Abstract
Hemin inhibits transcription of the tartrate resistant acid phosphatase (TRAP) gene. Using deletion mutagenesis of the mouse TRAP 5′-flanking region, we previously identified a 27-bp DNA segment containing a central GAGGC tandem repeat sequence (the hemin response element [HRE]), which bound nuclear proteins (hemin response element binding proteins [HREBPs]) from hemin-treated cells and appeared to be responsible for mediating transcriptional inhibition in response to hemin. We now have used affinity binding to HRE-derivatized beads to identify four HREBP components with apparent molecular masses of 133-, 90-, 80-, and 37-kD, respectively. The 80- and 90-kD components correspond to the p70 and p80/86 subunits of Ku antigen (KuAg) as documented by partial amino acid microsequencing of tryptic digests and immunologic reactivity. Based on reactivity of the HREBP gel shift band with antibodies to the redox factor protein (ref1) in shift Western experiments, it is shown that the 37-kD component represents ref1. The 133-kD component appeared to be a unique protein. KuAg participation in HREBP complexes was specific as it was present in HREBPs bound to HRE microcircles. Results of depletion/reconstitution experiments suggested that KuAg does not bind alone or directly to HRE DNA, but does so only in conjunction with the 133- and/or 37-kD proteins. We conclude that HREBP is a heterogeneous complex composed of KuAg, ref1, and a unique 133-kD protein. We speculate that the role of heme may be to promote interactions among these components, thereby facilitating HRE binding and downregulation of hemin responsive genes.
RECENTLY WE HAVE described inhibition of tartrate-resistant acid phosphatase (TRAP) gene transcription by hemin (ferric protoporphyrin IX).1,2 Using deletion mutagenesis of the mouse (m) TRAP gene 5′ flanking region, we identified a 27-bp DNA segment containing a central GAGGC tandem repeat sequence (the hemin response element [HRE]), which bound nuclear proteins (hemin response element binding proteins [HREBPs]) from hemin-treated cells and appeared to be responsible for mediating transcriptional inhibition in response to hemin.2 A GENINFO data base search showed several other mammalian genes with tandem GAGGC motifs in noncoding regions, providing the possibility that additional genes may be regulated in similar fashion by hemin at the level of transcription.2 We now report that the HREBP is a complex comprising up to four proteins and that both the Ku antigen (KuAg) (p70/p86)3-5 and the redox factor (ref1) protein6 are associated with this complex.
KuAg is a heterodimeric DNA binding protein consisting of p70 and p80/86 subunits originally described as an autoantigen recognized by antibodies from patients with systemic lupus erythematosus, overlap syndrome, and other autoimmune disorders.3-5,7,8 KuAg plays an important role in double-stranded break repair of DNA and V(D)J recombination, both functions dependent on the targeting to DNA by KuAg of the enzyme, DNA-dependent protein kinase (DNA-PK).9-14KuAg appears to recognize and bind DNA structures containing double-to- single-stranded DNA transitions, as well as double-stranded DNA ends.15-18 A unique feature of the KuAg-DNA interaction is that in vitro KuAg is able to translocate freely along DNA after its initial binding,19 although this translocation is blocked in vivo by nucleosomes.20 It has been suggested that KuAg may in this way serve as a general inhibitor of DNA-protein complex formation and of transcription by displacing specific transcription factors from DNA.21 In addition, Giffin et al22,23 have described and characterized sequence-specific KuAg binding to NRE1 (negative regulatory element 1), a DNA sequence element in the long terminal repeat (LTR) of mouse mammary tumor virus (MMTV). KuAg also has been described to possess DNA helicase activity.24
An interaction between KuAg and the ref1 protein has been previously described.25 Ref1 is a 37-kD protein that regulates the DNA binding activity of the Fos-Jun heterodimer by mediating reduction of conserved cysteine residues in the DNA-binding domains of these two proteins.6
Recently, a specific interest of this laboratory has been hemin-induced transcriptional inhibition of TRAP gene expression. We have shown that this process is mediated by binding of nuclear proteins, the HREBP, to a specific DNA element, the HRE. The aim of this study was to identify and characterize the nuclear proteins comprising the HREBP.
We now present data derived from affinity purification, microsequencing, immunoblotting, and gel mobility assays, which demonstrate complexing of KuAg, ref1, and a 133-kD protein with the HRE of the mTRAP gene. These data provide evidence for additional roles for KuAg and ref1 in transcriptional regulation of gene expression.
MATERIALS AND METHODS
Cell lines, antibodies, and oligonucleotides.
Human U937 cells,26 obtained from American Type Culture Collection (Rockville, MD), are maintained in our laboratory. Monoclonal antibodies recognizing KuAg components p70 (Ab-4, clone N3H10), p80 (Ab-2, clone 11D), or both p70/p80 (Ab-3, clone 162) were obtained from NeoMarkers, Fremont, CA.27 Dr Tom Curran (St Jude Children's Research Hospital, Memphis, TN) kindly provided affinity-purified rabbit anti-ref1 antibody. Native, scrambled, and biotinylated HRE 27-bp oligonucleotides were synthesized at GIBCO-BRL (Gaithersburg, MD). The native 27-mer sequence is as follows: 5′-ACCTTGGAGGCGAGGCGCAGGTAATGG-3′.2The tandem repeat HRE sequence is underlined. Scrambled oligonucleotides used in some experiments had the following sequence: 5′-GTTATAATGTGTACATCCATCACTGT-3′.
Affinity isolation of HREBPs.
U937 cells were incubated with hemin (50 μmol/L) for 48 hours. Nuclear proteins were extracted as previously described2 using the technique of Andrews and Faller.28 We used a modification of the methods described by Ren et al29 to isolate from the nuclear extracts proteins binding to the 27-mer HRE DNA sequence. Briefly, biotinylated authentic or scrambled oligonucleotides were coupled to streptavidin-coated magnetic beads (Dynabeads M-280; Dynal, Lake Success, NY). Beads were washed twice with 1% bovine serum albumin (BSA) in phosphate-buffered saline (PBS), twice with 1 mol/L NaCl in PBS, then twice with 10 mmol/L Tris-CL, 1 mmol/L EDTA, pH 8.0 (TE buffer). Oligonucleotides diluted in 0.5 mL TE buffer were added to the washed beads and incubated with rotation for 30 minutes at room temperature. Unbound oligonucleotides were removed by washing with TE buffer. Derivatized beads contained 250 pmol of oligonucleotide per 400 μL of beads (6 to 7 × 108 beads/mL). To isolate specific DNA binding proteins, nuclear extracts first were dialyzed against 25 mmol/L HEPES, pH 7.6, 40 mmol/L KCl, 0.1 mmol/L EDTA, 10% glycerol, 1 mmol/L dithiothreitol, 0.1 mmol/L phenylmethylsulfonyl fluoride, 0.1 μg/mL aprotinin, 0.5 μg/mL N-tosyl-L-phenylalanine chloromethyl ketone (TPCK) (dialysis buffer). Subsequent incubations were scaled up many fold compared with those performed by Ren et al,29 but buffers and proportions were the same. We used 400 μL of derivatized Dynabeads per 3 to 5 mL of nuclear proteins in dialysis buffer. Binding reactions were performed for 30 minutes at room temperature with rotation. For one part nuclear protein extract we used 10 parts 5× gel shift buffer (1× gel shift buffer = 20 mmol/L Tris-HCl, pH 7.0, 100 mmol/L NaCl, 5 mmol/L EDTA, 0.1% Nonidet P-40, 0.5 μg/mL BSA, 5% glycerol); 0.125 parts 1 mol/L MgCl2; and 37.8 parts dialysis buffer. After incubation, unbound proteins were removed by washing the beads with 1× gel shift buffer containing 2.5 mmol/L MgCl2 and 0.1% Nonidet P-40, but without BSA. Beads were collected using a Dynal magnetic particle concentrator and proteins were recovered by boiling in Laemmli sample buffer then separated by sodium dodecyl sulfate-polyacrylamide gel electropheresis (SDS-PAGE) in 10% acrylamide gels.30Proteins were transferred electrophoretically to polyvinylidene difluoride (PVDF) membranes for immunoblotting and to obtain material for microsequencing. Microsequencing was performed in the Institutional Protein Core Laboratory, UTHSCSA, directed by Dr Lynda Bonewald.
Immunodetection of KuAg.
Immunoblotting was performed as previously described31 with KuAg subunit specific monoclonal antibodies used at a dilution of 1:500. Secondary antibody was goat antimouse IgG F(ab′)2 conjugated to horseradish peroxidase (Pierce Chemical Co, Rockport, IL) used at a dilution of 1:4,000 followed by treatment with chemiluminescence solution (Pierce Supersignal Chemiluminescent, Rockford, IL) and visualization by autoradiography.
Gel mobility assays.
Gel mobility assays were performed as previously described2using nuclear extracts prepared according to Andrews and Faller.28 HRE oligonucleotide probes (10 pmol/L) were radiolabeled with 10 mCi/mL γ-32P (specific activity, 3,000 Ci/mmol) by reaction with T4 polynucleotide kinase for 30 minutes at 37°C. The reaction was stopped with 0.5 mol/L EDTA, and probes were purified by the Qia quick nucleotide removal kit (Qiagen, Valencia, CA) used according to the manufacturer's instructions. Probes were suspended in TE buffer and used in binding reactions (10,000 to 20,000 cpm) with 2 to 10 μg nuclear extract protein incubated at 37°C for 30 minutes with BSA and poly-(dI-dC). After incubation, reaction mixtures were analyzed by electrophoresis in a 4% low ionic strength native polyacrylamide gel with an acrylamide:bisacrylamide ratio of 80:1, followed by autoradiography.
For supershift experiments, incubations with oligonucleotide probes were performed as described, then followed by an additional incubation for 30 minutes with antibody, 10 μg per reaction mixture.
Construction of HRE microcircles and gel mobility assays with microcircles.
Tandem repeats of HRE containing complementary oligomers were synthesized (GIBCO-BRL) with BamHI restriction enzyme sites flanking the ends. Both strands were annealed and subcloned into pBluescript II SK+ vector at the corresponding site. Direct sequencing confirmed incorporation of the correct 27-bp HRE sequence. A 310-bp fragment containing the HRE sequences was excised from this plasmid construct (pHRE#5) by PVU II/Xho I restriction enzyme digestion and subjected to fill-in reaction in the presence of32P labeled deoxycytidine triphosphate (dCTP) using Klenow-enzyme. Alternatively, a 1.69-kb DNA fragment containing the HRE sequences was also excised from the pHRE#5 plasmid by Bgl I and end labeled in the presence of 32P-labeled γ ATP using T4 polynucleotide kinase. The radiolabeled fragments were self-ligated using T4 DNA ligase and the products were digested with 1 U each ofBal 31 and S 1 nucleases. The resulting HRE-containing microcircles resistant to these exonucleases were gel purified and used in gel mobility shift assays. Gel mobility assays with microcircles were performed using 1% agarose gels rather than the standard 4% polyacrylamide used for the same assays with double-stranded oligonucleotides because microcircles did not enter the polyacrylamide when incubated with nuclear extract.
Immunodepletion experiments.
Immunodepletion was used to remove KuAg from nuclear extracts before some gel mobility assays. In brief, KuAg specific monoclonal antibody, Ab-3, was bound to tosylactivated magnetic beads (Dynabeads M-280 Tosylactivated; Dynal) according to the manufacturer's instructions. Derivatized beads were incubated with nuclear extracts prepared as described above for 30 minutes at room temperature, then collected magnetically, and washed with 1× gel shift buffer. The captured proteins were eluted from the beads by incubation for 30 minutes at 4°C in high salt extraction buffer, 20 mmol/L HEPES-KOH, pH 7.9, 24% glycerol, 420 mmol/L NaCl, 1.5 mmol/L MgCl2, 0.2 mmol/L EDTA, 0.5 mmol/L dithiothreitol, 0.2 mmol/L phenylmethylsulfonyl fluoride at 4°C.
Shift-Western blotting.
The method of Demczuk et al32 was used to identify the protein components of gel mobility assays in combination with immunoblotting. After completion of gel mobility assays performed as described above, electrophoretic transfer was performed at room temperature in 48 mmol/L Tris/39 mmol/L glycine/20% methanol, pH 8.5. Stacked membranes were used with the first membrane below the gel being nitrocellulose (Millipore, Bedford, MA) to bind proteins, but not double-stranded DNA probes, followed by a polyvinylidene difluoride (PVDF) membrane (Millipore) to retain radiolabeled probe. The membrane components were separated with Whatman 3MM paper (Whatman, Hillsboro, OR) during transfer and all were preequilibrated in the transfer buffer. Subsequent to transfer, the radiolabeled probe was identified by autoradiography, and protein bands retained on the nitrocellulose membrane were analyzed by Western blotting as described above.
Ultraviolet (UV) cross-linking experiments.
To estimate sizes of DNA-binding proteins or protein complexes detected by gel mobility assays, UV cross-linking was performed as described by Chodosh.33 For these experiments, nuclear extracts and radiolabeled nucleotide probes were incubated under conditions used for gel mobility assays, then subjected to an additional 60-minute incubation with UV illumination (324 nm at 5 cm). After incubation, reaction mixtures were analyzed by PAGE and autoradiography.
RESULTS
Affinity isolation of HREBP.
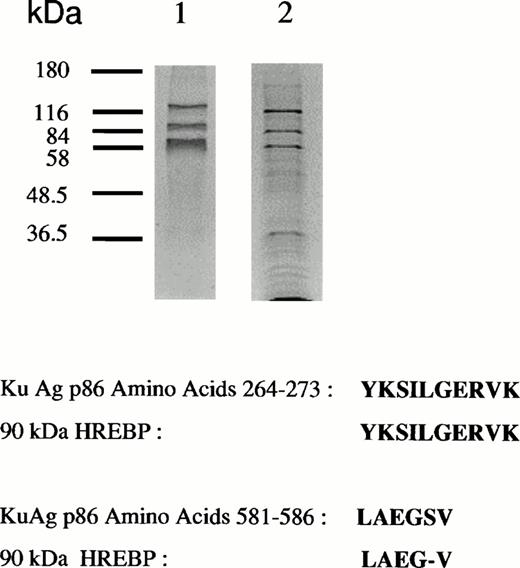
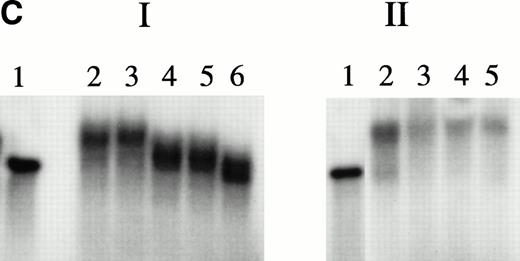
Figure 1 shows gel mobility assays performed with nuclear extracts from hemin-treated U937 cells before and after affinity depletion by beads derivatized with the 27-bp HRE in wild-type or scrambled configurations. Extracts subjected to multiple rounds of specific affinity depletion no longer produced a band shift, whereas those treated with the scrambled oligomer produced a pattern indistinguishable from that given by unmanipulated extracts. Therefore, specific depletion of HREBPs was accomplished. Protein components bound to HRE-derivatized beads were analyzed by SDS-PAGE (Fig 2A). Protein components of 133-, 90-, and 80-kD were regularly obtained. As illustrated by the results in Fig2, a component of 37 kD also was isolated from some, but not all, preparations. It is unknown why variable recovery of this component occurred despite identical affinity isolation conditions. Nuclear extracts derived from 2.5 × 108 cells contained approximately 5 mg of total protein from which was derived 500 ng of the 90-kD component.
Analyses of HREBPs in nuclear extracts by gel mobility assays. Gel mobility assays were performed as described in Materials and Methods using nuclear extracts from hemin-treated U937 cells and radiolabeled 27-mer HRE probes. For the experiments in lanes 2 and 4, extracts were preincubated x 5 with beads derivatized with irrelevant (scrambled) or wild-type 27-mer HREs, respectively. Lane 1, free probe. Lane 2, U937 extract preincubated with scrambled 27-mer beads. Lane 3, unmanipulated extract. Lane 4, extract preincubated with beads derivatized with wild-type 27-mer HRE sequence.
Analyses of HREBPs in nuclear extracts by gel mobility assays. Gel mobility assays were performed as described in Materials and Methods using nuclear extracts from hemin-treated U937 cells and radiolabeled 27-mer HRE probes. For the experiments in lanes 2 and 4, extracts were preincubated x 5 with beads derivatized with irrelevant (scrambled) or wild-type 27-mer HREs, respectively. Lane 1, free probe. Lane 2, U937 extract preincubated with scrambled 27-mer beads. Lane 3, unmanipulated extract. Lane 4, extract preincubated with beads derivatized with wild-type 27-mer HRE sequence.
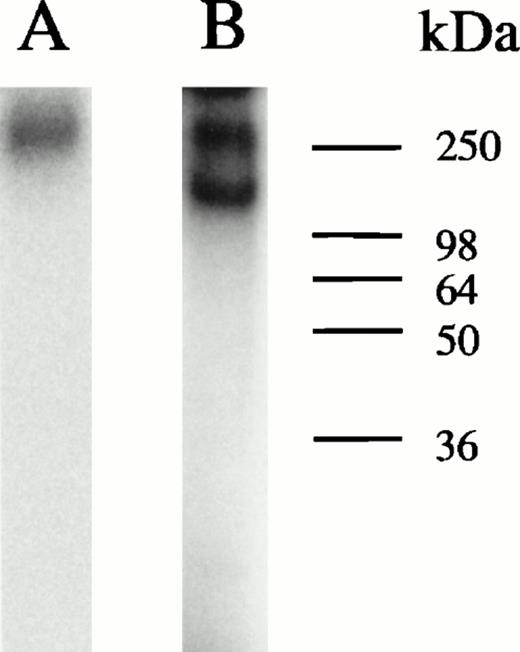
Analyses of protein bound to HRE-derivatized magnetic beads. (A) SDS-PAGE analysis. Nuclear extracts from hemin-treated U937 cells were incubated with magnetic beads derivatized with the 27-mer HRE DNA sequence as described in Materials and Methods. Bound proteins were released by boiling in Laemmli buffer, then separated by SDS-PAGE in 10% acrylamide gels. Protein bonds were visualized by silver or Coomassie blue staining. Lanes 1 and 2 depict results of two separate isolations. (B) Microsequence analysis of two peptides from the 90-kD HREBP band. Digests were sequenced on an Applied Biosystems 477A sequencer with an ABI 120 HPLC. KuAg p86 sequences are from the BLAST data base. Swiss-Prot accession:P13010; NCBI Seq ID:125731. (C) Immunoblot analyses of isolates in (A) with anti-KuAg monoclonal antibodies. Lane 1, Ab-3, clone 162 recognizing p70/p80; lane 2, Ab-4, clone N3H10 recognizing p70; lane 3, Ab-2, clone 11D recognizing p80.
Analyses of protein bound to HRE-derivatized magnetic beads. (A) SDS-PAGE analysis. Nuclear extracts from hemin-treated U937 cells were incubated with magnetic beads derivatized with the 27-mer HRE DNA sequence as described in Materials and Methods. Bound proteins were released by boiling in Laemmli buffer, then separated by SDS-PAGE in 10% acrylamide gels. Protein bonds were visualized by silver or Coomassie blue staining. Lanes 1 and 2 depict results of two separate isolations. (B) Microsequence analysis of two peptides from the 90-kD HREBP band. Digests were sequenced on an Applied Biosystems 477A sequencer with an ABI 120 HPLC. KuAg p86 sequences are from the BLAST data base. Swiss-Prot accession:P13010; NCBI Seq ID:125731. (C) Immunoblot analyses of isolates in (A) with anti-KuAg monoclonal antibodies. Lane 1, Ab-3, clone 162 recognizing p70/p80; lane 2, Ab-4, clone N3H10 recognizing p70; lane 3, Ab-2, clone 11D recognizing p80.
Identification of KuAg as a component of the HREBP complex.
Sequencing of the 90-kD HREBP yielded two amino acid sequences homologous to ATP-dependent DNA helicase II, the KuAg p80/86 subunit3 (Fig 2B). There was complete identity between a 10-amino acid sequence of the 90-kD HREBP and the p86 KuAg, amino acids 264 to 273; and another sequence of the HREBP was 83% homologous to the p86 KuAg sequence at amino acids 581 to 586.3 To confirm that KuAg was present in the HREBPs affinity-purified from hemin-treated U937 nuclear extracts, immunoblotting was performed with KuAg subunit specific monoclonal antibodies.27 Results are given in Fig 2C and indicated that both p80/86 and p70 KuAg components were recovered as HREBPs with the 90-kD band representing KuAg p80/86 and the 80-kD band representing KuAg p70.
Identification of KuAg and ref1 in gel mobility assays.
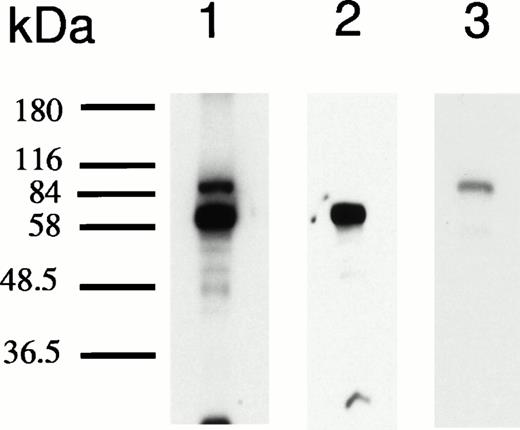
Gel mobility assays performed with nuclear extracts of hemin-treated cells and radiolabeled 27-mer HRE oligonucleotides produced a characteristic pattern2 (Fig 1). To determine if KuAg were involved in this gel mobility shift observed after hemin treatment, two types of additional experiments were performed: (1) supershift experiments with anti-Ku monoclonal antibodies, and (2) shift-Western blotting.31 Results of a representative supershift experiment using the monoclonal anti-Ku antibody, Ab-3 (reactive with both KuAg components), are illustrated in Fig 3A. The appearance of a gel mobility supershift in the presence of antibody provides evidence for presence of KuAg in the HREBP complex. For the shift-Western experiments, various KuAg subunit specific monoclonal antibodies27 were used to probe proteins retained on the nitrocellulose filters after electrophoretic transfer to stacked membranes was performed as described in Materials and Methods. Results in Fig 3B indicate that both the p80/86 and p70 KuAg components were present in the band shifts of gel mobility assays performed under standard conditions. To provide additional confirmation that KuAg contributed to the gel mobility shift results, nuclear extracts were prepared as usual, then immunodepleted of KuAg, as described in Materials and Methods. Immunoblotting was performed to determine the extent of KuAg depletion (Fig 3C). Using KuAg-depleted nuclear extracts in the standard gel mobility assay resulted in abrogation of the usual gel shift pattern compared with that given by undepleted extracts (Fig 3C). Therefore, we conclude that KuAg is a component of the HREBP involved in the gel mobility shift observed with nuclear extracts after hemin treatment of the cells.
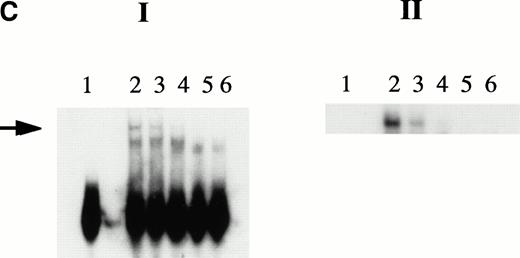
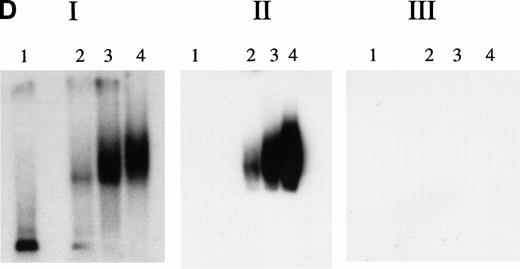
KuAg is bound to HREs in gel mobility assays with nuclear extracts from hemin-treated U937 cells. (A) Supershift experiment. Gel mobility assays with radiolabeled 27-bp HRE probes were performed as described in Materials and Methods. Supershift experiments included an additional 30-minute incubation with Ab-3. Lane 1, standard gel mobility assay; lane 2, parallel supershift experiment with Ab-3. The arrow indicates the supershifted band. (B) Shift-Western experiments. Gel mobility assays with HRE probes were performed, followed by electrophoretic transfer of proteins and probes to stacked nitrocellulose and PVDF membranes as described in Materials and Methods. Panel 1, autoradiography of PVDF membrane. Remaining panels are immunoblots of nitrocellulose membrane with the anti-KuAg monoclonals Ab-4 (anti-P70), panel 2; and Ab-2 (anti-p80), panel 3; or with the anti-ref1 antibody, panel 4. Each panel depicts duplicate lanes. The arrow indicates the location of the shift band. The band at the top of each lane corresponds to the position of the sample wells. (C) Gel mobility assays with Ku depleted extracts. Nuclear extracts were prepared from hemin-treated U937 cells, then depleted of KuAg by serial incubations with Ab-3–derivatized magnetic beads as described in Materials and Methods. Panel I depicts standard gel mobility assays with radiolabeled 27-bp HRE probe. Lane 1, free probe; lane 2, undepleted extract; lanes 3 to 6 extract after 1, 2, 3, or 4 incubations, respectively, with anti-KuAg beads. Panel II: immunoblot of KuAg depleted extracts using antibody Ab-3. Lanes correspond to those of panel I. The arrow indicates the position of the largest gel shift band.
KuAg is bound to HREs in gel mobility assays with nuclear extracts from hemin-treated U937 cells. (A) Supershift experiment. Gel mobility assays with radiolabeled 27-bp HRE probes were performed as described in Materials and Methods. Supershift experiments included an additional 30-minute incubation with Ab-3. Lane 1, standard gel mobility assay; lane 2, parallel supershift experiment with Ab-3. The arrow indicates the supershifted band. (B) Shift-Western experiments. Gel mobility assays with HRE probes were performed, followed by electrophoretic transfer of proteins and probes to stacked nitrocellulose and PVDF membranes as described in Materials and Methods. Panel 1, autoradiography of PVDF membrane. Remaining panels are immunoblots of nitrocellulose membrane with the anti-KuAg monoclonals Ab-4 (anti-P70), panel 2; and Ab-2 (anti-p80), panel 3; or with the anti-ref1 antibody, panel 4. Each panel depicts duplicate lanes. The arrow indicates the location of the shift band. The band at the top of each lane corresponds to the position of the sample wells. (C) Gel mobility assays with Ku depleted extracts. Nuclear extracts were prepared from hemin-treated U937 cells, then depleted of KuAg by serial incubations with Ab-3–derivatized magnetic beads as described in Materials and Methods. Panel I depicts standard gel mobility assays with radiolabeled 27-bp HRE probe. Lane 1, free probe; lane 2, undepleted extract; lanes 3 to 6 extract after 1, 2, 3, or 4 incubations, respectively, with anti-KuAg beads. Panel II: immunoblot of KuAg depleted extracts using antibody Ab-3. Lanes correspond to those of panel I. The arrow indicates the position of the largest gel shift band.
In other experiments, shift Western analyses using affinity purified rabbit ref1 antiserum34 demonstrated presence of ref1 in the HREBP complex (Fig 3B). This antiserum did not bind to the complex in the presence of specific ref1 blocking peptide (Santa Cruz Biotechnology, Santa Cruz, CA; data not shown).
Gel mobility assays with HRE microcircles.
The potential for KuAg to represent a confounding variable for the interpretation of gel mobility assays has been recognized.35 To avoid the potential problem of nonspecific KuAg binding to the free ends of radiolabeled oligonucleotide probes, Giffin et al22 23 constructed DNA microcircles containing NRE1 to demonstrate sequence-specific KuAg binding. We used a similar technique to prepare HRE microcircles, then used 32P-labeled microcircles in agarose gel mobility and immunoblot assays. Microcircles were resistant to S1 or Bal 31 nuclease digestion (Fig 4A). Gel shift patterns with nuclear extracts are illustrated in Fig 4B and C. Incubation of radiolabeled microcircles with nuclear extracts from hemin-treated U937 cells resulted in a gel mobility shift (Fig 4B). Concentrations of bovine serum albumin more than sixfold the concentration of nuclear extract used did not produce a shift band (Fig 4B). Cold competition experiments with authentic 27-bp HRE in linear form demonstrated specific dose-dependent inhibition by authentic HRE sequences (Fig 4C), thus confirming specificity of the gel shift pattern observed with microcircles. Parallel competition experiments with scrambled 27-bp HRE (Fig 4C) or with 27-bp HRE in which the tandem GAGGC repeat was replaced with a different 10-bp nucleotide sequence (——CCGGCTTCCC——) gave no inhibition (data not shown). Immunoblotting with anti-KuAg showed KuAg bound to microcircles in the shift band (Fig 4D). Persistence of a mobility shift pattern with the microcircles and identification of KuAg by immunoblotting establishes the specificity of this component for the HREBP complex.
Experiments with microcircles. (A) Nuclease digestion experiments. Radiolabeled linear 27-bp HRE linear probes or microcircles containing HRE sequences were incubated with S1 nuclease (1 U, 15 minutes, 37°C) or Bal 31 nuclease (1 U, 15 minutes, 37°C), then analyzed by PAGE and autoradiography. Lane 1, untreated linear probe. Lane 2, untreated microcircles after ligation reaction and before gel purification. The arrow indicates unincorporated residual linear probe. Lanes 3 and 4, linear probe after digestion with S1 nuclease or Bal 31 nuclease, respectively. Lanes 5 and 6, microcircles after digestion with S1 nuclease or Bal 31 nuclease, respectively. (B) Mobility shift assays with nuclear extracts. Radiolabeled HRE microcircles were incubated with nuclear extracts from hemin-treated U937 cells (12 μg) or with bovine serum albumin, 4 to 80 μg, then analyzed by agarose gel electrophoresis and autoradiography. Lane 1, free microcircle probe. Lanes 2 and 3, probe plus nuclear extracts. Lanes 4, 5, 6, and 7, probe plus BSA, 4, 8, 40, or 80 μg, respectively. (C) Cold competition experiments. Gel mobility shift assays were performed with radiolabeled HRE microcircle probes. Competition experiments include a 15-minute preincubation with increasing concentrations of unlabeled linear oligonucleotides, either authentic 27-bp HRE, or scrambled HRE sequences. Panel I: Competition study with authentic probe. Lane 1, free microcircle probe. Lanes 2 and 3, probe plus nuclear extracts. Lanes 4, 5, and 6, probe, nuclear extract plus 50-fold, 100-fold, or 250-fold excess of linear 27-bp HRE. Panel II: Competition study with irrelevant probe. Lane 1, free microcircle probe; Lane 2, probe plus nuclear extract. Lanes 3, 4, and 5, probe, nuclear extract, plus 50-fold, 100-fold, or 200-fold excess of linear scrambled HRE. (D) Immunoblot analyses of gel shift bonds. Radiolabeled HRE microcircles were incubated with 12, 40, or 80 μg of nuclear extract from hemin-treated U937 cells, then analyzed by agarose gel electrophoresis. Panel I: Analysis by autoradiography (48-hour exposure). Lane 1, free microcircle probe. Lanes 2, 3, and 4, probe plus 12, 40, or 80 μg of nuclear extract. Panel II: Immunoblot analysis with anti-KuAg (Ab-3) (chemiluminescence exposure, 2 minutes). Lanes 1 to 4, same as panel I. Panel III: Immunoblot analysis with mouse anti–c-myc IgG (chemiluminescence exposure, 2 minutes). Lanes 1 to 4, same as panels I and II.
Experiments with microcircles. (A) Nuclease digestion experiments. Radiolabeled linear 27-bp HRE linear probes or microcircles containing HRE sequences were incubated with S1 nuclease (1 U, 15 minutes, 37°C) or Bal 31 nuclease (1 U, 15 minutes, 37°C), then analyzed by PAGE and autoradiography. Lane 1, untreated linear probe. Lane 2, untreated microcircles after ligation reaction and before gel purification. The arrow indicates unincorporated residual linear probe. Lanes 3 and 4, linear probe after digestion with S1 nuclease or Bal 31 nuclease, respectively. Lanes 5 and 6, microcircles after digestion with S1 nuclease or Bal 31 nuclease, respectively. (B) Mobility shift assays with nuclear extracts. Radiolabeled HRE microcircles were incubated with nuclear extracts from hemin-treated U937 cells (12 μg) or with bovine serum albumin, 4 to 80 μg, then analyzed by agarose gel electrophoresis and autoradiography. Lane 1, free microcircle probe. Lanes 2 and 3, probe plus nuclear extracts. Lanes 4, 5, 6, and 7, probe plus BSA, 4, 8, 40, or 80 μg, respectively. (C) Cold competition experiments. Gel mobility shift assays were performed with radiolabeled HRE microcircle probes. Competition experiments include a 15-minute preincubation with increasing concentrations of unlabeled linear oligonucleotides, either authentic 27-bp HRE, or scrambled HRE sequences. Panel I: Competition study with authentic probe. Lane 1, free microcircle probe. Lanes 2 and 3, probe plus nuclear extracts. Lanes 4, 5, and 6, probe, nuclear extract plus 50-fold, 100-fold, or 250-fold excess of linear 27-bp HRE. Panel II: Competition study with irrelevant probe. Lane 1, free microcircle probe; Lane 2, probe plus nuclear extract. Lanes 3, 4, and 5, probe, nuclear extract, plus 50-fold, 100-fold, or 200-fold excess of linear scrambled HRE. (D) Immunoblot analyses of gel shift bonds. Radiolabeled HRE microcircles were incubated with 12, 40, or 80 μg of nuclear extract from hemin-treated U937 cells, then analyzed by agarose gel electrophoresis. Panel I: Analysis by autoradiography (48-hour exposure). Lane 1, free microcircle probe. Lanes 2, 3, and 4, probe plus 12, 40, or 80 μg of nuclear extract. Panel II: Immunoblot analysis with anti-KuAg (Ab-3) (chemiluminescence exposure, 2 minutes). Lanes 1 to 4, same as panel I. Panel III: Immunoblot analysis with mouse anti–c-myc IgG (chemiluminescence exposure, 2 minutes). Lanes 1 to 4, same as panels I and II.
UV cross-linking experiments.
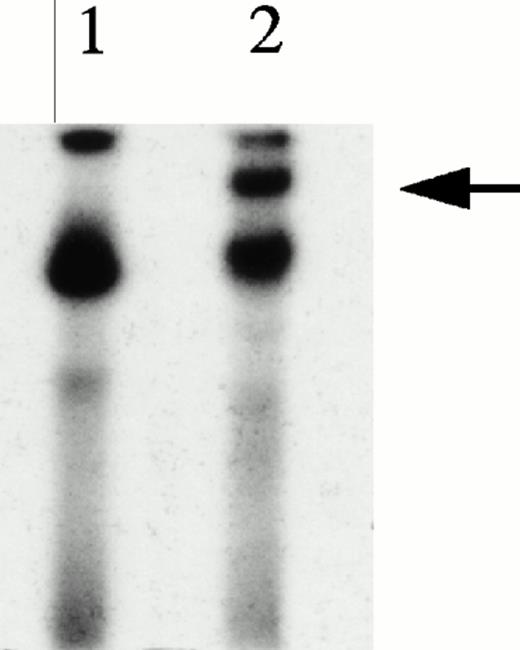
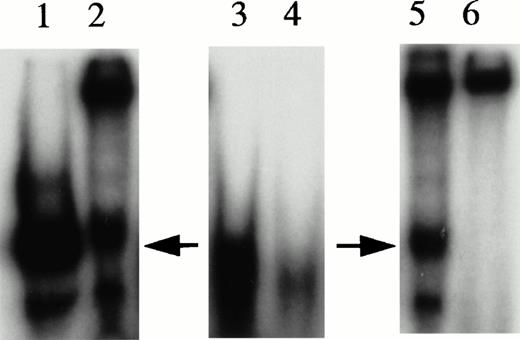
UV cross-linking experiments were performed to estimate molecular mass of protein complexes bound to the 27-mer HRE oligonucleotides during gel mobility assays and to serve as an indication as to whether KuAg was present alone or complexed with other proteins. UV cross-linking was performed as described in Materials and Methods34 and products were analyzed on denaturing gels. Results are presented in Fig 5. Two species of approximately 300 and 174 kD were present, with the former predominant in unmanipulated nuclear extracts. Conversely, in extracts first depleted of KuAg by incubation with antibody-coated beads, the predominant species was the 174-kD species. These data suggest that the KuAg may not itself interact directly with the HRE, but may bind only in association with other protein components, which in turn, may provide the DNA binding specificity. Based on estimated molecular weight (MW) of the HRE binding complex from Ku-depleted extract, DNA recognition and binding specificity appears to be provided by the 133-kD protein (p133) and/or the ref1 protein present in the affinity isolates.
UV cross-linking experiments. UV cross-linking experiments with hemin-treated U937 nuclear extracts and radiolabeled HRE probes were performed as described in Materials and Methods. Autoradiographs of the PAGE analyses are illustrated. Lane A, undepleted extract; lane B, KuAg-depleted (5 rounds of depletion) extract.
UV cross-linking experiments. UV cross-linking experiments with hemin-treated U937 nuclear extracts and radiolabeled HRE probes were performed as described in Materials and Methods. Autoradiographs of the PAGE analyses are illustrated. Lane A, undepleted extract; lane B, KuAg-depleted (5 rounds of depletion) extract.
Depletion/reconstitution experiments.
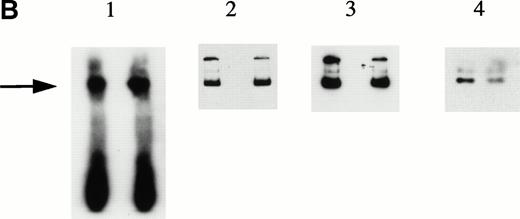
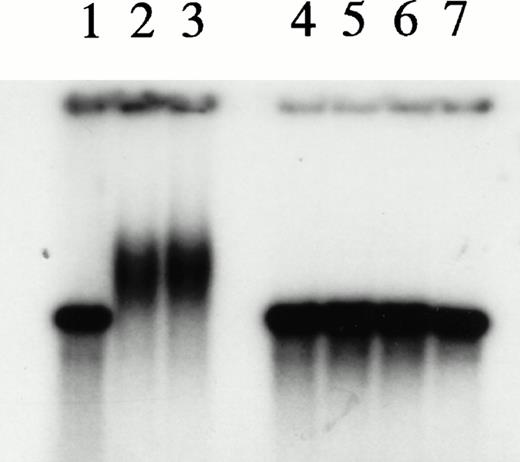
To explore further the relationships among the components of the HREBP complex, we prepared nuclear extracts depleted of HREBP by extensive sequential absorptions with HRE-derivatized, then anti–KuAg-derivatized beads. As expected, the usual gel shift pattern was abolished in assays using the depleted extract (Fig 6). The emergence of a lower molecular weight gel shift band in assays with depleted extracts most likely represents a low-affinity HRE binding component masked in the presence of HREBP. We next prepared nuclear extract fractions A and B. Fraction A was the 0.42 mol/L NaCl eluate from anti– KuAg-derivatized beads and contained predominantly KuAg components. Fraction B was the 0.42 mol/L NaCl eluate from HRE-derivatized beads to which first had been applied a KuAg-depleted nuclear extract. Fraction B contained predominantly non-KuAg HRE components. A series of reconstitution experiments were then performed in which depleted nuclear extract was mixed with Fraction A, Fraction B, or Fractions A+B. Results are shown in Fig 6. Under the experimental conditions, reconstitution with Fraction A containing predominantly KuAg did not produce the usual gel mobility shift pattern. However, either Fraction B alone or together with Fraction A added to depleted extract did partially reconstitute the pattern. Reconstitution with Fraction B alone produced a major lower MW and minor high MW bands consistent with some contamination of Fraction B with KuAg. These data are additional evidence that KuAg requires interactions with the other components of the HREBP complex to interact with the HRE DNA sequence.
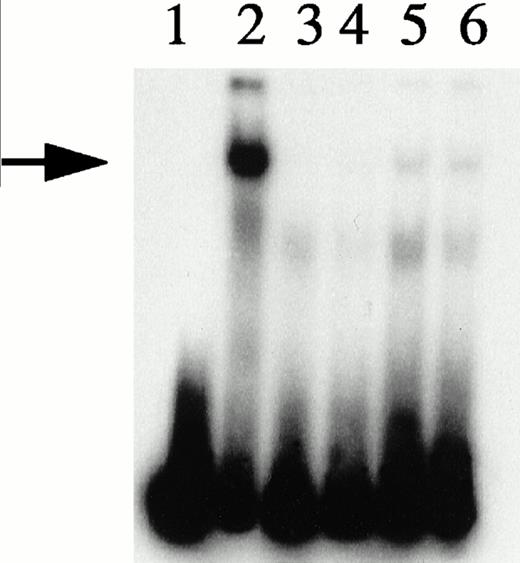
Depletion/reconstitution experiments. Nuclear extracts were prepared from hemin-treated U937 cells, then depleted of KuAg by affinity absorption to KuAg-derivatized beads followed by serial incubations with HRE-derivatized beads to remove non-KuAg HREBPs. Fraction A (primarily KuAg) was the eluate from KuAg-derivatized beads. KuAg-depleted extract was incubated with HRE-derivatized beads to obtain Fraction B, the eluate from these beads comprising predominantly non-KuAg HREBPs. Gel mobility assays were then performed with radiolabeled HRE probes and these various extracts alone, or reconstituted with fraction A, B, or A+B. Lane 1, free probe; lane 2, unmanipulated extract; lane 3, depleted extract; lane 4, depleted extract + Fraction A; lane 5, depleted extract + Fraction B; lane 6, depleted extract + Fractions A + B. The arrow indicates the position of the major gel shift band.
Depletion/reconstitution experiments. Nuclear extracts were prepared from hemin-treated U937 cells, then depleted of KuAg by affinity absorption to KuAg-derivatized beads followed by serial incubations with HRE-derivatized beads to remove non-KuAg HREBPs. Fraction A (primarily KuAg) was the eluate from KuAg-derivatized beads. KuAg-depleted extract was incubated with HRE-derivatized beads to obtain Fraction B, the eluate from these beads comprising predominantly non-KuAg HREBPs. Gel mobility assays were then performed with radiolabeled HRE probes and these various extracts alone, or reconstituted with fraction A, B, or A+B. Lane 1, free probe; lane 2, unmanipulated extract; lane 3, depleted extract; lane 4, depleted extract + Fraction A; lane 5, depleted extract + Fraction B; lane 6, depleted extract + Fractions A + B. The arrow indicates the position of the major gel shift band.
DISCUSSION
Previously we have identified a novel mechanism of transcriptional inhibition mediated by hemin.2 Induction of TRAP mRNA in cultured peripheral mononuclear cells was inhibited by hemin. This inhibition was dependent on nucleotide sequences in the mTRAP 5′-flanking region localized by deletion mutagenesis experiments to a 27-bp sequence at −1815 to −1789 bp relative to ATG.2 A tandem repeat sequence, GAGGC; GAGGC, contained within this DNA segment was shown to be involved in specific binding of HREBP in response to hemin. Highly homologous sequences have been identified in the 5′-flanking region of the hTRAP gene.36 A 607-bp segment of the mTRAP 5′-flanking region containing the HRE and adjacent sequences conferred hemin regulation on the viral SV40 promoter.2 Southwestern blotting experiments probing nuclear extracts of hemin-treated U937 cells with the 27-mer HRE sequence identified two protein bands at 37 and 133 kD representing candidate HREBPs.2
The present studies begin to elucidate the composition of HREBPs. Using affinity isolation techniques, we have identified four protein components binding specifically to immobilized HRE DNA sequences (Fig2). The 80- and 90-kD components correspond to the p70 and p80/86 subunits of KuAg, respectively, as documented by partial amino acid microsequence data and by immunologic reactivity with KuAg specific mouse monoclonal antibodies (Fig 2). The two components of 133 and 37 kD may correspond to the two components previously identified by Southwestern blotting.2 The demonstration by shift Western blotting that HREBP complexes contain the ref1 protein (Fig 3B) makes it likely that the 37-kD component represents ref1. Limited amino acid sequence data indicate that p133 may be a unique protein (O. Alcantara and D.H. Boldt, unpublished results, October 1997).
Because KuAg may be capable of nonspecific binding to double-stranded DNA ends,21,35 it is important to exclude this type of nonspecific interaction as an explanation for the findings. Several lines of evidence argue against this. First, KuAg was not removed from nuclear extracts by a scrambled double-stranded 27-mer HRE oligonucleotide used as a control for the affinity isolation procedure (Fig 1). Second, the shift Western blotting experiments demonstrated that KuAg was present in specific HREBP complexes identified by gel mobility assays (Fig 3). The specificity of these complexes for binding to the HRE tandem repeat GAGGC sequence has been established previously.2 Third, the Southwestern blotting experiments previously reported demonstrated no interaction of double-stranded 27-mer HRE probes with either p70 or p80/86 KuAg components.2 Fourth, KuAg alone was unable to reproduce the characteristic gel shift pattern when added to nuclear extracts first depleted of specific HRE-binding components (Fig 6). Finally, experiments with HRE microcircles confirmed the presence of KuAg in HREBP complexes (Fig 4). The use of microcircles to overcome potential problems of nonspecific KuAg-DNA interactions has been previously documented.22,23 37
Although the results of our experiments establish the specificity of the KuAg participation in HREBP binding to DNA, they do not establish the determinants of that specificity. Which component or components of HREBP binds directly to DNA and the sequence and manner of complex assembly remain to be determined, as does also the role of hemin induction. However, the UV cross-linking experiments (Fig 5) and the depletion/reconstitution experiments (Fig 6) strongly suggest that KuAg does not directly bind to HRE, but interacts only in the context of the HREBP complex.
Recent reports have highlighted the capability of KuAg to mediate sequence specific DNA binding.22,23 In those reports, KuAg was identified as a transcription factor capable of binding specifically to NRE1. When bound, KuAg recruited DNA-PK and the resulting complex inhibited glucocorticoid-induced gene transcription via phosphorylation and inactivation of glucocorticoid receptors.22 Although a consensus DNA sequence required for KuAg recognition has not been determined rigorously, limited data suggest that a polypurine/polypyrimidine core represents a minimal requirement.23 In the study of Giffin et al,23this characteristic plus substantial sequence identity to NRE1 were common features of DNA sequences interacting with KuAg. Neither are features of HRE.2 Therefore, this is additional evidence that binding of the HREBP complex to DNA is not mediated directly by KuAg. Additional experiments will be required to determine this point directly.
A number of other potential sequence-specific KuAg DNA binding sites have been proposed, but in general, these lack clear homology to NRE1 and potential confounding variables have not been rigorously excluded.38-42 It has been suggested that accumulation of KuAg at these sites may result from translocation after initial binding of KuAg to free ends, nicks, or other DNA structural features.23
In at least two instances, some data suggest that the interaction of KuAg with additional proteins may confer unique DNA binding specificities.25,41 Of particular interest to the findings in this report is the suggestion that interaction of KuAg with ref1 facilitates protein complex interaction with the DNA sequence comprising the negative calcium-responsive element (nCaRE) resulting in regulation of gene expression.25 Further experimentation will be required to determine whether such an interaction is responsible for the hemin-induced transcriptional inhibition we have observed. Although shift-Western experiments demonstrated ref1 in HREBP complexes, we were unable to confirm the presence of ref1 by immunoreactivity in complexes bound to HRE microcircles (data not shown). Although this result was observed repeatedly, these experiments were all performed with one U937 cell nuclear extract. We observed earlier that the 37-kD component could not be isolated from every nuclear extract examined (Fig 2A). The data provide the possibility that ref1 may participate in HRE binding, but its precise role remains equivocal.
The observations summarized above and the data in this report emphasize the variety of potential KuAg-dependent transcriptional regulatory mechanisms and also the potentially great flexibility inherent in a system in which DNA binding specificity might vary according to the specific protein partner complexing with KuAg. In this context, it is intriguing to speculate that hemin may function by promoting interaction(s) between KuAg and p133 and/or ref1 to produce the HREBP complex and lead to downregulation of hemin responsive genes. Such a role has recently been proposed for heme in regulating transcription of the CYP2B1/B2 gene in rat liver.43
In summary, we have demonstrated that the HREBP complex consists of the Ku antigen, a unique 133-kD protein, and the ref1 protein. Because binding of this complex to the HRE mediates transcriptional inhibition of TRAP gene expression in response to hemin treatment, future studies will be directed to understanding the potential role of hemin in complex assembly and/or DNA interaction and how binding of the complex functions to block transcription. Additional studies are in progress to clone and characterize the 133-kD protein.
ACKNOWLEDGMENT
The authors thank Jue Yang for technical assistance and Cheryl Muzzi Adams for secretarial support. We are grateful to Dr Tom Curran for providing anti-ref1 antibody.
Supported by Grant No. DK 50425 from the National Institute of Diabetes and Digestive and Kidney Diseases and Grant No. CA 40035 from the National Cancer Institute, National Institutes of Health, Bethesda, MD.
Address reprint requests to D.H. Boldt, MD, Medicine/Hematology, University of Texas Health Science Center, 7703 Floyd Curl Dr, San Antonio, TX 78284-7880.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal