Abstract
We previously reported that retroviral vectors displaying epidermal growth factor (EGF) as part of a chimeric envelope glycoprotein are sequestered upon binding to EGF receptor (EGFR)-positive target cells, leading to loss of infectivity. In the current study, we have displayed stem cell factor (SCF) on β-galactosidase-transducing ecotropic and amphotropic retroviral vector particles as a factor Xa protease-cleavable N-terminal extension of the envelope glycoprotein. Viral incorporation of the SCF chimeric envelopes was demonstrated by immunoblotting of pelleted virions and their specific attachment to Kit receptors was demonstrated by flow cytometry. Gene transfer studies showed that when SCF was displayed on an amphotropic envelope, the infectivity of the SCF-displaying vectors was selectively inhibited on Kit-expressing cells, but could be restored by adding soluble SCF to block the Kit receptors or by cleaving the displayed SCF domain from the vector particles with factor Xa protease. The host range properties of EGF-displaying and SCF-displaying vectors were then compared in cell mixing experiments. When EGFR-positive cancer cells and Kit-positive hematopoietic cells were mixed and exposed to the different engineered vector particles, the cancer cells were selectively transduced by the SCF-displaying vector and the hematopoietic cells were selectively transduced by the EGF-displaying vector. Retroviral display of polypeptide growth factors can therefore provide the basis for a novel inverse targeting strategy with potential use for selective transduction of hematopoietic or nonhematopoietic cells (eg, cancer cells) in a mixed cell population.
PLURIPOTENT hematopoietic stem cells (HSC) are attractive targets for gene therapy of a wide variety of hereditary and acquired disorders and several studies have shown that committed human hematopoietic progenitor cells can be efficiently transduced by retroviral vectors in ex vivo transduction protocols.1-4 However, long-term follow-up of patients who have been reconstituted with genetically marked progenitor cells shows that pluripotent HSC are highly resistant to retroviral transduction,5-7 in part because their relative quiescence8 makes them relatively resistant to retroviral transduction9 and in part because the cell surface density of amphotropic retroviral receptors on human HSC is very low.10 A major deficiency of current retroviral vectors is that they lack specificity for particular target cell types and are therefore unsuitable for protocols requiring selectivity of gene transfer to HSCs in mixed cell populations. Thus, vector targeting is highly desirable for certain ex vivo stem cell gene therapy protocols and will be a prerequisite for the more distant goal of in vivo transduction of mitotically active HSCs.
Murine leukemia virus (MLV)-based vectors are the most common retroviral vectors in clinical use. Their host range is determined by their envelope glycoproteins, which mediate attachment to specific receptors on target cells. The N-terminal domain of the surface (SU) glycoprotein confers receptor specificity and and exhibits a high degree of conservation between MLVs with different host ranges.11 The Moloney (Mo) MLV envelope binds to a murine cationic amino acid transporter Rec-1 and has an ecotropic host range infecting only mouse cells.12 The amphotropic or 4070A MLV envelope binds to a ubiquitous phosphate transporter RAM-1, which is conserved throughout many mammalian species.13 It thus infects both human and murine cells.
Towards the goal of targeting MLV-based vectors, we previously described a strategy for the display of polypeptide binding domains on retroviral vectors as N-terminal extensions of their envelope SU glycoproteins.14 Subsequent work has shown that various chimeric envelopes can be efficiently incorporated into retroviral vector particles and that vector attachment can be redirected via the displayed binding domains.15-17 However, such redirected vectors do not always result in gene transfer. The choice of receptor through which vectors are targeted has been shown to be an important determinant of gene transfer efficiency following redirected attachment. Ecotropic vectors displaying a domain that targeted their attachment to the amphotropic retrovirus receptor (RAM-1) gave a relatively high efficiency of targeted gene transfer on human cells,17 whereas ecotropic vectors displaying antibodies to the CD3 complex, or to various antigens on human colonic cells, gave a very low efficiency of gene delivery to the targeted cells.16 In addition, it was found that amphotropic vectors incorporating chimeric envelopes in which epidermal growth factor (EGF) was the displayed domain were able to actively and selectively interfere with gene delivery to EGF receptor-positive cells.15 18 Thus, the EGF-displaying vectors gave very low efficiency of gene transfer to EGF receptor-positive cells, but were fully infectious on EGF receptor-negative cells.
In the current study, we sought to determine whether stem cell factor (SCF), a large glycosylated growth factor domain that recognizes a receptor present on HSC, could be displayed on retroviral vectors and to determine what influence this would have on the host range properties of the chimeric vector particles. The SCF receptor, Kit, is a tyrosine kinase receptor, which is known to be expressed on hematopoietic progenitor cells,19 and plays an important role in hematopoietic cell biology. Kit is therefore a potentially suitable receptor target for selective stem cell transduction. SCF exists as both soluble and membrane bound forms and binds to the Kit receptor with an affinity variously reported to be between 27 and 240 picomolar.20,21 The soluble form is released from the 248 amino acid membrane anchored species by proteolytic cleavage to yield a 165 amino acid glycosylated extracellular domain,22-24which has an approximate serum concentration of 3.4 ng/mL.25
We therefore generated chimeric envelopes in which SCF was fused to the SU glycoproteins of the ecotropic Mo MLV whose receptor is absent on human cells and to the amphotropic 4070A MLV, which promiscuously infects both murine and human cells. Our results show that, like EGF, a virally displayed SCF domain can actively and selectively interfere with gene transfer on Kit receptor-positive cells. We therefore performed cell mixing experiments to determine whether receptor-mediated sequestration of vector particles displaying EGF or SCF could provide the basis for a novel ‘inverse’ targeting strategy to achieve selective transduction of receptor-negative target cells in a mixed cell population.
MATERIALS AND METHODS
Construction of chimeric envelope expression plasmids.
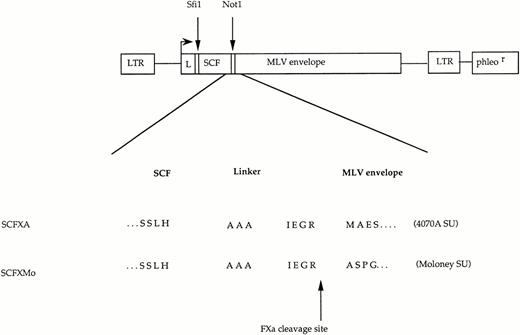
The unmodified envelopes of 4070A MLV and MoMLV were encoded by the expression plasmids FB4070ASALF and FBMoSALF.15 Plasmids were constructed encoding chimeric envelopes in which SCF was fused to the first codon of the Moloney (ecotropic) and 4070A (amphotropic) MLV SU envelope glycoproteins by a factor Xa cleavable linker. The construct SCFXA was derived from the construct EXA, in which EGF is fused to amino acid +1 of the 4070A MLV SU by a linker consisting of three alanines (Not 1 site ) or three alanines and the IEGR factor Xa cleavage site.18 The cDNA coding for the extracellular domain of SCF was polymerase chain reaction (PCR) amplified and tailed with Sfi 1 and Not 1 restriction sites using primers SCFforNot and SCFbackSfi. The PCR products were cloned into EXA after digestion with the restriction enzymes Sfi1 and Not 1.
The construct SCFXMo was derived from the EXMo,17 in which EGF is fused to amino acid +1 of the MoMLV SU, also by a Not 1site and the IEGR (single-letter amino acid code) factor Xa cleavage signal. The cDNA coding for the extracellular domain of SCF was PCR amplified and tailed with Sfi 1 and Not 1 restriction sites using primers SCFforNot and SCFbackSfi. The PCR products were cloned into EXMo after digestion with the restriction enzymes Sfi 1 and Not 1. The sequences of the portions of constructs generated by PCR were confirmed between theSfi 1 and Not 1 sites by dideoxysequencing. Oligonucleotides used were as follows (with restriction enzyme sites underlined) SCFforNot: GCAAATCTGCGGCCGCGTGTAGGCTGGAGTCTCCAGG; SCFBackSfi: GTCCATGCGGCCCAGCCGGCCGAAGGGATGCAGGAATCG.
Production of viruses.
The chimeric envelope and control (Mo and 4070A) envelope expression constructs were expressed in TELCeB.6 complementing cells, which express MLV gag-pol core particles and a nlsLacZ retroviral vector.26 Envelope expression plasmids were transfected by calcium phosphate coprecipitation into the TELCeB.6 cells.27 Transfected cells were selected with phleomycin (50 μg/mL) and stable transfectants were expanded and pooled. Cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS). Viral supernatants were harvested after overnight incubation in either serum-free DMEM or DMEM containing 10% FCS and filtered with a 0.45 μm filter for immediate use in infection or binding experiments. For immunoblotting, viral supernatants were filtered (0.45 μm) and then pelleted by ultracentrifugation at 30,000 rpm in a SW40 rotor (Beckman, Palo Alto, CA) for 1 hour at 4°C. The pelleted viral particles were resuspended in 100 μL of phosphate-buffered saline (PBS) and stored at −20°C.
Target cells.
The murine cell line NIH 3T3 and the human (Kit-negative) cell line A431 were grown in DMEM supplemented with 10% FCS. The human SCF receptor (Kit)-expressing cell line HMC-1, kindly provided by Dr J.H. Butterfield,28 was grown in Iscove's modified Dulbecco's Eagle medium (IMDM) supplemented with 10% FCS and monothioglycerol. Jurkat (Kit-negative ) cells were grown in RPMI supplemented with 10% FCS. TF-1 (Kit-positive) human cells were grown in RPMI supplemented with 10% FCS and 1 ng/mL recombinant human granulocyte-macrophage colony-stimulating factor (rHuGM-CSF). The Kit receptor status of all cell lines had been previously determined29 30 and was verified using fluorescence-activated cell sorting (FACS) analysis with an anti-Kit antibody (Sigma Biosciences, St Loius, MO) and a secondary antimouse fluorescein isothiocyanate (FITC)-conjugated IgG (Santa Cruz Biotechnology, Santa Cruz, CA).
Immunoblots.
Ten microlitres of the pelleted viral particles was separated on a 10% polyacrylamide gel under reducing conditions and subsequently transferred onto nitrocellulose. For demonstration of factor Xa cleavage, 10 μL of the pelleted virus was incubated with 4 μg/mL of factor Xa protease (New England Biolabs, Beverly, MA) in the presence of 2.5 mmol/L CaCl2. The viral SU proteins were detected using a first layer goat antienvelope antiserum raised against Rausher murine leukemia virus envelope glycoproteins (Quality Biotech, Camden, NJ). Blots were developed using a secondary rabbit antigoat antibody conjugated to horseradish peroxidase (Dako, Glostrup, Denmark) and an enhanced chemiluminesence kit (Amersham Life Science, Bucks, UK).
Infections.
Target cells were plated into six-well plates at approximately 105 cells per well and incubated overnight at 37°C (adherent cells) or plated into six-well plates at approximately 106 cells per well 1 hour before infection (suspension cells). Filtered viral supernatant in serum-free medium was added to the target cells and incubated for 2 to 4 hours in the presence of 8 μg/mL polybrene.
The retroviral supernatant was then removed from the target cells, the medium was replaced with the usual medium, and the cells were placed at 37°C for a further 48 to 72 hours. X gal staining for detection of β-galactosidase activity was performed as previously described.31 Viral titer was calculated by counting blue stained colonies microscopically and expressed as enzyme-forming units per mL for the adherent cells. For suspension cells, 300 to 500 individual cells were counted and the percentage of blue stained cells was calculated. For infections involving factor Xa cleavage, the virus was incubated with 4 μg/mL of factor Xa protease in the presence of 2.5 mmol/L CaCl2 for 90 minutes before infection. The competition experiments were performed as above after preincubation of the target cells for 2 hours at 37°C with 100 nmol/L rHuSCF (Amgen, Thousand Oaks, CA, a kind gift from Stephen Devereux, University College, London, UK).
Binding assays.
Target cells were washed with 2% bovine serum albumin (BSA)/PBS and aliquots of approximately 106 cells were incubated with viral supernatant at 32°C for 45 minutes. The cells were then washed again and incubated with 100 μL of 83A25 monoclonal anti-SU antibody32 at 4°C for 45 minutes. After a further wash, dichlorotriazinyl amino fluorescein (DTAF)-conjugated affinity purified F(ab)2 fragment goat antirat IgG (Immunotech, Marseille, France) was added and the incubation continued at 4°C. Cells were counterstained with 10 μg/mL propidium iodide 1 to 2 minutes before the final washes. Fluorescence of live cells was analyzed using a fluorescent-activated cell sorter (FACSCalibur or FACScan; Becton Dickinson, San Jose, CA).
RESULTS
SCF chimeric envelope expression constructs were made in which a cDNA coding for the extracellular portion (165 aa) of human SCF was fused to the amino terminal codon of both the Moloney and 4070A MLV envelope glycoproteins via a factor Xa cleavable linker (Fig 1). For incorporation into β-galactosidase transducing retroviral vectors particles, the chimeric envelope expression constructs and control Moloney and 4070A unmodified envelope expression constructs were stably transfected into the TELCeB6 complementing cell line. Phleomycin-resistant colonies were pooled and the pooled transfectants were used as a source of vector for subsequent analysis.
The chimeric envelope expression constructs. Plasmid constructs encoding SCFXMo and SCFXA. The general format is shown diagrammatically and the amino acid sequence surrounding the site of fusion between the displayed ligand and the envelope protein is shown in detail. LTR, long terminal repeat; L, envelope signal peptide; phleor, phleomycin resistance gene. The Sfi 1 andNot 1 cloning sites are shown.
The chimeric envelope expression constructs. Plasmid constructs encoding SCFXMo and SCFXA. The general format is shown diagrammatically and the amino acid sequence surrounding the site of fusion between the displayed ligand and the envelope protein is shown in detail. LTR, long terminal repeat; L, envelope signal peptide; phleor, phleomycin resistance gene. The Sfi 1 andNot 1 cloning sites are shown.
The SCF chimeric envelopes are incorporated into retroviral vector particles and are susceptible to factor Xa cleavage.
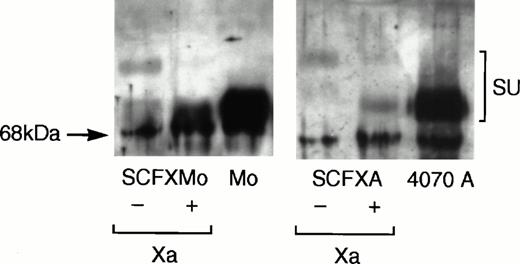
Viral supernatant was harvested from confluent plates of pooled TELCeB6 transfectants and pelleted viral particles were immunoblotted. Figure 2 shows that the chimeric envelopes were incorporated into virions and that their mobility on the immunoblot was retarded relative to that of the wild-type envelope glycoproteins. Chimeric envelope incorporation into virions is reduced approximately 10-fold compared with the wild-type envelope incorporation. Upon factor Xa cleavage of the pelleted virions, the mobility of the chimeric envelopes SCFXMo and SCFXA become indistinguishable from the mobility of the unmodified Moloney and 4070A envelope glycoproteins. Thus, the FXa signals in the interdomain linkers of SCFXMo and SCFXA are correctly recognized and cleaved by factor Xa.
Immunoblotting shows that the chimeric envelopes are incorporated into retroviral vector particles and are susceptible to factor Xa cleavage. Vectors incorporating the SCF-Moloney and SCF-4070A chimeric envelopes were pelleted by ultracentrifugation and subjected to denaturing sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. Vectors incorporating the wild-type Moloney and 4070A envelopes were used as a control. After transfer to nitrocellulose, the blots were probed with an anti-SU antiserum and developed using enhanced chemiluminescense. The mobility of the chimeric envelopes is retarded relative to that of the unmodified Moloney and 4070A envelope glycoproteins, but becomes indistinguishable from them upon factor Xa cleavage.
Immunoblotting shows that the chimeric envelopes are incorporated into retroviral vector particles and are susceptible to factor Xa cleavage. Vectors incorporating the SCF-Moloney and SCF-4070A chimeric envelopes were pelleted by ultracentrifugation and subjected to denaturing sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. Vectors incorporating the wild-type Moloney and 4070A envelopes were used as a control. After transfer to nitrocellulose, the blots were probed with an anti-SU antiserum and developed using enhanced chemiluminescense. The mobility of the chimeric envelopes is retarded relative to that of the unmodified Moloney and 4070A envelope glycoproteins, but becomes indistinguishable from them upon factor Xa cleavage.
The SCF-Moloney chimeric envelope binds to the SCF receptor, Kit.
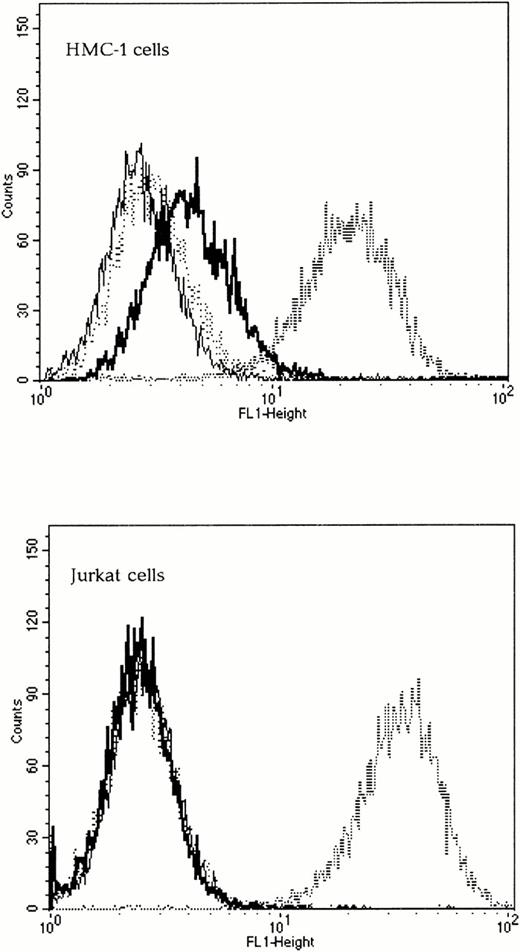
To determine if the chimeric envelopes could bind to Kit, SU binding assays were performed by FACS analysis. The wild-type 4070A SU was capable of binding to both Kit-positive and Kit-negative human cells and was used as a positive control. The Kit receptor status of all cell lines was verified using FACS analysis with an anti-Kit antibody (Santa Cruz Biotechnology) and a secondary antimouse FITC-conjugated IgG (Sigma, Poole, UK), data not shown. As expected, the wild-type Moloney ecotropic SU did not bind to any of the human cells tested, whereas SCFXMo SU was able to bind to Kit-positive, but not to Kit-negative human cells (Fig 3). To confirm the specificity of binding, the target cells were preincubated with 100 nmol/L SCF for 30 minutes and the subsequent viral binding step was performed in the presence of SCF. Under these conditions, the binding of SCFXMo SU was competitively inhibited (Fig 3). Thus, the Moloney SCF chimeric SU glycoproteins are capable of specific binding to Kit.
The SCF-Moloney chimeric envelope binds to the SCF receptor, Kit. Binding assays were performed on both Kit-positive (HMC-1) and Kit-negative (Jurkat) human cell lines using unmodified Moloney and 4070A envelopes and the SCFXMo chimeric envelope. The fine line represents Moloney binding (negative control), the dashed line, 4070A binding (positive control), the thick line, SCFXMo binding, and the dotted line, SCFXMo binding in the presence of exogenous rHuSCF. SCFXMo binds only to the Kit-positive cell line and the binding is competitively inhibited by the addition of rHuSCF.
The SCF-Moloney chimeric envelope binds to the SCF receptor, Kit. Binding assays were performed on both Kit-positive (HMC-1) and Kit-negative (Jurkat) human cell lines using unmodified Moloney and 4070A envelopes and the SCFXMo chimeric envelope. The fine line represents Moloney binding (negative control), the dashed line, 4070A binding (positive control), the thick line, SCFXMo binding, and the dotted line, SCFXMo binding in the presence of exogenous rHuSCF. SCFXMo binds only to the Kit-positive cell line and the binding is competitively inhibited by the addition of rHuSCF.
The SCF-Moloney chimeras retain the ability to infect mouse cells, but are unable to infect Kit-positive or Kit-negative human cells.
The vectors were titrated on murine NIH3T3 cells and on human Kit-positive (HMC-1 and TF-1) and Kit-negative (A431) cells. Vectors incorporating the SCFXMo chimeric envelopes gave a titer of 2.4 × 106 cfu/mL on NIH3T3 cells. This was slightly lower than the titer of 7.2 × 106 cfu/mL seen with the vectors incorporating unmodified Moloney envelope, in keeping with the reduced envelope incorporation. Vectors incorporating the SCFXMo chimeric envelopes did not infect human Kit-positive cells; there was no detectable β-galactosidase gene transfer when HMC-1 cells were infected with 1 mL of SCFXMo viral supernatant. Thus, retroviral vectors displaying SCF as an extension of the Moloney envelope glycoprotein are able to bind to human Kit, but subsequent steps in the virus entry pathway are not permitted.
Amphotropic vectors displaying SCF are competitively sequestered by Kit receptors on Kit-expressing cells.
The host-range properties of the amphotropic vectors incorporating the chimeric envelope SCFXA were then tested on Kit-expressing and nonexpressing cell lines. The EXA and wild-type 4070A vectors were used as controls. The titer of SCFXA was greatly reduced compared with EXA and 4070A on the Kit-positive cell lines HMC-1 and TF-1. All of these vectors were then treated with factor Xa protease to cleave off the displayed binding domains before infection. Preincubation with 4 μg/mL FXa had no effect on the titer of the EXA or wild-type vectors, but restored the titer of SCFXA towards that of the wild-type vector on the Kit-positive cell lines. The titer of SCFXA was unaffected by factor Xa cleavage on the Kit-negative cell lines. These data show that infectivity of the SCFXA vector on Kit-positive cells is blocked by the display of SCF and can be restored by cleavage of the displayed domain (Table 1).
Titration of Vectors on Different Cell Lines Before and After Factor Xa Cleavage
| . | Kit-Negative Cells . | Kit-Positive Cells (%) . | ||||||
|---|---|---|---|---|---|---|---|---|
| NIH 3T3 . | A431 . | HMC-1 . | TF-1 . | |||||
| −Xa . | +Xa . | −Xa . | +Xa . | −Xa . | +Xa . | −Xa . | +Xa . | |
| SCFXA | 3.6 × 105 | 5 × 105 | 4.2 × 105 | 6.7 × 105 | 4.2 | 40 | 2 | 38 |
| EXA | 5 × 105 | 5 × 105 | 2 × 102 | 1 × 106 | 40 | 36 | 45 | 46 |
| 4070A | 5.4 × 105 | 5 × 105 | 2.5 × 106 | 2.6 × 106 | 63 | 64 | 77 | 75 |
| . | Kit-Negative Cells . | Kit-Positive Cells (%) . | ||||||
|---|---|---|---|---|---|---|---|---|
| NIH 3T3 . | A431 . | HMC-1 . | TF-1 . | |||||
| −Xa . | +Xa . | −Xa . | +Xa . | −Xa . | +Xa . | −Xa . | +Xa . | |
| SCFXA | 3.6 × 105 | 5 × 105 | 4.2 × 105 | 6.7 × 105 | 4.2 | 40 | 2 | 38 |
| EXA | 5 × 105 | 5 × 105 | 2 × 102 | 1 × 106 | 40 | 36 | 45 | 46 |
| 4070A | 5.4 × 105 | 5 × 105 | 2.5 × 106 | 2.6 × 106 | 63 | 64 | 77 | 75 |
The host-range properties of the amphotropic vectors incorporating the chimeric envelope SCFXA were tested on Kit-expressing and nonexpressing cell lines with EXA and wild-type 4070A vectors as controls. All of these vectors were also tested after treatment with 4 μg/mL factor Xa protease before infection. The titer of SCFXA was reduced considerably compared with that of EXA and 4070A on the Kit-positive cell lines, but was restored by factor Xa protease cleavage. The titer of SCFXA was unaffected by factor Xa cleavage on the Kit-negative cell lines. These data show that infectivity of the SCFA vector on Kit-positive cells is blocked by the display of SCF and can be restored by cleavage of the displayed domain. Titers are shown as enzyme-forming units per mL (adherent cells) or percentage blue cells (suspension cells).
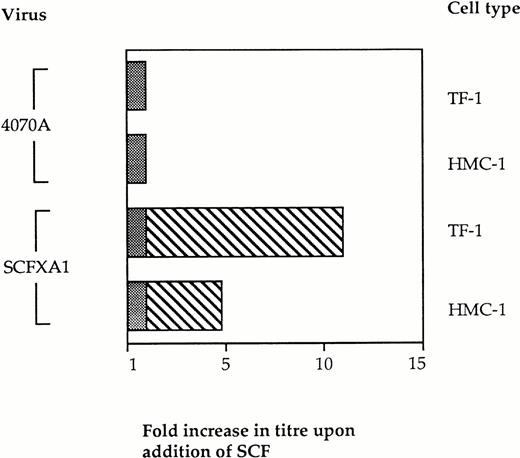
We then investigated the specificity of this block to infectivity on Kit-positive cells by retroviral display of SCF. If binding of the vector particles via the displayed domain prevents infection, the addition of exogenous SCF at the time of infection would be expected to restore infectivity by introducing competition with the vector particles for binding to Kit. Vector binding to Ram-1 will be favored under these conditions, thus enhancing infection and gene delivery. Infection experiments were therefore performed in the presence of soluble SCF as competitor. Kit-positive cells were preincubated with 100 nmol/L rHuSCF (Amgen, a kind gift from Stephen Devereux) for 2 hours before performing the infections with SCFXA1 and 4070A wild-type virus, as previously described. The infectivity of the SCFXA chimera was increased fivefold on HMC-1 cells and 11-fold on TF-1 cells (Fig 4). No increase in infectivity was seen with the wild-type control vectors. This indicates that the reduced ability of the chimeric vectors to infect Kit-positive cells is a consequence of targeted binding to SCF receptors.
The infectivity of vectors incorporating SCFXA chimeric envelopes is increased by the addition of exogenous rHuSCF. Infection of two different Kit-positive cells lines was performed in the presence of 100 nmol/L SCF. There was no change in the titer of wild-type 4070A vectors, but the titer of the vectors incorporating SCFXA chimeric envelopes increased fivefold on HMC-1 cells and 11-fold on TF-1 cells. The plain bars represent the background level of infectivity and the striped bars represent the fold increase in titer on addition of SCF.
The infectivity of vectors incorporating SCFXA chimeric envelopes is increased by the addition of exogenous rHuSCF. Infection of two different Kit-positive cells lines was performed in the presence of 100 nmol/L SCF. There was no change in the titer of wild-type 4070A vectors, but the titer of the vectors incorporating SCFXA chimeric envelopes increased fivefold on HMC-1 cells and 11-fold on TF-1 cells. The plain bars represent the background level of infectivity and the striped bars represent the fold increase in titer on addition of SCF.
Inverse targeting by SCF and EGF displaying retroviruses.
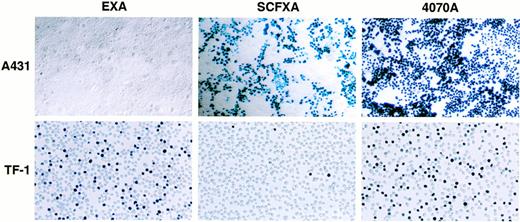
To determine whether EGF and SCF displaying vectors are capable of discriminating between receptor-positive and receptor-negative cell types in a cell mixture, cell mixing experiments were performed. Two milliliters of EXA, SCFXA, or 4070A viral supernatant was used to infect a mixed population of adherent A431 cells (kit-negative, EGF-R–positive) and either HMC-1 or TF-1 suspension cells (kit-positive, EGF-R–negative). After infection, the cells were separated and propagated in their respective media for 72 hours before X-gal staining. Infection with the wild-type control vector gave high titers on both cell types with 100% of the EGF-R–positive, kit-negative A431 cells being infected and 52% or 51% of the kit-positive, EGF-R–negative TF-1, or HMC-1 cells, respectively. In contrast, the chimeric vectors could efficiently discriminate between the different cell types as shown by the big difference between the cell types in the percentage of cells infected. The SCFXA vector preferentially infected the Kit-negative A431 cells, infecting 50% of the A431 cells, but only 3.6% and 4.3% of the TF-1 and HMC-1 cells, respectively, whereas EXA vectors preferentially infected the EGF-R–negative TF-1 or HMC-1 cells (42% and 37%, respectively), but infected less than 1% of the A431 cells. These data show that EXA and SCFXA vectors can be used to target diferent cell types in a cell mixing experiment. Pooled data showing the average percentage of cells infected from three separate experiments are shown in Table 2. A photomicrgraph of a representative experiment is shown in Fig5.
Inverse Targeting of Receptor-Negative Cells in a Mixed Population
| . | Experiment No. . | EXA (%) . | SCFXA (%) . | 4070A (%) . |
|---|---|---|---|---|
| A431 | 1 | <1 | 50 | 100 |
| 2 | <1 | 50 | 100 | |
| 3 | <1 | 50 | 100 | |
| TF-1 | 1 | 72 | 3 | 92 |
| 2 | 20 | 2 | 53 | |
| 3 | 35 | 6 | 56 | |
| A431 | 1 | <1 | 50 | 100 |
| 2 | <1 | 50 | 100 | |
| 3 | <1 | 50 | 100 | |
| HMC-1 | 1 | 61 | 3 | 66 |
| 2 | 20 | 2 | 29 | |
| 3 | 29 | 8 | 57 |
| . | Experiment No. . | EXA (%) . | SCFXA (%) . | 4070A (%) . |
|---|---|---|---|---|
| A431 | 1 | <1 | 50 | 100 |
| 2 | <1 | 50 | 100 | |
| 3 | <1 | 50 | 100 | |
| TF-1 | 1 | 72 | 3 | 92 |
| 2 | 20 | 2 | 53 | |
| 3 | 35 | 6 | 56 | |
| A431 | 1 | <1 | 50 | 100 |
| 2 | <1 | 50 | 100 | |
| 3 | <1 | 50 | 100 | |
| HMC-1 | 1 | 61 | 3 | 66 |
| 2 | 20 | 2 | 29 | |
| 3 | 29 | 8 | 57 |
Two milliliters of EXA, SCFXA, or 4070A viral supernatant was used to infect a mixed population of adherent A431 cells (kit-negative, EGF-R–positive) and either HMC-1 or TF-1 suspension cells (kit-positive, EGF-R–negative). After infection, the cells were separated and propagated in their respective media for 72 hours before X-gal staining. Infection with the wild-type control virus gave high titers on both cell types. In contrast, the chimeric vectors could discriminate between cell types. SCFXA vectors preferentially infected the Kit-negative A431 cells, whereas EXA vectors preferentially infected the EGF-R–negative TF-1 or HMC-1 cells. Data showing the individual titers from 3 separate experiments are shown in this table. Titers are shown as percentage blue cells.
Inverse targeting by SCF and EGF displaying retroviruses. When a mixed population of A431 (Kit-negative, EGF-R–positive) and TF-1 cells (Kit-positive, EGF-R–negative) was infected by vectors incorporating either the SCFXA or the EXA chimeric envelopes, the SCFXA vectors preferentially infected the Kit-negative cells and the EXA vectors preferentially infected the EGF-R–negative cells. A photomicrograph of a representative experiment is shown here. Upon X-gal staining, infected cells turn blue.
Inverse targeting by SCF and EGF displaying retroviruses. When a mixed population of A431 (Kit-negative, EGF-R–positive) and TF-1 cells (Kit-positive, EGF-R–negative) was infected by vectors incorporating either the SCFXA or the EXA chimeric envelopes, the SCFXA vectors preferentially infected the Kit-negative cells and the EXA vectors preferentially infected the EGF-R–negative cells. A photomicrograph of a representative experiment is shown here. Upon X-gal staining, infected cells turn blue.
DISCUSSION
We have shown that SCF can be displayed on retroviral vector particles as an N-terminal extension of the Moloney (ecotropic) and 4070A (amphotropic) MLV SU glycoproteins. The MLV envelope glycoproteins are incorporated into virions as a homotrimeric complex,33,34whereas, at least at high concentrations, soluble SCF exists as a glycosylated noncovalently associated dimer.35Theoretically, dimerization of an N-terminal SCF domain on the chimeric MLV envelopes might be expected to interfere with trimer assembly, leading to protein aggregation in the endoplasmic reticulum, failure of transport to the cell surface, and reduced incorporation into the vector particles. However, at physiologic concentrations, SCF dimers are known to dissociate such that the majority of circulating SCF is probably in monomeric form.36 In the current study, the SCF chimeric envelopes were incorporated into virions with approximately 10-fold reduced efficiency compared with the incorporation of wild-type unmodified envelopes. However, we have no direct evidence that the virally displayed SCF domains are associating into dimers at the tips of the envelope glycoprotein subunits on which they are displayed, and we have observed a similar reduction in the efficiency of viral incorporation of chimeric envelopes displaying an N-terminal interleukin-2 (IL-2) domain, which is of a similar size to SCF, but is not known to be capable of dimer formation (A.K. Fielding, F.J. Morling, and S.J. Russell, unpublished data, September 1995).
The extracellular domain of SCF is a large (165 amino acid) heavily glycosylated structure, which might be expected to interfere with the ability of the N-terminal domain of the underlying viral SU glycoprotein (to which it is grafted) to bind to its specific receptor. NIH3T3 cells express Rec-1 (ecotropic) and RAM-1 (amphotropic) receptors, but do not express Kit receptors. Therefore, infectivity on NIH3T3 cells reflects the interaction (attachment and fusion triggering) between the chimeric envelopes and the natural virus receptors. It is therefore notable that the vectors incorporating the SCF chimeric envelopes (SCFXA and SCFXMo) retain their capacity to infect NIH3T3 cells. Although they show slightly reduced infectious titers on these cells compared with wild-type controls, these reductions in titer are fully in keeping with the reduced levels of SCF chimeric envelope incorporation in the vector particles. This suggests that the displayed SCF in the chimeric envelopes does not strongly hinder attachment of the MLVs to their natural receptors, or interfere with subsequent fusion triggering. Nevertheless, a small part of the reduction in titer on NIH3T3 is probably due to a weak steric blocking effect because there was a slight (twofold) increase in infectious titer on these cells when the displayed SCF domain was first cleaved from the vector particles by factor Xa protease.
Our results show that the retrovirally displayed SCF domains can bind specifically to Kit receptors, but that this does not lead to efficient gene delivery into Kit-expressing cells. Thus, vector particles incorporating SCFXMo chimeric envelopes could bind to Kit receptors on human cells, but this did not lead to detectable transfer of the β-galactosidase gene. This result is fully consistent with previous experiments from several laboratories that have reported very low infectivity of ecotropic vectors, which have been retargeted to different human cell surface receptors.37-43
The results obtained with the amphotropic 4070A-SCF chimeric envelope show that, in the same way that EGF-displaying vectors are actively sequestered away from their entry pathway by binding to EGF receptors on EGF receptor-positive cells,18 SCF-displaying viruses are sequestered by SCF receptors on Kit-expressing cells. The SCFXA vectors could readily infect Kit-negative human cells, but gave greatly reduced titers on Kit-positive human cells, unless the displayed SCF domain was first cleaved from the vector particles using factor Xa protease. Moreover, their infectivity on the Kit-expressing cells was greatly enhanced when soluble SCF was added to the culture medium at the time of infection, indicating that the chimeric vector particles were being actively sequestered by the Kit receptors.
This is the first study to show that a displayed growth factor domain other than EGF can actively inhibit retrovirus infectivity by a receptor-dependent mechanism. It will be interesting to determine whether receptor-mediated sequestration of ligand-displaying retroviral vectors can be mediated by other tyrosine kinase growth factor receptors or by other classes of cell surface receptor. These experiments are ongoing.
Receptor-mediated sequestration of ligand-displaying retroviral vectors can provide the basis for a novel inverse targeting strategy allowing selective transduction of receptor-negative cells (or nontransduction of receptor-positive cells) in a mixed cell population. The cell mixing experiments presented in this study convincingly show the feasibility of the inverse targeting strategy. When the SCF-displaying vector was used to infect a mixture of carcinoma cells and hematopoietic cells, the Kit-negative carcinoma cells were efficiently transduced, whereas the Kit-positive hematopoietic cells were very poorly transduced (Table2). Conversely, when the EGF-displaying vector was used to infect the same cell mixture, the hematopoietic cells (which lack EGF receptors) were efficiently transduced, whereas the EGF receptor-positive carcinoma cells were hardly infected at all.
Inverse targeting can now be added to a growing list of retroviral targeting strategies. The classical approach is to extend the host range of vectors that do not naturally bind to human cells by domain substitution.37-40,42 Indeed, when erythropoietin37 or a single chain variable fragment against the low-density lipoprotein receptor41 were displayed on the Moloney MLV envelope and coincorporated into retroviral particles with unmodified envelopes, targeted entry to human cells was observed. Although these studies establish the principle of targeting, the very low titers seen abrogate the use of this approach for clinical applications. A recent attempt to target gene transfer to Kit-positive cells using a plasmid DNA vector conjugated to SCF also showed a small transient increase in reporter gene activity over a control vector in the Kit-positive cells.44 Unlike targeted retroviral vectors, entry into the endosomal pathway is the predominant route for these vectors to enter the cell. For retroviral vectors, entry into the endosomal pathway probably reduces the likelihood of viral fusion with the cell membrane and hence leads to failure to infect the cell. Molecular conjugate vectors however remain unsuitable for applications involving HSC, as they do not permit integration of a therapeutic gene into the cellular DNA. An alternative approach using retroviral vectors has been to generate protease-activatable vectors whose infectivity is dependent on the presence of a specific protease.18,45 46 In contrast to these previously described targeting strategies, inverse targeting is ideally suited to protocols in which it is important to avoid infecting a specific population of cells (see below).
The strength of the use of EGF-displaying retroviruses for ‘inverse targeting’ of hematopoietic cells lies in the fact that EGF receptors are widely expressed on many different cell types. A high-density of EGF receptors has been found on many nonhematologic malignancies,47 but there are no EGF receptors on hematopoietic cells.48 EGF displaying retroviral vectors will be preferable to the nontargeted vectors that are currently being used in clinical studies for the transduction of hematopoietic stem cells with chemotherapy resistance genes such as the multiple drug resistance-1 (MDR-1) gene.1,49 In this strategy, a drug resistance gene is introduced into HSCs of patients undergoing high-dose therapy with stem cell rescue for the treatment of malignancy. After engraftment, it is envisaged that the transduced hematopoietic cells will have increased resistance to cytotoxic chemotherapy, allowing multiple cycles of intensive therapy to be delivered with minimal myelotoxicity to patients with chemosensitive tumors, such as carcinoma of the breast. A major weakness in the approach is that, even in patients with no overt evidence of bone marow involvment by tumor, it has been shown that bone marrow and peripheral blood stem cell harvests may be contaminated by malignant cells.50 51 Avoidance of transduction of contaminating tumor cells with the MDR-1 gene is clearly desirable and, for EGF receptor-positive solid tumors such as breast carcinoma, might conveniently be achieved by using an EGF-displaying vector.
Similarly, vectors incorporating SCF chimeric envelopes may find application in situations where it is desirable to transduce Kit-negative tumor cells, eg, with ‘suicide genes’ such as the HSV-TK gene52 and avoid transduction of hematopoietic stem cells.
ACKNOWLEDGMENT
We thank Dr C. Casimir for the SCF cDNA and the TF-1 cell line, Dr S. Devereux and Amgen UK for the rHuSCF, Dr J.H. Butterfield for the HMC-1 cell line, and Dr L. Evans for the 83A25 monoclonal antibody-producing cell line.
Supported by the Leukaemia Research Fund of Great Britain (London, UK) and by the Medical Research Council (London, UK).
Address reprint requests to Stephen J. Russell, MD, Centre for Protein Engineering, MRC Centre, Hills Rd, Cambridge CB2 2QH, UK.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal