Abstract
Porphyrias, a group of inborn errors of heme synthesis, are classified as hepatic or erythropoietic according to clinical data and the main site of expression of the specific enzymatic defect. Hereditary coproporphyria (HC) is an acute hepatic porphyria with autosomal dominant inheritance caused by deficient activity of coproporphyrinogen III oxidase (COX). Typical clinical manifestations of the disease are acute attacks of neurological dysfunction; skin photosensitivity may also be present. We report a variant form of HC characterized by a unifying syndrome in which hematologic disorders predominate: harderoporphyria. Harderoporphyric patients exhibit jaundice, severe chronic hemolytic anemia of early onset associated with hepatosplenomegaly, and skin photosensitivity. Neither abdominal pain nor neuropsychiatric symptoms are observed. COX activity is markedly decreased. In a first harderoporphyric family, with three affected siblings, a homozygous K404E mutation has been previously characterized. In the present study, molecular investigations in a second family with neonatal hemolytic anemia and harderoporphyria revealed two heterozygous point mutations in the COX gene. One allele bore the missense mutation K404E previously described. The second allele bore an A→G transition at the third position of the donor splice site in intron 6. This new COX gene mutation resulted in exon 6 skipping and the absence of functional protein production. In contrast with other COX gene defects that produce the classical hepatic porphyria presentation, our data suggest that the K404E substitution (either in the homozygous or compound heterozygous state associated with a mutation leading to the absence of functional mRNA or protein) is responsible for the specific hematologic clinical manifestations of harderoporphyria.
HUMAN PORPHYRIAS ARE a group of inborn errors of heme biosynthesis that are classified as hepatic or erythropoietic according to clinical data and the main site of expression of the specific enzymatic defect.1 Hereditary coproporphyria (HC) is an autosomal dominant acute hepatic porphyria with incomplete penetrance due to a partial deficiency of coproporphyrinogen III oxidase (COX; EC 1.3.3.3). COX is a mitochondrial enzyme2,3 that catalyzes the sixth step in heme biosynthesis, the decarboxylation of coproporphyrinogen III to protoporphyrinogen IX.4 Typical clinical manifestations of the disease resemble two other forms of inherited acute hepatic porphyria, acute intermittent porphyria (AIP), and variegate porphyria (VP).5 These porphyrias are characterized by acute attacks of neurologic dysfunction with abdominal pain, hypertension, tachycardia, and peripheral neuropathy. Skin photosensitivity may also be present in HC and VP. Excretion of large amounts of coproporphyrin III, mostly in feces and in urine, is observed.1 COX activity is decreased to 50% of normal controls in all tissues from coproporphyric patients as well as from asymptomatic carriers of the gene defect.1 Human cDNA encoding COX has been sequenced,6,7 and the COX gene structure has been determined.8,9 To date, eight different mutations have been characterized, which are distributed all over the COX gene. These mutations, either in the heterozygous (n = 7) or homozygous state (n = 1), are responsible for typical HC.3 9-13
Harderoporphyria is an erythropoietic variant form of HC that is biochemically characterized by marked overproduction in the erythrocytes and increased fecal excretion of the tricarboxylic porphyrin called harderoporphyrin and a markedly decreased lymphocyte COX activity. Harderoporphyria was first diagnosed in three siblings from healthy nonconsanguineous parents mainly on the basis of neonatal hemolytic anemia and skin photosensitivity.14 Molecular studies in the family identified a lysine to glutamic acid susbtitution (K404E) produced by a homozygous A to C transition at position 1210 in exon 6 of the COX gene.11 In the present study, we describe and investigate a new case of harderoporphyria bringing new insights into the clinical and molecular basis of the disease.
MATERIALS AND METHODS
Case report.
The patient was born at term of healthy, nonconsanguineous French parents. Shortly after birth, he developed severe jaundice. Physical findings included hepatosplenomegaly and hypospadias. The total serum bilirubin level was 243 μmol/L. The hemoglobin level was 11.9 g/dL. The nucleated cell count was 160 × 109/L, of which 85% were erythroblasts. After four exchange transfusions, performed between the 10 and 91 hours of life, partial regression of hepatosplenomegaly and resolution of icterus were observed. At 3 months of age, the child was investigated. A blood smear showed 14% erythroblasts and basophilic stippling. Hemoglobin electrophoresis, erythrocyte enzyme activities, globin chain synthesis, and immulogic investigations were normal. Bone marrow aspirate was normal. Hepatic biopsy showed significant iron storage in hepatocytes without any other abnormality. At 2 and 7 years of age, the patient was again examined. Erythrocyte thermal sensitivity was normal and osmotic fragility increased. Spectrin examination findings were normal. Another bone marrow examination showed hyperplastic marrow with 50% erythroblasts without dyserythropoiesis. Perls' Prussian blue stain showed 46% sideroblasts without ring sideroblasts. Ultrastructural bone marrow morphology was normal. The hematologic data are summarized in Table 1. Growth and development remained normal despite persistent hemolytic anemia and mild splenomegaly. At 8 years of age, skin fragility, thickening, and erosion of the back of both hands appeared intermittently without evidence of precipitating factors.
Hematologic Data From the Proband and His Family
| . | Age . | Hb (g/dL) . | Erythro blastosis (%) . | MCV (fL) . | Reticulocyte Count (%) . | Basophilic Stippling . | Bone Marrow Examination . |
|---|---|---|---|---|---|---|---|
| Patient | Birth | 11.2-150 | 85 | ||||
| 3 mo | 6-151 | 14 | 64 | 84,000 | + | ||
| 7% | |||||||
| 2 yr | 9 | 50 | 500,000 | Hyperplasia | |||
| 15% | Sideroblasts-152: 46% | ||||||
| 6 yr | 9.2 | 59 | 285,000 | Hyperplasia | |||
| 8% | Sideroblasts-152: 30% | ||||||
| 19 yr | 11 | 60 | 169,000 | ||||
| 5% | |||||||
| Father | 50 yr | 14.2 | 85 | 25,000 | |||
| <0.6% | |||||||
| Mother | 49 yr | 13.9 | 80 | 37,000 | |||
| <0.6% | |||||||
| Sister | 22 yr | 13.8 | 78 | 32,000 | |||
| <0.6% |
| . | Age . | Hb (g/dL) . | Erythro blastosis (%) . | MCV (fL) . | Reticulocyte Count (%) . | Basophilic Stippling . | Bone Marrow Examination . |
|---|---|---|---|---|---|---|---|
| Patient | Birth | 11.2-150 | 85 | ||||
| 3 mo | 6-151 | 14 | 64 | 84,000 | + | ||
| 7% | |||||||
| 2 yr | 9 | 50 | 500,000 | Hyperplasia | |||
| 15% | Sideroblasts-152: 46% | ||||||
| 6 yr | 9.2 | 59 | 285,000 | Hyperplasia | |||
| 8% | Sideroblasts-152: 30% | ||||||
| 19 yr | 11 | 60 | 169,000 | ||||
| 5% | |||||||
| Father | 50 yr | 14.2 | 85 | 25,000 | |||
| <0.6% | |||||||
| Mother | 49 yr | 13.9 | 80 | 37,000 | |||
| <0.6% | |||||||
| Sister | 22 yr | 13.8 | 78 | 32,000 | |||
| <0.6% |
Abbreviations: Hb, hemoglobin; MCV, mean corpuscular volume.
Exchange transfusions were performed.
A transfusion was performed.
No ring sideroblasts.
Hepatic porphyria was suspected only when the patient was 18 years old because of skin lesions associated with chronic hemolytic anemia. The cutaneous lesions, characterized by the formation of vesicles and bullae up to 2 cm in diameter, which crusted over and took several weeks to heal, were localized on light-exposed areas of the backs of the hands and on the arms and face. The patient had increased skin fragility, but no hypertrichosis, alopecia, or porphyrin-rich gall stones were found.
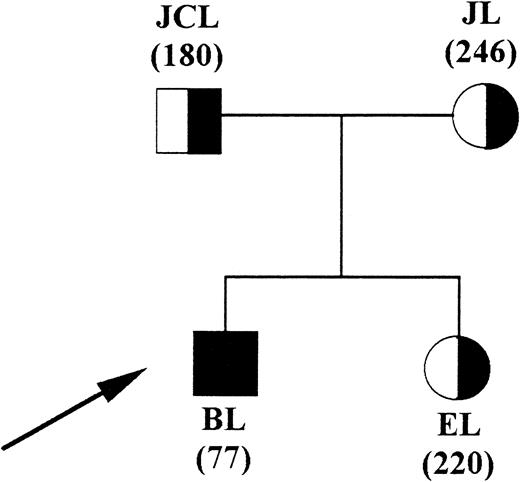
The patient is the second of two siblings (Fig 1). In both parents and his sister, hematologic data were normal (Table 1). The proband and his relatives never exhibited abdominal and/or neurologic symptoms typical of acute hepatic porphyrias.
Family pedigree (solid symbols, patient). In parenthesis is the lymphocyte COX activity expressed as picomoles of protoporphyrin per hour per milligram of protein at 37°C (normal control value, 350 ± 80; mean ± 2 SD).
Family pedigree (solid symbols, patient). In parenthesis is the lymphocyte COX activity expressed as picomoles of protoporphyrin per hour per milligram of protein at 37°C (normal control value, 350 ± 80; mean ± 2 SD).
Porphyrin synthesis investigations (Table 2).
Erythrocyte, urinary, and fecal porphyrins were determined using standard methods.15,16 Lymphocyte COX activity was measured as described.17
Porphyrin Concentration in Urine, Feces, and Erythrocytes of Patient and His Parents
| Subject . | Age (yr) . | Erythrocytes . | Urine . | Feces . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Proto (nmol/L) . | ALA (μmol/L) . | PBG (μmol/L) . | URO (nmol/L) . | Copro (nmol/L) . | Total Porphyrins (nmol)* . | Copro (%) . | Hardero (%) . | Proto (%) . | ||
| Father | 50 | 425 | 27 | 8 | 39 | 395 | 202 | 17 | ND | 83 |
| Mother | 49 | 402 | 38 | 8 | 35 | 247 | 104 | 22 | ND | 71 |
| Patient | 19 | 3,340 | 40 | 9 | 120 | 1,820 | 1,159 | 6.2 | 90 | 3 |
| Normal controls | <1,900 | <38 | <5 | <50 | <200 | <200 | <22 | ND | <75 | |
| Subject . | Age (yr) . | Erythrocytes . | Urine . | Feces . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Proto (nmol/L) . | ALA (μmol/L) . | PBG (μmol/L) . | URO (nmol/L) . | Copro (nmol/L) . | Total Porphyrins (nmol)* . | Copro (%) . | Hardero (%) . | Proto (%) . | ||
| Father | 50 | 425 | 27 | 8 | 39 | 395 | 202 | 17 | ND | 83 |
| Mother | 49 | 402 | 38 | 8 | 35 | 247 | 104 | 22 | ND | 71 |
| Patient | 19 | 3,340 | 40 | 9 | 120 | 1,820 | 1,159 | 6.2 | 90 | 3 |
| Normal controls | <1,900 | <38 | <5 | <50 | <200 | <200 | <22 | ND | <75 | |
Abbreviations: ALA, aminolevulinic acid; PBG, porphobilinogen; Copro, coproporphyrin; Hardero, harderoporphryin; URO, uroporphyrin; Proto, protoporphyrin; ND, not detected.
Results are expressed per gram dry weight.
DNA preparation and amplification by polymerase chain reaction (PCR).
Genomic DNA from the proband, his parents, and his sister was extracted from peripheral blood according to a standard protocol.18Genomic DNA fragments of interest were amplified by PCR using primers selected from the published COX sequence.9 12 Twenty picomoles of each set of primers was mixed in 50 μL of PCR solution containing 1 U of Taq polymerase (Beckmann Inc, Fullerton, CA), 50 mmol/L KCl, 10 mmol/L Tris-HCl, pH 8.5, 1.5 mmol/L MgCl2, and 200 mmol/L of each dNTP. Reactions were performed in a DNA thermocycler (Hybaid, Teddington, UK) as follows: 35 cycles of denaturation at 94°C for 30 seconds, annealing at specific temperature for 30 seconds, and elongation at 72°C for 1 minute.
Reverse transcription PCR (RT-PCR).
DNA sequencing.
All exons and exon/intron boundaries of the COX gene were amplified using previously selected primers.9 12 PCR products were purified with the Wizard PCR preps DNA purification system (Promega-Biotech, Madison, WI). Genomic DNA and cDNA fragments were directly sequenced using 35S-dATP and the fMol DNA sequencing kit (Promega-Biotech).
Construction and prokaryotic expression of normal and mutated human COX cDNA.
Normal human cDNA was expressed using the pGEX-2T expression vector (Pharmacia LKB Biotechnology Inc, Uppsala, Sweden) as already described.11 To study mutated cDNA with the exon 6 deletion, site-directed mutagenesis was performed using normal cloned COX cDNA (pGEX-2T:COX) as template. We used the Transformer site-directed mutagenesis kit (Clontech Laboratories, Palo Alto, CA), which is based on the long primer-unique site elimination mutagenesis method described by Deng and Nickoloff.20 Briefly, long primers were generated by PCR. 5′-Phosphorylated sense oligonucleotide (mutagenic primer), which bypasses exon 6 (105 bp), has 22-bp and 20-bp matching sequences, respectively, flanking the 5′ and 3′ sides of the deleted exon. An antisense oligonucleotide which mutates a single BsaAI restriction site in the pGEX-2T plasmid (selection primer) was used. The sequences of these primers are as follows: mutagenic primer (Del.exon6), 5′GGCAGCAGCT CAGAAGAGGACG↓ ATGGGAGTACATGCATTCAC (the arrow indicates the exon 6 bypass); and selection primer, 5′ACACTCCGCTATCGCTCCGCGACTGGGTCATGGCT (mutated bases abolishing the single BsaAI restriction site are in bold and underlined).
Standard DNA elongation, ligation, and two-step digestion/transformation of mutated plasmids in mutS Escherichia coli and E coli DH5α strains were performed according to the manufacturer's recommendations. The entire sequence of the mutated plasmid was verified by sequencing. The recombinant bacteria (E coli DH5α) were grown and COX activities in bacteria lysates were determined as previously described.21
RESULTS
The patient displayed symptoms and signs of severe hemolytic anemia with splenomegaly and compensatory hyperactive bone marrow features. In this proband, an atypical profile of porphyrin excretion was found in feces with massive accumulation of harderoporphyrin (Table 2). COX activity compared with control values was decreased by 78% in the patient's lymphocytes and by 50% and 30% in those of the father and mother, respectively (Fig 1).
Sequencing of the seven exons and intron-exon boundaries of the COX gene from the patient showed two mutations in the heterozygous state, each on a different allele. The numbering of the mutations is based on the first base of the initiation codon described by Delfau-Larue et al.9
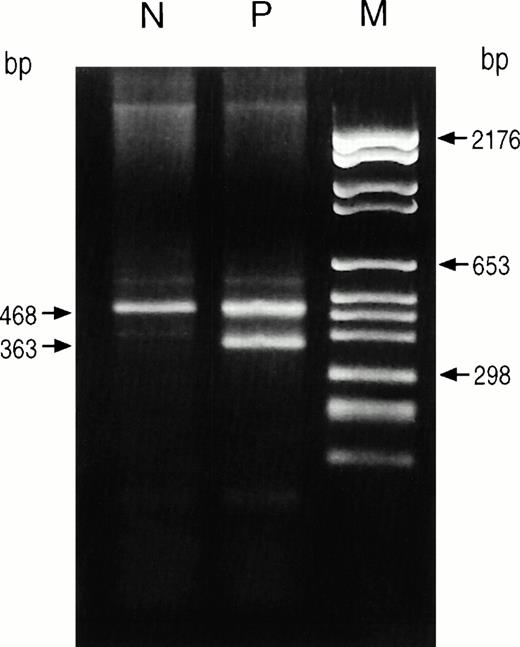
The first mutation, an A to G transition at nucleotide 1210, had been identified in the first reported harderoporphyric patients.11 This mutation resulted in a lysine to glutamate substitution at position 404 in the abnormal protein (K404E). The second mutation was found to be an A to G transition at the third position of the donor splice site in intron 6 (1277+3A→G). This mutation is responsible for exon 6 skipping. After amplification of cDNA from the proband, PCR products showed two bands, one of the expected size and the other corresponding to a 105-bp deletion in accordance with exon 6 skipping (Fig 2). Sequencing of the cDNA confirmed the exon 6 deletion that corresponds to an in-frame deletion of 35 amino acids in the abnormal protein. Procaryotic expression studies of the exon-6–deleted cDNA are summarized in Table 3. The enzymatic activity of the K404E mutated COX protein had already been investigated.11 In the proband, no other abnormality was found in the coding sequence.
Analysis of RT-PCR products from lymphocyte mRNAs. Amplified fragments encompassing exon 5, exon 6, and the coding part of exon 7 were obtained from lymphocyte cDNAs by RT-PCR using primers HUCO-2-Bio-A and HUCO-10S (9) and analyzed on 2% agarose gel. Two amplified products were obtained from the heterozygous harderoporphyric patient (P), the 468-bp fragment containing the K404E missense mutation and the 363-bp fragment resulting from the mRNA with complete deletion of exon 6. Amplification of control mRNA (N) showed only the normal 468-bp fragment. M, molecular size markers.
Analysis of RT-PCR products from lymphocyte mRNAs. Amplified fragments encompassing exon 5, exon 6, and the coding part of exon 7 were obtained from lymphocyte cDNAs by RT-PCR using primers HUCO-2-Bio-A and HUCO-10S (9) and analyzed on 2% agarose gel. Two amplified products were obtained from the heterozygous harderoporphyric patient (P), the 468-bp fragment containing the K404E missense mutation and the 363-bp fragment resulting from the mRNA with complete deletion of exon 6. Amplification of control mRNA (N) showed only the normal 468-bp fragment. M, molecular size markers.
Expression of the Exon-6–Deleted COX cDNA in E coli
| . | Construct . | COX Activity* . |
|---|---|---|
| Normal control | pGEX-2T:COX/sense | 15,870 |
| Negative control | pGEX-2T:COX/antisense | 1,317 |
| Exon-6–deleted cDNA | pGEX-2T:COX/del. exon 6 | 629 |
| . | Construct . | COX Activity* . |
|---|---|---|
| Normal control | pGEX-2T:COX/sense | 15,870 |
| Negative control | pGEX-2T:COX/antisense | 1,317 |
| Exon-6–deleted cDNA | pGEX-2T:COX/del. exon 6 | 629 |
Expression vector: pGEX-2T. Values are expressed as the mean of two duplicate experiments.
Activity given as picomoles of protoporphyrin per hour per milligram at 37°C.
Direct sequencing of exon 6 and its intron junctions from the proband's relatives' genomic DNAs showed that the father was heterozygous for the splice site mutation (1277+3A→G), whereas the mother and the sister were heterozygous for the K404E missense mutation.
DISCUSSION
In this study, we report clinical and molecular investigations in a second family with harderoporphyria. The proband had an early onset porphyria with severe neonatal hemolytic anemia. The pattern of porphyrin excretion showed that the major part of fecal porphyrin was harderoporphyrin, while a large amount of coproporphyrin was found in urine; in addition, protoporphyrin was increased in erythrocytes. Enzymatic studies of COX activity in lymphocytes showed a markedly decreased activity compatible with a homozygous deficient COX gene. Harderoporphyric patients reported to date (this case and Nordmann et al14) exhibited strictly identical clinical symptoms characterized by early onset of hemolytic anemia associated with chronic cutaneous manifestations. It must be emphasized that abdominal pain and neurologic symptoms, suggestive of acute hepatic porphyrias, have not been seen in harderoporphyric patients. In both harderoporphyric families, the parents were clinically asymptomatic but exhibited slightly abnormal fecal porphyrin excretion and an approximately 50% reduction in COX lymphocyte activity.
In the first harderoporphryic cases, molecular studies showed a homozygous point mutation (A to G transition at nucleotide 1210 in exon 6 of the COX gene) resulting in a lysine to glutamic acid substitution (K404E).11 The mutated K404E protein expressed in a procaryotic system showed abnormal kinetics with reduced affinity, less stability, and a decreased residual activity (25% of control). It has been suggested that the two decarboxylation steps catalyzed by COX take place at the same catalytic site and that this mutation was localized at this active site of the enzyme.11 Consistent with this hypothesis, COX from harderoporphyric patients has been found to have a similarly increased Km for both coproporphyrinogen and harderoporphyrinogen substrates.14
In this study, we show that harderoporphyria, occurring in a second family, resulted from compound heterozygous mutations affecting the COX gene. The previously reported K404E mutation was found in the heterozygous state in the proband, his mother, and his sister. The second mutation, an exon 6 skipping mutation caused by an A to G transition at position 1277+3, is a new mutation that resulted in an in-frame deletion of 35 aminoacids in the mutated protein. This mutation was found in the heterozygous state in the proband and his father. The father did not exhibit acute porphyria syndrome typical of HC, probably because of the low penetrance of the disease, especially in males. Expression studies showed that the truncated protein encoded by the exon-6–deleted COX mRNA had virtually no residual enzymatic activity (Table 3). Interestingly, a previous exon 6 skipping mutation (not assessed with expression studies) has been reported in a patient of Czech origin with a heterozygous G to A transition at position 1277, the last position of the splice donor site of exon 6.9 This patient had a typical clinical form of HC. He repeatedly exhibited neurologic symptoms with paresis, and the diagnosis was made during hospitalization after treatment with barbiturates.9 All these data indicate that the residual enzyme activity found in compound heterozygous harderoporphyric patients results exclusively from the K404E mutated COX protein. Therefore, the specific harderoporphyric symptoms appear directly related to this point mutation in the COX gene.
To date, eight different mutations in the COX gene have been characterized. They were responsible for typical HC.3 Only one homozygous form of HC has been reported,10 but its clinical and biologic presentation was completely different from harderoporphyria. The patient had a clinical history of severe acute attacks of hepatic porphyria, without chronic hemolytic anemia, a large accumulation in feces of coproporphyrin with harderoporphyrin being absent, and a profound defect of COX activity in lymphocytes.22 Molecular investigations showed an arginine to tryptophan substitution (R231W) in exon 5 of the COX gene.10
It has been hypothesized that the active COX protein acts as a homodimer of approximatively 70 to 74 kD.23,24 Because of the lack of crystallographic data, little structural information about the human COX enzyme is available. Recently, a histidine residue at position 258 has been shown to be a highly conserved region of aerobic COX; it could be involved in COX catalytic activity through a hypothetic and controversial interaction with Cu2+.25,26 Our studies on harderoporphyria show that the lysine residue at position 404 is also important for catalytic activity of the enzyme: the K404E mutation is probably responsible for accumulation of harderoporphyrinogen, an intermediate in the oxidative decarboxylation of coproporphyrinogen. It has been suggested that this intermediate would leave the abnormal enzyme more easily and, after spontaneous oxidation to harderoporphyrin, would accumulate in the patient.11 Moreover, comparison of nucleotide deduced amino acid sequences from humans, Saccharomyces cerevisiae,Salmonella typhimurium, E coli,26-29 and mouse30 showed that the K404E mutation occurred in a region highly conserved throughout evolution (Table 4). Our data and the high percentage of conserved aminoacids suggest that exon 6 may play an important role in the catalytic activity and/or maintenance of the active conformation of the enzyme.
Comparison of Amino Acid Sequences Deduced From Nucleotides Sequences of the Human (HC), From Codon 387 to 448, Mouse (MC), Saccharomyces cerevisiae (SC), E coli (EC), Salmonella typhimurium (ST), and Soybean (GM)
| K404E | |||
| ↓ | |||
| HC | LRRG | RYVEFNLLYDRGTKFGLFTPGSRIESILMSLPLT | ARWEYMHSPSENSKEAEILEVLRHPRDWV |
| MC | LRRG | RYVEFNLLYDRGTKFGLFTPGSRIESILMSLPLT | ARWEYMHSPSENSKEAEILEVLRHPRDWV |
| SC | IRRG | RYVEFNLIYDRGTQFGLRTPGSRVESILMSLPEH | ASWLYNHHPAPGSREAKLLEVTTKPREWV |
| EC | YRRG | RYVEFNLVWDRGTLFGLQT-GGRTESILMSMPPL | VRWEYDYQPKDGSPEAALSE-FIKVRDWV |
| ST | YRRG | RYVEFNLVWDRGTLFGLQT-GGRTESILMSMPPL | VRWEYDWQPEAGSPEAALSE-FIQVRDWI |
| GM | LRRG | RYVEFNLVYDRGTTFGLKT-GGRIESILVSLPLT | ARWEYDHKPEEGSEEWKLLDACINPKEWI |
| *** | ******* **** *** * * * **** * * | *** * * * * |
| K404E | |||
| ↓ | |||
| HC | LRRG | RYVEFNLLYDRGTKFGLFTPGSRIESILMSLPLT | ARWEYMHSPSENSKEAEILEVLRHPRDWV |
| MC | LRRG | RYVEFNLLYDRGTKFGLFTPGSRIESILMSLPLT | ARWEYMHSPSENSKEAEILEVLRHPRDWV |
| SC | IRRG | RYVEFNLIYDRGTQFGLRTPGSRVESILMSLPEH | ASWLYNHHPAPGSREAKLLEVTTKPREWV |
| EC | YRRG | RYVEFNLVWDRGTLFGLQT-GGRTESILMSMPPL | VRWEYDYQPKDGSPEAALSE-FIKVRDWV |
| ST | YRRG | RYVEFNLVWDRGTLFGLQT-GGRTESILMSMPPL | VRWEYDWQPEAGSPEAALSE-FIQVRDWI |
| GM | LRRG | RYVEFNLVYDRGTTFGLKT-GGRIESILVSLPLT | ARWEYDHKPEEGSEEWKLLDACINPKEWI |
| *** | ******* **** *** * * * **** * * | *** * * * * |
Deleted aminoacid sequence deduced from exon 6 deletion nucleotides sequence is boxed. Mutated amino acid (K404) encoded by A1210G allelic mutation is represented above the human sequence. Asterisks indicate identical amino acids.
The pathogenesis of the hematologic symptoms in harderoporphyria is not yet fully understood. However, as observed in erythropoietic porphyrias (Günther's disease, erythro-hepatic porphyria), harderoporphyric patients exhibit splenomegaly. The spleen is the major site for removal of damaged or hemolyzed erythrocytes31; hence, the splenomegaly observed in harderoporphyria could be presumed to be secondary to this process. Extrinsic abnormalities of erythrocytes seem unlikely, because the direct Coomb's test was negative and the survival time of normal erythrocytes transfused into harderoporphyric patients was normal. The overproduction of porphyrins in harderoporphyria may account for the hemolytic symptoms. The elevated level of erythrocyte protoporphyrin found in all the harderoporphyric patients, in contrast with classical HC, provides evidence favoring the bone marrow as a source of elevated porphyrins in this disease and could be involved, at least in part, in the hemolytic process. Hemolysis of erythrocytes may also result from photolysis as porphyrin-laden cells are exposed to light in the dermal capillaries. Light wavenlengths suitable for porphyrin photoactivation are known to penetrate the skin to a depth sufficient to produce this phenomenon and photohemolysis has also been demonstrated in vitro.32 33
In conclusion, this study suggests for the first time the existence of a phenotype/genotype relationship in the human COX gene. In contrast with other COX gene defects responsible for HC, the K404E mutation in the homozygous state or associated with a deleterious allele (exon 6 skipping in this case) induces harderoporphyria. The abnormal kinetic pattern with reduced affinity, less stability, and the decreased (25%) residual activity of the mutated K404E is responsible for the unusual accumulation of harderoporphyrin and the specific hematologic and clinical symptoms of harderoporphyria. Harderoporphyria is a unifying syndrome of childhood onset with clinical features quite different from those observed in other hepatic porphyrias. It is characterized by jaundice, hemolytic anemia, hepatosplenomegaly, skin photosensitivity, and a marked increase in harderoporphyrin in urine and feces.
Supported by grants from INSERM (U409), University Paris VII in collaboration with Charles University of Praha (Czech Republic), and Association Française contre les Myopathies.
Address reprint requests to Y. Nordmann, MD, Laboratoire de Biochimie, Hôpital Louis Mourier, 92701 Colombes Cedex, France.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal