Abstract
Myoepithelial sialadenitis (MESA) is the reactive salivary gland lymphoid infiltrate that occurs in patients with Sjögren's syndrome. Although it is well established that mucosa-associated lymphoid tissue (MALT)-type lymphomas may develop from MESA, the issue of whether monoclonal B-cell populations in early MESA-associated lesions represent MALT lymphomas or more benign types of expansions has been very controversial. In addition, it is unknown whether antigen stimulation plays a role in the development or growth of MESA-associated clones. To investigate these issues, we have analyzed the Ig VH genes used by MESA-associated clones in sequential biopsies obtained from contralateral sites of seven different patients. In three cases, single clones were identified in the follow-up biopsies that were distinct from the single clones identified in the initial specimens, whereas in three other cases, the same clone was identified in both the initial and subsequent specimens. In the remaining case, two clones were identified in the second biopsy specimen, one of which was distinct from the initial clone. Of the 11 distinct clones identified in the 14 specimens that were analyzed, 8 were derived from a V1-69 VH gene segment, whereas the other 3 were derived from a V3-7 VH gene segment. In addition, the MESA clones also showed conserved amino acids sequence motifs in their third complementarity-determining regions (CDR3), some of which were encoded by N nucleotides. The marked VH gene restriction along with the similar CDR3 sequences suggests that MESA-associated clones even from different patients may bind the same or similar antigens and are selected for clonal expansion on that basis. The high rates of ongoing VH gene mutation observed in some of the cases futher suggest that the growth of early MESA clones is still dependent on antigen stimulation. In addition, our finding that different biopsies from the same patient may contain distinct clones indicates that some MESA-associated clones have not yet evolved to malignant lymphomas.
MOST PRIMARY SALIVARY gland lymphomas are of mucosa associated lymphoid tissue (MALT) type and are thought to arise from a reactive infiltrate termed myoepithelial sialadenitis (MESA) that is also known as benign lymphoepithelial lesion.1-3 MESA is found in patient's with Sjögren's syndrome and is responsible for the salivary gland dysfunction that occurs in this disease but can also be seen in patients who may not have Sjögren's syndrome. The transition from reactive MESA to monoclonal lymphoma is not well understood, although there has been speculation that chronic stimulation by exogenous or autoantigens is playing an important role by driving the proliferation of specific B cells and increasing the frequency of their transformation.4-8 Reports that the proportion of B cells expressing the 17.109 and G6 idiotypes are increased in salivary gland biopsies from patients with Sjögren's syndrome compared with controls suggest that the B-cell infiltration early on represents a selected population.5-7 Antigen selection of specific B cells for transformation was also suggested by our earlier study of salivary gland MALT lymphomas, in which 3 of the 5 cases examined used the 51p1 VH and Humkv325 VL genes that encode the G6 and 17.109 idiotypes, respectively.8 A role for antigen stimulation in the growth of MALT lymphomas has also been proposed for those that occur in the stomach that are thought to develop out ofHelicobacter pylori-associated gastritis.9Remarkably, several studies have now shown that the majority of low-grade gastric MALT lymphomas may be curable using only antibiotics that eliminate the H pylori infection.10 11
It is well recognized that early salivary gland MALT lymphomas can be very difficult to distinguish from reactive MESA, which is also invariably present.1 Histologic criteria to make this distinction are controversial and have evolved with the recognition that MALT-type lymphomas represent a distinct type of extranodal lymphoma.1,12,13 The significance of finding monoclonal B-cell populations by polymerase chain reaction (PCR) or Southern blot analysis is also unclear, because they have been detected in MESA-associated salivary gland lesions that histologically appear benign.13,14 Although it is possible these clones represent very early lymphomas, as some have argued,15,16 clones in histologically benign MESA or early low-grade MALT lymphomas often behave clinically in a very indolent fashion and can remain localized to the salivary glands for years without evidence of other spread.13,17 Moreover, it is well recognized that patients with MESA and Sjögren's syndrome often have monoclonal paraproteins or cryoglobulins that are thought to reflect a stable nonmalignant lymphoproliferative disorder.18
To further evaluate the malignant potential and evolution of B-cell clones in MESA-associated salivary gland lesions, we studied the clonal VH genes in successive biopsies from seven patients that had been obtained from contralateral sites. Sequence analysis of VH genes can provide considerable information regarding how different clones may be related and whether antigen selection may be playing a role in clonal evolution.19-21 Although both MESA and MALT lymphomas frequently show bilateral involvement of salivary glands, the relationship, if any, among clones from such lesions has not been extensively studied.17 22 Finding that different biopsies of MESA or low-grade MALT lymphoma from the same patient can contain different clones supports a model of lymphomagenesis in which the emergence of monoclonality in the salivary gland precedes the onset of malignant lymphoma. Moreover, the restricted use of VH gene segments and CDR3 sequences we found strongly suggest that the B-cell clones associated with MESA lesions in the salivary gland arise through the process of antigenic selection.
MATERIALS AND METHODS
Patient material and DNA isolation.
The 16 biopsy specimens were obtained from patients seen at the University of Pittsburgh Medical Center and satisfied histologic criteria for MESA, MESA with halos of monocytoid cells, or low-grade MALT lymphoma as described.13 DNA was isolated from the three fresh tissue biopsy specimens (HA #1 and #2, OR #1) as described.8 For the remaining 13 biopsies, for which only paraffin-embedded tissue was available, DNA was isolated using a different method, as described.23
Amplification of rearranged VH genes.
DNA extracted from the biopsy specimens was initially amplified as described24 for 37 cycles with a consensus primer specific for the 3′ end of VH gene framework three (FW3) regions and the consensus JH1 primer (Table 1). Approximately one third of the resultant products were electrophoresed in 8% acrylamide gels, and prominent bands indicative of B-cell clones were identified by visual inspection after staining with ethidium bromide. Rearranged VH genes were further amplified from the biopsy specimens in which clonal bands were identified using a variety of 5′ primers specific for framework one (FW1) regions (Table 1) in conjunction with a consensus JH primer in individual reactions containing a single FW1-JH primer combination. After 35 cycles of amplification, 0.5 μL of the resultant products was reamplified with internal FW1 or JH primer for 15 cycles. Typically, the internal JH2 primer would be substituted for JH1, although in some cases in which the JH2 primer did not appear to bind because products were not generated, internal FW1 primers were used in the secondary reactions. This nested approach was necessary with the paraffin-extracted DNA templates to generate easily identifiable VH bands after electrophoresis in 1.5% agarose and ethidium bromide staining or to obtain sufficient quantities of DNA for subsequent cloning.
Primers Used for VH Gene Amplification
| Name . | Sequence (5′-3′) . |
|---|---|
| VH1FW* | CAGGTTCAGCTGGTGCAGTCTGG |
| VH2FW | CAGGTCACCTTGAAGGAGTCTGG |
| VH3FW | GAGGTGCAGCTGGTGGAGTCTGG |
| VH4aFW | CAGGTGCAGCTGCAGGAGTCGGG |
| VH4bFW | CAGGTGCAGCTACAGCAGTGGGG |
| VH6FW | CAGGTACAGCTGCAGCAGTCAGG |
| VH1FWI | CCTCAGTGAAGGT(C,T)TCCTGCAAGG |
| VH2FWI | CAGACCCTCAC(A,G)CTGACCTGCACC |
| VH3FWI | GGTCCCTGAGACTCTCCTGTGCAG |
| VH4FWI | TCGGAGACCCTGTCCCTCACCTGC |
| VH5FWI | GTGAAAAAGCCCGGGGAGTCTCTG |
| VH6FWI | CTCACTCACCTGTGCCATCTCCGG |
| FW3 | GACACGGC(C,T)(A,G)TGTATTACTG |
| JH1 | ACCTGAGGAGACGGTGACC |
| JH2 | CCA(G,T)GGT(C,G,T)CCTTGGCCCCAG |
| Name . | Sequence (5′-3′) . |
|---|---|
| VH1FW* | CAGGTTCAGCTGGTGCAGTCTGG |
| VH2FW | CAGGTCACCTTGAAGGAGTCTGG |
| VH3FW | GAGGTGCAGCTGGTGGAGTCTGG |
| VH4aFW | CAGGTGCAGCTGCAGGAGTCGGG |
| VH4bFW | CAGGTGCAGCTACAGCAGTGGGG |
| VH6FW | CAGGTACAGCTGCAGCAGTCAGG |
| VH1FWI | CCTCAGTGAAGGT(C,T)TCCTGCAAGG |
| VH2FWI | CAGACCCTCAC(A,G)CTGACCTGCACC |
| VH3FWI | GGTCCCTGAGACTCTCCTGTGCAG |
| VH4FWI | TCGGAGACCCTGTCCCTCACCTGC |
| VH5FWI | GTGAAAAAGCCCGGGGAGTCTCTG |
| VH6FWI | CTCACTCACCTGTGCCATCTCCGG |
| FW3 | GACACGGC(C,T)(A,G)TGTATTACTG |
| JH1 | ACCTGAGGAGACGGTGACC |
| JH2 | CCA(G,T)GGT(C,G,T)CCTTGGCCCCAG |
*Number in VH primer name refers to the VH family. The VH1FW primer will also recognize FW1 region of VH5 family. Primers ending with “I” are specific for VH FW1 regions 3′ (internal) to the others.
Cloning and sequencing of PCR products.
After electrophoresis in 8% acrylamide, clonal FW3-JH1 bands were isolated from gels and incubated for 24-72 hours in 500 μL of TE at 37°C. After centrifugation, the TE solution was precipitated with sodium acetate, and the resulted DNA pellet was resuspended in H2O. Appropriately sized VHFW1-JH PCR products were isolated from 1.5% low melt agarose gels and further purified using Wizard DNA preps (Promega, Madison, WI). Approximately one seventh of the purified DNAs were cloned using the PCR-Script kit (Stratagene, LaJolla, CA). Plasmid DNA was isolated from overnight 1.5-mL cultures of randomly selected colonies using Wizard mini-preps (Promega). Diodeoxy sequencing was performed with Sequenase (US Biochemical, Cleveland, OH) following the manufacturer's protocol using approximately one fourth of the isolated plasmid DNA. Clones were sequenced in both directions using the M13 forward and reverse primers. In several cases, the small FW3-JH1 products were directly sequenced without cloning. Single-stranded templates for these sequencing reactions were generated by subjecting one third of the purified DNAs to 20 cycles of amplification with either the FW3 or JH1 primer. After precipitation with ammonium acetate, sequencing was performed as described above using one half of the precipitated single-stranded DNA with the primer not used to generate the templates.
Analysis of VH gene mutations.
Expected numbers of replacement (R) and silent (S) mutations in the first and second compementarity-determining regions (CDR) and VH framework regions (FWR) were calculated as described by Chang and Casali.25 Any shared mutations found in several clonally related VH sequences were counted only once. The calculated mutation values were rounded to the nearest integer.
RESULTS
Identification of B-cell clones.
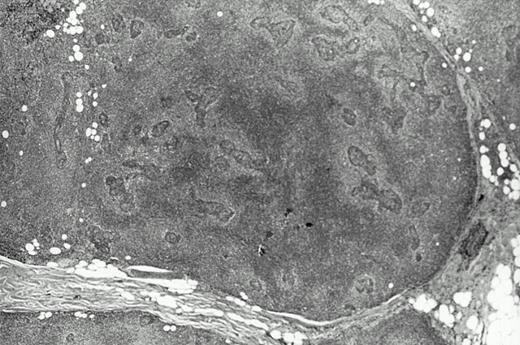
Two contralateral biopsy specimens of MESA-associated lesions from eight different patients were analyzed by a standard PCR technique for detecting clonal B cells. In seven of the eight cases, both of the contralateral biopsies, which are further described in Table 2, contained clones. The biopsies from one case showed only polyclonal B cells and were not further studied. With the exception of entries for patient HA, most of the other information in Table 2 can be found in our earlier study (see Table 7 in Quintana et al13) along with other clinical data for these patients. Biopsies with material available for analysis in this study were largely from parotid glands and the time intervals between the first and second biopsies ranged from several months to more than 10 years. Histologically, most of the biopsies met criteria considered to be diagnostic of low-grade MALT lymphoma by showing confluent areas of pale monocytoid cells (Fig 1). However, several of the biopsies showed only MESA with small nonconfluent halos of monocytoid cells around the lymphoepithelial lesions, which has been proposed to represent a precursor lesion or the earliest histologic sign of lymphoma. In one case (MA), both of the biopsies that were analyzed histologically showed only reactive MESA.
Features of Biopsies With B-Cell Clones
| Patient . | Age/Sex . | History . | Biopsy site . | Time (mo) . | Histology . |
|---|---|---|---|---|---|
| MA | 52/F | SS | Left parotid | 0 | MESA |
| Right parotid | 3 | MESA | |||
| LE | 58/M | — | Right parotid | 0 | MESA-Halos |
| Left parotid | 20 | MESA-Halos | |||
| JA | 55/F | — | Left parotid | 0 | Lymphoma |
| Right parotid | 26 | MESA-Halos | |||
| PO | 70/M | DM | Left neck LN | 0 | Lymphoma |
| Right parotid | 6 | Lymphoma | |||
| HA | 62/F | SS | Right parotid | 0 | Lymphoma |
| Left lacrimal | 20 | Lymphoma | |||
| BA | 66/M | RA | Left parotid | 0 | Lymphoma |
| Right parotid | 85 | Lymphoma | |||
| OR | 58/F | SS | Left submandib | 0 | Lymphoma |
| Right parotid | 122 | Lymphoma |
| Patient . | Age/Sex . | History . | Biopsy site . | Time (mo) . | Histology . |
|---|---|---|---|---|---|
| MA | 52/F | SS | Left parotid | 0 | MESA |
| Right parotid | 3 | MESA | |||
| LE | 58/M | — | Right parotid | 0 | MESA-Halos |
| Left parotid | 20 | MESA-Halos | |||
| JA | 55/F | — | Left parotid | 0 | Lymphoma |
| Right parotid | 26 | MESA-Halos | |||
| PO | 70/M | DM | Left neck LN | 0 | Lymphoma |
| Right parotid | 6 | Lymphoma | |||
| HA | 62/F | SS | Right parotid | 0 | Lymphoma |
| Left lacrimal | 20 | Lymphoma | |||
| BA | 66/M | RA | Left parotid | 0 | Lymphoma |
| Right parotid | 85 | Lymphoma | |||
| OR | 58/F | SS | Left submandib | 0 | Lymphoma |
| Right parotid | 122 | Lymphoma |
Abbreviations: SS, Sjögren's syndrome; DM, diabetes mellitus; RA, rheumatoid arthritis.
Histologic features of the low grade salivary gland MALT lymphoma from patient JA. Note the halos of pale monocytoid cells around lymphoepithelial lesions on the right that become more confluent particularly on the left.
Histologic features of the low grade salivary gland MALT lymphoma from patient JA. Note the halos of pale monocytoid cells around lymphoepithelial lesions on the right that become more confluent particularly on the left.
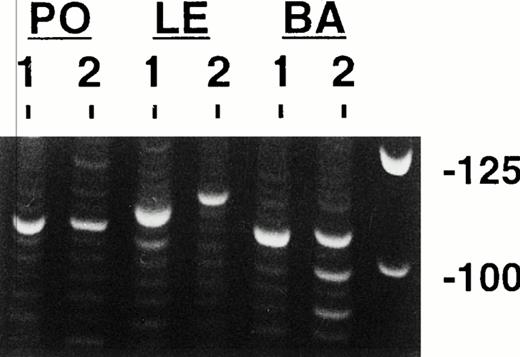
Representative clonal bands after electrophoresis and ethidium bromide staining of FW3-JH PCR products are shown in Fig 2 for three of the cases. Observe that for case PO, single clonal bands of the same size are present in both biopsy specimens, whereas case LE shows single clonal bands in both biopsies that have different sizes. In case BA, at least two prominent monoclonal bands can be seen in the second specimen, with one being the same size as the single band in the first specimen. The clonal bands were purified and sequenced, which in all cases yielded a unique in frame clonal CDR3 sequence (Fig 3). For three of the cases (JA, PO, and HA), the same single clone was present in both biopsy specimens, because each had identical or nearly identical CDR3 sequences. However, for three other cases (MA, LE, and OR), the single clones identified in the two biopsies were different and had distinct CDR3s due to the use of different D segments and N nucleotides. In case BA, two different potentially functional clones were identified in the second biopsy, one of which was the same as the initial clone.
PCR clonality analysis. Shown are representative products produced by amplifying paraffin-extracted DNAs from three cases with the consensus FW3 and JH1 primers after electrophoresis and ethidium bromide staining. Single clonal bands are seen in all of these specimens except in the second biopsy from patient BA, which shows multiple clonal bands. Molecular weight markers are shown in the left most lane. Each lower rung of the polyclonal background ladders represents 3 nucleotides or one amino acid.
PCR clonality analysis. Shown are representative products produced by amplifying paraffin-extracted DNAs from three cases with the consensus FW3 and JH1 primers after electrophoresis and ethidium bromide staining. Single clonal bands are seen in all of these specimens except in the second biopsy from patient BA, which shows multiple clonal bands. Molecular weight markers are shown in the left most lane. Each lower rung of the polyclonal background ladders represents 3 nucleotides or one amino acid.
CDR3 nucleotide sequences. The clonal CDR3 sequences in the second biopsies are compared with those identified in the initial biopsies. Whether the sequences were derived from directly sequencing FW3-JH1 PCR products or from sequence analysis of individual clones of FW3-JH1 products is indicated in parentheses along with the number of related clones to total clones analyzed. N nucleodies are shown in lower case, whereas D and J segments are shown as boldface capitols (J segments left most). Nucleotide differences from the proposed germline D and J segments are underlined. (The names of the D and J segments are given in Table 3.)
CDR3 nucleotide sequences. The clonal CDR3 sequences in the second biopsies are compared with those identified in the initial biopsies. Whether the sequences were derived from directly sequencing FW3-JH1 PCR products or from sequence analysis of individual clones of FW3-JH1 products is indicated in parentheses along with the number of related clones to total clones analyzed. N nucleodies are shown in lower case, whereas D and J segments are shown as boldface capitols (J segments left most). Nucleotide differences from the proposed germline D and J segments are underlined. (The names of the D and J segments are given in Table 3.)
Characterization of clonal VH genes.
To further study these clones, rearranged VH genes were amplified from the specimens using a variety of FW1-JH primer combinations, and the products from some of these reactions were cloned and sequenced. Complete or nearly complete VH gene nucleotide sequence information was obtained for all of the B-cell clones identified previously with the FW3-JH1 primers, because the VH genes had the same CDR3 sequences. The two VH genes from biopsies HA#1 and OR#2 were described in an earlier study and were amplified using VH leader primers.8Comparing the VH sequences to those in the VBASE and GeneBank directories indicated that the 11 clonal VH genes we had identified with distinct CDR3s were most closely related to germline VH gene segments from only two loci, V1-69 and V3-7 (Fig 4 and Table 3). Of the eight distinct V1-69 genes, five are most homologous to the 51p1 gene and three are most homologous to the closely related allelic variant 7M27, which only differs from 51p1 by one nucleotide in FW3.26 In addition to the frequent use of specific VH gene segments, comparing the clonal CDR3 sequences to known germline elements also showed evidence of restricted use of J and D segments. For the eight distinct V1-69 genes, seven appear to use a J4 segment and five of these use D segments from the DXP family, D21-9 (4 genes) and DXP ′1 (1 gene), in the same second reading frame. For the three V3-7 genes, all use a J3 segment and D segments from the DXP family, D21-9 (2 genes), and DXP ′1 (1 gene) also in the second reading frame. Some of the CDR3s also show evidence of shared amino acid residues encoded by N-nucleotides at the VH-D junction. All three of the V3-7 CDR3s start with glycine and aspartate residues (GD single letter code,) whereas four of eight V1-69 genes have glutamate (E single letter code) as the first amino acid (Fig 4).
Deduced amino acid sequences of the clonal VH genes. Differences from the germline gene segments are shown as upper case letters or as underlined letters in the CDR3s. The locations of silent mutations in the nucleic acid sequences are indicated with lower case letters. Only mutations that were present in all of the PCR clones are shown, ie, the common mutations. The VH genes derived from 2m27 (JA-1, BA-1, and OR-1) do not have an E to K mutation in the eighth FW3 position, because this represents the single nucleotide difference between the 2m27 and 51p1 VH gene segments. Sequence gaps in the CDR3s identifies the J segments start. These sequence data are available from Genebank under accession nos. AF038442-AF038450 and U79591-U79592.
Deduced amino acid sequences of the clonal VH genes. Differences from the germline gene segments are shown as upper case letters or as underlined letters in the CDR3s. The locations of silent mutations in the nucleic acid sequences are indicated with lower case letters. Only mutations that were present in all of the PCR clones are shown, ie, the common mutations. The VH genes derived from 2m27 (JA-1, BA-1, and OR-1) do not have an E to K mutation in the eighth FW3 position, because this represents the single nucleotide difference between the 2m27 and 51p1 VH gene segments. Sequence gaps in the CDR3s identifies the J segments start. These sequence data are available from Genebank under accession nos. AF038442-AF038450 and U79591-U79592.
Analysis of Clonal VH Sequences
| Patient . | Specimen . | No. Analyzed . | Most Similar Germline Segments . | Differences From Germline % (no.) . | |||
|---|---|---|---|---|---|---|---|
| V* . | D . | J . | Common† . | Noncommon . | |||
| MA | 1 | 2 | V3-7 | D21-9 | J3 | 0.3 (1) | 2.5 (16) |
| 2 | 3 | — | DXP′1 | — | 1.0 (3) | 1.2 (12) | |
| LE | 1 | 2 | 51p1 | DXP′1 | J4b | 5.9 (18) | 0.3 (2) |
| 2 | 2 | V3-7 | D21-9 | J3 | 2.9 (9) | 4.0 (26) | |
| JA | 1 | 3 | 7m27 | D21-9 | J4a | 0.94 (3) | 2.5 (25) |
| 2 | 4 | — | — | — | 1.9 (6) | 0.9 (12) | |
| PO | 1 | 3 | 51p1 | DA5 | J4a | 7.3 (20) | 0 |
| 2 | 3 | — | — | — | 6.6 (18) | 0 | |
| HA | 1 | 10 | 51p1 | D21-9/DIR3/DXP4 | J4b | 2.0 (6) | 0.2 (8) |
| 2 | 5 | — | — | — | 3.7 (12) | 0.3 (5) | |
| BA | 1 | 4 | 7m27 | DK4 | J3 | 3.7 (11) | 2.2 (28) |
| 2 | 5 | — | — | — | 7.6 (21) | 0 | |
| 2 | 3 | 51p1 | D21-9 | J4a | 4.1 (12) | 0.11 (1) | |
| OR | 1 | 6 | 7m27 | DIR4 | J4a | 7.2 (18) | 0 |
| 2 | 7 | 51p1 | D21-9 | J4a | 1.6 (5) | 0 | |
| Patient . | Specimen . | No. Analyzed . | Most Similar Germline Segments . | Differences From Germline % (no.) . | |||
|---|---|---|---|---|---|---|---|
| V* . | D . | J . | Common† . | Noncommon . | |||
| MA | 1 | 2 | V3-7 | D21-9 | J3 | 0.3 (1) | 2.5 (16) |
| 2 | 3 | — | DXP′1 | — | 1.0 (3) | 1.2 (12) | |
| LE | 1 | 2 | 51p1 | DXP′1 | J4b | 5.9 (18) | 0.3 (2) |
| 2 | 2 | V3-7 | D21-9 | J3 | 2.9 (9) | 4.0 (26) | |
| JA | 1 | 3 | 7m27 | D21-9 | J4a | 0.94 (3) | 2.5 (25) |
| 2 | 4 | — | — | — | 1.9 (6) | 0.9 (12) | |
| PO | 1 | 3 | 51p1 | DA5 | J4a | 7.3 (20) | 0 |
| 2 | 3 | — | — | — | 6.6 (18) | 0 | |
| HA | 1 | 10 | 51p1 | D21-9/DIR3/DXP4 | J4b | 2.0 (6) | 0.2 (8) |
| 2 | 5 | — | — | — | 3.7 (12) | 0.3 (5) | |
| BA | 1 | 4 | 7m27 | DK4 | J3 | 3.7 (11) | 2.2 (28) |
| 2 | 5 | — | — | — | 7.6 (21) | 0 | |
| 2 | 3 | 51p1 | D21-9 | J4a | 4.1 (12) | 0.11 (1) | |
| OR | 1 | 6 | 7m27 | DIR4 | J4a | 7.2 (18) | 0 |
| 2 | 7 | 51p1 | D21-9 | J4a | 1.6 (5) | 0 | |
*7m27 (also termed hv1051K) and 51p1 are closely related V1-69 alleles (see text). A dash indicates identity with the above sequence.
Common differences are those present in all of the VH sequences, whereas noncommon differences are present in only some of the VH sequences. Comparisons were made to the proposed germline V, D, and J segments.
Mutation analysis.
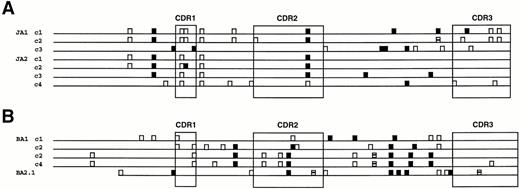
Numerous single nucleotide differences or mutations between the VH genes and the proposed germline sequences are evident in all of the clones (Fig 4 and Table 3). Some of these mutations were seen in all of the clonally related sequences analyzed from a given specimen, the common mutations, whereas others were only present in some of the clonal VH sequences, the noncommon mutations. In the four cases in which the initial clone was also identified in the second contralateral biopsy (PO, JA, HA, and BA), unique common mutations from germline were present in both clones, indicating that each site contained a distinct subclone. The number of noncommon mutations identified in many of the clonal VH genes was substantial and in some of the clones greater than the number of common mutations. The locations and type of noncommon mutations in individual clones for the two VH genes with the greatest number of noncommon mutations are depicted in Fig 5. Observe that many of the noncommon mutations are shared between different VH gene clones, indicating their sequential development or stepwise evolution. The noncommon mutations in these two VH genes are also tabulated in Table 4 and compared with the expected distributions of R and S mutations, assuming the mutations occurred by chance alone. As can be appreciated, the CDRs as well as FWRs have fewer R mutations and lower R/S ratios than expected, which suggests that there is selection against R mutations in both of these regions. Findings similar to these were also observed in the other clonal MESA VH genes that showed significant intraclonal heterogeneity.
Diagramatic representation of the noncommon mutations in the different VH clones from patients JA (A) and BA (B), the two VH genes with the greatest number of noncommon mutations. Silent (S) mutations are shown as open boxes, whereas replacement (R) mutations are indicated by solid boxes. The four codons that contained both S and R mutations are indicated by partially solid boxes.
Diagramatic representation of the noncommon mutations in the different VH clones from patients JA (A) and BA (B), the two VH genes with the greatest number of noncommon mutations. Silent (S) mutations are shown as open boxes, whereas replacement (R) mutations are indicated by solid boxes. The four codons that contained both S and R mutations are indicated by partially solid boxes.
Distribution of Noncommon Mutations in the Two VH Genes With the Greatest Number of Noncommon Mutations
| Gene . | Location* . | Observed . | Expected . | ||||
|---|---|---|---|---|---|---|---|
| R . | S . | R/S . | R . | S . | R/S . | ||
| JA | CDR | 3 | 4 | 0.75 | 5 | 1 | 3.6 |
| FWR | 11 | 10 | 15 | 5 | |||
| BA1 | CDR | 3 | 7 | 0.43 | 7 | 2 | 3.6 |
| FWR | 13 | 15 | 22 | 7 | |||
| Gene . | Location* . | Observed . | Expected . | ||||
|---|---|---|---|---|---|---|---|
| R . | S . | R/S . | R . | S . | R/S . | ||
| JA | CDR | 3 | 4 | 0.75 | 5 | 1 | 3.6 |
| FWR | 11 | 10 | 15 | 5 | |||
| BA1 | CDR | 3 | 7 | 0.43 | 7 | 2 | 3.6 |
| FWR | 13 | 15 | 22 | 7 | |||
*Included are CDR1 and 2 and FWR 1, 2, and 3. Shared mutations are counted only once.
DISCUSSION
The significance of finding monoclonal B cells in MESA-associated infiltrates by PCR or Southern blot analysis has been controversial. Fishleder et al14 as well as Knowles et al27suggested that these clones may represent nonmalignant expansions, because they can be frequently found in lesions that histologically appear benign and often remain localized for prolonged periods of time. However, these studies were completed several years before MALT lymphomas were generally recognized as distinct entities and the histologic features of their cases were not described in detail. More recently, Isaacson and others16 22 have argued that MESA-associated monoclonal B-cell populations represent malignant lymphomas, because they are usually associated with identifiable histologic features (halos or confluent areas of monocytoid B cells around lymphoepithelial lesions) and their propensity to remain localized simply reflects the unusually indolent behavior that is characteristic of low-grade MALT type lymphomas.
To help clarify the nature of clonal MESA-associated lymphoid infiltrates, we analyzed VH clones in sequential contralateral biopsies from seven different patients to determine how they may be related to one another. It is important to emphasize that the PCR technique we used to initially identify the clones is very specific even with DNA extracted from paraffin-embedded tissue that is often of poor quality because the amplification products are relatively small consisting mostly of CDR3 regions.28 29 Although false-positive clonal bands can occasionally be detected with this method from samples containing very few B cells when the DNA is of poor quality, this was not a problem even from our paraffin-extracted templates, because all demonstrated abundant B cells as shown by the prominent polyclonal signals that were seen along with the clonal bands. In terms of sensitivity, previous dilutional studies with polyclonal DNA indicated that a clone making up approximately 2% of a B-cell population represented our lower limit of detection, which is close to the detection limit of very sensitive Southern blot analysis. It is likely, therefore, that all of the clones we identified by PCR could also have been detected by Southern blot analysis if fresh tissue had been available. Indeed, for the three fresh tissue biopsies that were analyzed, Southern blot and our PCR clonality test gave concordant results.
In three of the seven cases that were studied, a single VH clone was identified in the follow-up biopsy that appeared to be clonally unrelated to the single VH clone identified in the initial biopsy, because it had a distinct CDR3 sequence with different D segments and N nucleotides. Because all of the clonal VH sequences we identified were in frame and free of stop codons, each could correspond to a distinct B-cell clone. Thus, finding distinct VH clones in the contralateral biopsies cannot be readily explained by differential amplification at each site of the productive and nonproductive allele from the same B cell. Similarly, in the one case in which two VH clones were identified in the follow-up biopsy, one of which was clonally distinct from the initial clone, each appears to correspond to a separate B-cell clone as opposed to the productive and nonproductive allele of the same clone. The possibility that our finding different clones resulted from a specimen labeling error was formally ruled out by PCR analysis of the androgen receptor gene, which gave a pattern of bands that was unique for each patient because of the variable number of CAG repeats30 but identical between paired specimens (not shown). Finding distinct B-cell clones in different biopsy specimens from the same patient strongly argues that monoclonal MESA-associated expansions are not necessarily malignant lymphomas. Moreover, in two of the patients with different clones in the second biopsies, the MESA lesions were also not histologically diagnostic of lymphoma.
Evidence that different B-cell clones may be present in sequential biopsies of MESA-related lymphoid infiltrates was also reported by Fishleder et al.14 In that study, Southern blot analysis showed that the salivary gland biopsies obtained 2 years apart from one patient contained different clonal heavy chain and light chain rearrangements. However, because the Ig gene somatic hypermutation mechanism can be active in MALT lymphomas,8,31 it was still possible that the different rearrangements did not represent separate clones but resulted from the creation of new restriction endonuclease sites in the same clone, similar to what has been described for follicular lymphomas, which also mutate their Ig genes during clonal expansion.32 A recent study by Diss et al22that used a PCR clonality technique similar to ours found all of the clonal bands to be of similar size in the sequential salivary gland biopsies from the six patients that were analyzed. However, in that study, the PCR bands were not sequenced, so it is possible that some may have been different clones similar to two of our cases in which the different clones had PCR bands that were the same sizes. Our greater success in identifying different clones may also be related to all of our sequential biopsies being from contralateral locations and to our inclusion of early MESA-associated lesions that histologically were not diagnostic of MALT lymphoma.
This study further establishes that MESA-associated clones, even from different patients, are not completely unrelated but often express VH genes that are remarkably similar to one another. One aspect of the VH gene similarity results from biased use of certain VH gene segments. Recent estimates indicated that there are approximately 50 different functional VH gene segments in the human genome.33Therefore, finding that the 11 MESA clones with distinct CDR3s described in this study use either V1-69 (8 clones) or V3-7 (3 clones) represents a highly nonrandom VH gene repertoire. Besides the biased use of VH gene segments, different MESA clones also contained similar amino acid sequence motifs in their CDR3s. Some of this CDR3 similarity can be explained by the frequent use of D-segments from the DXP1 family in the second reading frame in 8 of 11 clones, the use of J4 segments by seven of eight V1-69–derived clones, and the use of J3 segments by three of three V3-7 genes. In addition, identical amino acids were also noted at the 5′ end of many CDR3s (GDY in 3 of 3 V3-7 genes and E in 4 of 8 V1-69 genes), which appear to correspond to residues encoded by N nucleotides. These findings, along with the VH gene restriction noted above, strongly suggest that MESA-associated clones represent a highly restricted B-cell subset that has been selected for expansion through their Ig molecules.
The VH gene similarities between the two different clones identified in case MA are so striking that a cursory analysis might suggest they were clonally related to one another. Both used the same VH and JH segments and also had CDR3s that were the same sizes that only differed by 3 of 15 aminoacids, two of which were conservative glycine to serine (G to S) substitutions. However, it is unlikely that the G to S substitutions that were located in the D segments resulted from somatic mutations in the initial clone, because there was near perfect homology with substantial portions of the two different but related D21-9 and DXP'1 D segments that were proposed to be used with matches at 21 of 21 and 18 of 19 nucleotides, respectively, and the G to S substitutions represent germline areas of these D segments. In addition, no mutations were identified in any of the 12 individual PCR clones that were analyzed that corresponded to the 8 nucleotide differences in the CDR3 sequences between these genes. Further evidence that the two MA clones are indeed independent comes from consideration of the V3-7 MESA-associated clone we identified in our previous study that also had a CDR3 that was remarkably similar to the initial MA clone differing by only two D to E and H to I substitutions excluding its one additional amino acid of length.8 Moreover, the additional V3-7 MESA clone that was identified in this study also has a CDR3 sequence that is remarkably similar to those used by the MA clones. The very high degree of VH and CDR3 sequence similarity seen between different MESA-associated clones, especially those using V3-7, suggests that some may bind the same epitope or antigen.
Although the antigen specificity of the MESA clones is unknown, antibodies encoded by V1-69 and V3-7 have been described that display reactivity toward a variety of autoantigens, including IgG and DNA.34,35 Further support that the MESA clones may have anti-IgG or rheumatoid factor activity comes from their CDR3 sequences, because rheumatoid factors that use V1-69 or V3-7 typically have highly distinctive CDR3s compared with randomly chosen V1-69– or V3-7–derived VH genes.34 Specifically, like the MESA clones, rheumatoid factors derived from V3-7 usually have CDR3s that start with the amino acids glycine-aspartate-tyrosine (GDY), frequently use the D21-9 and J3 gene segments, and are typically 14 to 15 amino acids in length. In addition, the MESA clones using V1-69 also resemble the V1-69 rheumatoid factors in having CDR3s that often start with glutamate (E), frequently use the J4 segment, and use asparagine-proline (NP) residues at several D-J junctions. The possibility that MESA-associated clones frequently have rheumatoid factor activity is also consistent with the increased salivary gland production of rheumatoid factors reported in patients with Sjögren's syndrome.36
It is interesting that V1-69 VH genes are also frequently expressed by chronic lymphocytic leukemia (CLL), a low-grade malignancy of CD5 positive B cells, being found in approximately 20% of cases.21 This raises the possibility that CLL and salivary gland MALT lymphomas may be related in terms of pathogenesis and derived from the same selected B-cell subset. However, from analysis of these few cases, the restricted types of CDR3 sequences frequently used by V1-69–derived MESA and CLL VH genes appear to be quite different. For example, J6, which was not used by any of our eight MESA V1-69 sequences, was used by 12 of 26 CLL V1-69 genes described in a recent study by Johnson et al,37 whereas D21-9, identified in five of our eight MESA V1-69 sequences, was used by only 1 of these 26 CLL sequences. Because residues encoded by CDR3 frequently affect antigen binding, these observations suggest that, although CLL and MESA-associated clones may be highly selected populations, the V1-69 Igs from each will have different antigen specificities.
Antigen selection of the MESA clones is also supported by analyses of the numerous mutations that were found the VH genes. It is well established that VH genes expressed by MALT lymphomas are typically mutated from germline and often show considerable intraclonal variation indicative of ongoing Ig gene mutation.8,31 Replacement (R) mutations in the CDRs, which encode residues that form the conventional antigen binding site, can be selected for (positively selected) if they increase binding affinity or selected against (negative selection) if they lead to lower antigen binding affinity.38 In evaluating mutations for evidence of clonal selection, mutations that are known to have been acquired as the clone expands are potentially the most informative. These correspond to mutations that are present in only some of the clonally related VH sequences, the noncommon mutations, as opposed to mutations present in all of the VH sequences, the common mutations, which could have occurred before expansion or in a B-cell located in a different location.19 Analyses of noncommon mutations was facilitated in this study by finding four MESA VH genes with very high ongoing mutation rates of greater than 2%, which is considerably higher than what has been previously reported for MALT lymphomas.8,31,39 Instead of high numbers of noncommon R mutations in the CDRs indicative of positive selection, the MESA-associated clonal VH genes with significant intraclonal heterogeneity all contained fewer noncommon R mutations in the CDRs than expected by chance, indicating there is negative selection or selection against R mutation. Moreover, the ratios of R to S mutations in the CDRs were all less than 1.5, whereas ratios of R to S mutations in the V1-69 and V3-7 genes expected without selection should be 3.6:1 and 3.9:1, respectively. Negative selection suggests that clonal expansion is dependent on the preservation of residues in the CDRs that are important for antigen binding and has been previously described for rheumatoid factors that use the 51p1 gene.40 Negative selection of R mutations also appears to be operating in the FWRs, because they also have fewer noncommon R mutations and lower R/S ratios than expected by chance. Because the maintenance of Ig function requires preservation of many FWR amino residues, this further suggests that functional Ig is important for clonal proliferation.38Although our earlier study also provided evidence of negative selection of R mutations in the CDR and FWR of several salivary gland MALT lymphoma VH genes,8 that analysis was limited to mostly common mutations that may be less informative, as described above.
The reason for the variability in ongoing mutation rates among these clonal MESA-associated VH genes, which ranged from 0% to 4.0%, is unclear. However, one possibility is that those clones with low or nondetectable ongoing mutation are less dependent on antigen stimulation, because they have acquired additional oncogenic events that could substitute for an Ig-mediated growth signal. This idea is also consistent with our finding that the average ongoing mutation rate in clonal VH genes from more benign MESA-associated lesions twice the rate found in clones from lesions that histologically were diagnostic of MALT lymphomas (1.8 v .74). With the high rates of ongoing mutation in some of the clonal VH genes, it may also be significant that no stop codons were identified in any of the 86 total individual VH PCR clones or 120 noncommon mutations that were analyzed. This further supports the concept that expression of functional Ig molecules is important for the survival and proliferation of early MESA-associated clones.
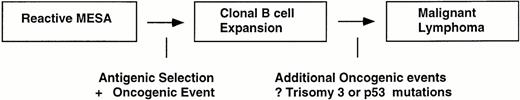
In summary, the highly restricted VH gene repertoire and ongoing mutation analysis indicates that Ig is playing an important role in the development and subsequent expansion of MESA-associated clones. The most straightforward model to propose is that B cells with the appropriate VH structures are selected for expansion through surface Ig stimulation by binding a common antigen (Fig 6). At some point, a genetic event probably occurs in one of the expanding clones that gives it a growth advantage over the others, because it is unlikely antigen stimulation by itself could generate a clonal population that would be large enough to be detected by PCR analysis. However, the development of clonality in MESA may be aided by immune system dysregulation, which is part of an autoimmune disease process. Because patients with MESA typically have involvement of multiple glands, independent clones can sometimes develop at different sites in the same patient, as we have shown. It is unlikely that clones that first become detectable by PCR are malignant, because no patients have been described with disseminated lymphomas in different organs that are clonally distinct.32,39,41Moreover, it is well established that not all monoclonal lymphoid proliferations behave in a malignant fashion.42-44 We propose, therefore, that additional oncogenic events after the emergence of monoclonality are usually required before MESA clones become malignant lymphomas capable of dissemination and growth outside of the salivary gland. It is possible that trisomy 3, a cytogenetic abnormality frequently found in MALT lymphomas,45represents such an event or mutations in the P53 gene that have been associated with histologic progression of MALT lymphomas.46Although the exact point at which lymphoma develops remains to be more precisely defined, our data suggest that the early nonmalignant clones are more likely to be found in MESA-associated lesions that are histologically more benign in appearance. From a practical standpoint, therefore, patients with a histologically benign or borderline MESA lesion that contains a clonal population may not require local antineoplastic therapies, as has been suggested,15 given the potential morbidity. Moreover, the rationale for local therapy might also be questioned in this situation, because additional clones may be present in a contralateral salivary gland.
Schematic model of MALT lymphomagenesis in the salivary gland. Clones detectable by PCR or Southern blot analysis develop in reactive MESA as a result of direct stimulation by a limited number of antigens and the occurrence of a premalignant oncogenic event. Initially, the clones are still dependent on antigen stimulation for growth and remain localized, although some may be capable of spreading to other salivary glands that are also involved by MESA. At some point, which may be many years after a clone first appears, additional oncogenic events occur that result in the clones becoming malignant lymphomas capable of widespread dissemination and growth in other organs.
Schematic model of MALT lymphomagenesis in the salivary gland. Clones detectable by PCR or Southern blot analysis develop in reactive MESA as a result of direct stimulation by a limited number of antigens and the occurrence of a premalignant oncogenic event. Initially, the clones are still dependent on antigen stimulation for growth and remain localized, although some may be capable of spreading to other salivary glands that are also involved by MESA. At some point, which may be many years after a clone first appears, additional oncogenic events occur that result in the clones becoming malignant lymphomas capable of widespread dissemination and growth in other organs.
ACKNOWLEDGMENT
The authors thank John Miklos for technical assistance and Diana Winters for secretarial assistance.
Supported by the Pathology Education and Research Foundation (University of Pittsburgh) and Grant No. IRG-58-34 from the American Cancer Society.
Address reprint requests to David W. Bahler, MD, PhD, Department of Pathology, Presbyterian University Hospital C604, 200 Lothrop St, Pittsburgh, PA 15213.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal