digests in case no. 6 (Table 1 and Fig 6). Southern blot detected two additional bands in Bgl II and Xba I digests, showing that the breakpoint fell within the X665 probe itself. These data indicated that the 3′ UTR within the BCL9 gene may contain a breakpoint cluster region. However, in this case, although the G-banding pattern of one copy of chromosome 1q was abnormal, no cytogenetic abnormality of 1q21 could be detected by either regular cytogenetics or by FISH using a BCL9 YAC. Furthermore, hybridization with a chromosome 1 paint showed that all of the abnormal chromosome material telomeric to BCL9 was derived from chromosome 1. These data therefore indicate that these rearrangements probably represented an internal chromosome 1q21 deletion or insertion. Molecular cloning of the rearrangements is being undertaken.
Southern blot analysis of DNA from case no. 6 and normal control using probe X665. Two rearranged signals were seen inBgl II and Xba I digests, indicating that the breakpoint fell within the region of this probe in the 3′ UTR ofBCL9.
Southern blot analysis of DNA from case no. 6 and normal control using probe X665. Two rearranged signals were seen inBgl II and Xba I digests, indicating that the breakpoint fell within the region of this probe in the 3′ UTR ofBCL9.
We then analyzed 17 cases and 3 cell lines by FISH using the nonchimeric CEPH YAC 882B3, which contains the BCL9 gene, as a probe, to assess the extent of involvement of BCL9 in cases in which breakpoints fell outside the range of conventional DNA blot (Table 1). Four cases and 1 cell line exhibited reciprocal translocations involving 1q21. Case no. 2, a case of follicular lymphoma with the translocation t(14;18) exhibiting the additional translocation t(1;22)(q21;q11) involving the IGλ locus at 22q11 (I.W., unpublished observations), showed rearrangement involving the genomic region of 850 kb covered by this YAC (Fig 2B). An additional case (case no. 7) that was initially described as having add(1)(q42) and der(19)t(1;19)(q21;p13) showed amplification of the YAC signal on the der(1) chromosome with apparently normal signals on both the normal 1 and the der(19) chromosomes. The abnormal derivative 1 chromosome, carrying the amplified signal, also showed the presence of additional material between the centromere of chromosome 1 and the hybridization signal, suggesting the presence of an insertion within the long arm of chromosome 1 (Fig 2C).
Results in the other cases showed a heterogeneity of breakpoints at 1q21, breaks occurring both telomeric and centromeric of BCL9with equal frequency (10 each). However, 12 cases exhibited unbalanced abnormalities of chromosome 1; therefore, breakpoints falling within the YAC may not have been readily detectable. Additional experiments with YAC clones flanking BCL9 are currently being performed to assess the location of breakpoints in these cases.
DISCUSSION
In B-cell malignancy, a number of recurrent translocations involving the IG loci have been characterized. However, a number of other recurrent cytogenetic abnormalities that only rarely involve theIG loci have also been identified, some of which are of prognostic significance. Among these are breakpoints involving 1q21-q23 and 1p32-36, which are associated with a poor prognosis and appear to exhibit promiscuity in the translocation partners with which they rearrange.16 In this, both loci are comparable to 3q27, the location of the BCL6 gene, which has been shown to be translocated to a large number of loci other than the IGloci, eg, the t(3;4)(q27;p13) involving the TTF gene at 4p1325 and the t(3;11)(q27;q23) involving the BOB1gene at 11q23.26
Using a PCR-based method for cloning IGHJrearrangements,14 we have identified a novel gene,BCL9, at chromosome 1q21 through the cloning of a translocation t(1;14)(q21;q32) in a precursor B-cell ALL cell line. This gene coded for a large proline-rich protein that contained a potential nuclear localization signal but no other significant motifs and no homology with known proteins. One region of BCL9 showed homology over a region of 30 amino acids to an EST cloned from a Drosophila cDNA library. The significance of this homology remains to be determined: further sequencing of the full-length Drosophila cDNA clone is currently being undertaken. We have shown that one consequence of the t(1;14)(q21;q32) in the CEMO-1 cell line was the overexpression ofBCL9. This gene is normally expressed at very low levels in B cells, and the pathological consequences of such overexpression remain to be determined. Expression of a 1.6-kb transcript was seen in spleen, thymus, and small intestine, suggesting that this isoform may be differentially involved in cell proliferation. Interestingly, this isoform was not seen in CEMO-1.
We have also demonstrated that breaks within the region around BCL9were found in some cases of B-NHL with abnormalities of 1q21. In one case, the breakpoint was within the 3′ UTR of BCL9, similar to that found in CEMO-1, indicating that this may be a recurrent breakpoint. Furthermore, FISH analysis using an 850-kb YAC indicated that, in 1 of 17 other cases with abnormalities of 1q21, the breakpoint fell close to the BCL9 gene. This case exhibited the translocation t(1;22)(q21;q11) involving the IGλ locus, which would appear to be a variant of the t(1;14)(q21;q32), similar to the variant translocation t(8;22)(q24;q11) involving MYC andIGλ. By analogy with rearrangements of BCL1, BCL2,MYC, or PAX-5 loci in other cases of B-NHL in which deregulation by IG transcriptional enhancers can occur at distances of several hundred kilobases,4,12 27-29 it is feasible that BCL9 expression could be deregulated at this distance by regulatory elements within the IG loci. In case no. 6, both conventional cytogenetics and FISH failed to show any abnormality of the BCL9 locus, suggesting the presence of a submicroscopic chromosomal rearrangement. In case no. 7, there was clear evidence of amplification of the YAC hybridization signal on an abnormal 1q chromosome. Whether these abnormalities resulted in increased BCL9 expression could not be determined due to a lack of suitable material.
However, it is clear that not all rearrangements of 1q21 in B-cell malignancy involve BCL9. We failed to find evidence of gene rearrangement and/or BCL9 overexpression in 13 cell lines of various subtypes of B-cell malignancy and in 19 of 22 cases of ALL/B-NHL with abnormalities of 1q21. FISH analysis showed a surprising heterogeneity of 1q21 breakpoints: these fell both centromeric and telomeric of BCL9 with equal frequency, although in many of these cases the translocations were unbalanced and therefore rearrangement of the YAC signal would not have been clearly seen, and RNA was not available for analysis of BCL9 expression.
Abnormalities of the long arm of chromosome 1, and in particular the 1q21 region, are seen in a variety of preneoplastic and neoplastic diseases, including both hematopoietic malignancies and solid tumors,30-32 and in some cases occur as an early event. Further work will be required to determine what role the BCL9gene has to play in these other diseases and to identify other genes of pathological importance in B-cell malignancy.
ACKNOWLEDGMENT
The authors thank Rifat Hamoudi, Rachel Jackson, Bina Desai, and Samantha Dibley (Gene Cloning Facility, ICR) for their help with automated sequencing. We also thank Prof Gilbert Lenoir for kindly providing the BL lines used in this study and Prof Antonino Carbone (Centro Regionale di Riferimento Oncologico, INRCCS, Aviano, Italy) for kindly providing the AS283A cell line.
Supported in part by grants from the Leukaemia Research Fund (T.G.W.), the Kay Kendall Leukaemia Research Trust (L.J.A.C., D.M.J., and M.J.S.D.), and the Louise Buchanan-Fulbright Scholarship (M.J.S.D.). I.R.Z. and M.L.M.S. acknowledge the support of the International Outreach Program of St Jude's Children's Hospital (Memphis, TN).
Address reprint requests to M.J.S. Dyer, MD, DPhil, Academic Department of Haematology and Cytogenetics, Haddow Laboratories, Institute of Cancer Research, Sutton, Surrey SM2 5NG, UK.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal