Abstract
Rare individuals who lack all of the Rh blood group antigens are called Rhnull and may be classified as “regulator” or “amorph” types. The suppression of Rh antigen expression for regulator types may be attributed to mutations of the RH50gene, which is independent of the RH locus. The RH50 gene encodes a glycoprotein that interacts with the Rh proteins to form a functional complex within the red blood cell membrane. This report describes an RH50 gene mutation for a previously unclassified Rhnull donor. Sequencing cDNA clones from Rh50 mRNA revealed a single base change (G836A) yielding a missense and nonconservative mutation (Gly279Glu) within a predicted hydrophobic domain for this membrane protein. Genomic DNA studies using polymerase chain reaction (PCR) restriction analysis and sequencing showed that the Rhnull propositus was a composite heterozygote for this mutation, carrying two alleles with the A and G at nucleotide 836, respectively. In contrast, cDNA studies showed that only the A836 sequence was present, suggesting that the second allele with G836 was apparently silent (no transcript detected). Family studies showed that the mutant RH50 allele (836A) was inherited maternally, whereas the silent RH50 allele (836G) was from paternal transmission. These findings provide further evidence that rare but diverse genetic alterations may occur along the RH50 gene where the Rhnull syndrome of the regulator type occurs. The single amino acid change (Gly to Glu) provides insight into the critical value of these residues for assembly of the Rh antigen complex within the membrane.
RARE INDIVIDUALS WHOSE red blood cells lack all of the known Rh antigens found within the Rh blood group system are defined as Rhnull.1 These donors exhibit a mild clinical syndrome, called Rh-deficiency syndrome, characterized by a chronic hemolytic anemia in which the red blood cells have a stomatocytosis morphology, an increased osmotic fragility, an altered ion transport system, and abnormal membrane phospholipid organization.2 3 Family studies show that two classes of Rhnull types exist and arise from independent genetic events. The “amorph” type is caused by homozygosity for a silent allele at the RH locus, whereas the more common “regulator” type is apparently caused by homozygosity for an autosomal suppressor gene (Xor) that is genetically independent of the RH locus.
The RH locus is located on chromosome 1p34.3-1p36.1 and contains two highly homologous genes, D and CE, which have been cloned and sequenced.4,5 The RhD antigen polymorphism generally arises from complete absence of the D gene, and individuals therefore may carry zero, one, or two copies of theD gene, although the CE gene is almost invariably present in two copies.6 Sequencing studies have shown that the coding regions for RH genes are completely normal not only for Rhnull individuals of the regulator type, as expected, but also for one sample of the amorph type, suggesting that the abnormalities in the latter arise from a transcriptional defect.7
The D and CE genes encode two major Rh proteins, designated Rh30, that carry D and C/c, E/e specificities,8although minor species arising by alternative splicing may also occur.9 Within the red blood cell membrane, Rh30 proteins interact with a number of other proteins that include a glycoprotein of 50 to 100 kD (designated Rh50), CD47, glycophorin B, and the glycoproteins carrying the LW and Fy antigens.4,5 Protein defects of the Rhnull cell membranes include the lack of Rh30 and LW polypeptides and the absence or severe reduction of Rh50 and CD47.3 In addition, a reduced expression of the S/s antigens on glycophorin B and of other blood group antigens (Fy5, U, and Duclos) has been documented.10 The gene encoding the Rh50 protein has been cloned and sequenced and maps to chromosome 6p11-p21.1.11,12 Both the RH50 and CD47 genes are candidates for the postulated gene suppressing Rh antigen expression in Rhnull of the regulator type. In support of this, three distinct abnormalities have now been described along the RH50 gene for four regulator Rhnull.12
This report describes a novel single point mutation along theRH50 gene for an Rhnull individual, Y.T., for which the regulator or amorph type has never been formally documented, although the donor's cells were used in several biochemical studies.7 13-16 Preliminary family studies showed that functional D and C antigens were transmitted from Y.T. to three children, suggesting that Y.T. belonged to the regulator type. The following molecular studies show the pattern of inheritance of the novel mutation and also show that Y.T. was a composite heterozygous donor, carrying one mutant RH50 allele and one transcriptionally silent RH50 allele.
MATERIALS AND METHODS
Blood samples and Rh typing.
Blood samples were collected from the Rhnull propositus Y.T., her husband, their three children, and Y.T.'s parents. The parents are not related, and this is supported by the family history. Y.T.'s paternal grandfather was English and the paternal grandmother was French. They met in France during World War I and migrated to Australia. Y.T.'s maternal great-grandparents both migrated from Ireland and Scotland to a separate region of Australia. Y.T.'s parents therefore grew up in these separate regions. The propositus Y.T., aged 49, is in good health. She was diagnosed with non–insulin-dependent diabetes in 1995. Her hemoglobin level (114 to 134 g/L between 1984 and 1991 and 105 to 116 g/L between 1995 and 1997) is at or below the lower range of normal (normal range for females, 115 to 165 g/L). For Rh phenotyping, standard serology techniques were used. D gene dosage was performed by D genotyping using DNA amplification of exon 7 of the D gene as previously described.17
Flow cytometric analysis of Rh D antigen site density.
A suspension of 5% red blood cells in phosphate-buffered saline (PBS) was prepared from frozen cells. Monoclonal anti-D IgG (Diagast, Lille, France) was diluted 1:7.5 in PBS. The 5% (vol/vol) red blood cell suspensions (50 μL) were mixed with 100 μL anti-D or with 5% bovine serum albumin (BSA). The latter served as control. These mixtures were incubated for 1 hour at 37°C and then washed three times in saline. The anti-human Ig F(ab)2 fraction conjugated to fluorescein (Silenus, Hawthorn, Victoria, Australia) was diluted 1:20 in saline; 100 μL of this mixture was mixed with the washed and sensitized cells and with the respective negative control for the cell blocked with BSA. Reaction mixtures were incubated at room temperature for 1 half-hour, washed three times, and resuspended in 2 mL PBS before determining the fluorescence. A relative measure of D antigen expression was established by determining the mean channel fluorescence signals on a Coulter Epics XL-MCL Flow Cytometer (Coulter Electronics, Sydney, Australia). The controls were samples for which the absolute D gene dosage measurement was described previously.18
Reverse transcription coupled with PCR amplification.
Reticulocyte RNAs from 30 mL peripheral blood from the propositus Y.T. were extracted by selectively lysing red blood cells by the Orskov reaction.19 Total RNAs from different members of Y.T.'s family were extracted from 10 mL fresh EDTA whole blood using the RNA Isolation Kit (Stratagene, La Jolla, CA). One microgram of RNAs was reverse-transcribed in a total volume of 33 μL using the First Strand cDNA synthesis kit (Pharmacia, Uppsala, Sweden) following the manufacturer's instructions. Five microliters of cDNA products were then subjected to PCR in a 50-μL reaction mixture containing 1× Taq buffer (50 mmol/L KCl, 20 mmol/L Tris (pH 8.3), 1.5 mmol/L MgCl2, and 0.01% gelatin), 0.2 mmol/L of the four dNTPs, 50 pmol of each primer, and 2.5 U Taq polymerase (Perkin-Elmer-Cetus, Norwalk, CT). RH50 cDNA from the propositus Y.T. was amplified using the primer pair X and Y (Table1) spanning the full coding sequence. PCR conditions were as follows: denaturation for 1 minute at 94°C, annealing for 1 minute at 55°C, and extension for 90 seconds at 72°C for 30 cycles. Relevant PCR fragments were purified on a 1% low-melting agarose gel followed by a Wizard PCR preps minicolumn (Promega, Madison, WI), and subcloned into a PCR II vector using the TA cloning kit (Invitrogen, San Diego, CA). cDNA products prepared from the family members of Y.T. were further amplified between D and C primers (Table 1). PCR conditions were as follows: denaturation for 5 minutes at 94°C, and 30 cycles of denaturation for 30 seconds at 94°C, annealing for 45 seconds at 54°C, and extension for 30 seconds at 72°C. Primers D and C amplify sequences within an exon; therefore, with this primer combination, the mRNA extract was treated with DNAse before cDNA preparation to avoid genomic contamination.
Sequence of Primers Used in the PCRs
| Primer . | Sequence (5′ → 3′)* . | Position (nt)-151 . |
|---|---|---|
| A | AATGCCACCCTTGCTGG (sense) | 820-836 |
| B | AATGCCACCCTTGCTGA (sense) | 820-836 |
| C | GCAATGCTCCCAATAATCA (antisense) | 908-890 |
| D | gattacgaattcAATGCCACCCTTG (sense) | 820-832 |
| X | GCCTCTGTCCTTTGCCAC (sense) | −22 to −5 |
| Y | TGGTCACCATGTCCATGGAA (antisense) | 1,262-1,243 |
| Primer . | Sequence (5′ → 3′)* . | Position (nt)-151 . |
|---|---|---|
| A | AATGCCACCCTTGCTGG (sense) | 820-836 |
| B | AATGCCACCCTTGCTGA (sense) | 820-836 |
| C | GCAATGCTCCCAATAATCA (antisense) | 908-890 |
| D | gattacgaattcAATGCCACCCTTG (sense) | 820-832 |
| X | GCCTCTGTCCTTTGCCAC (sense) | −22 to −5 |
| Y | TGGTCACCATGTCCATGGAA (antisense) | 1,262-1,243 |
*Coding sequence is in capitals; lowercase letters indicate a 12-base extension tail.
nt + 1 is taken as the first nt of the translation initiation codon.
Genomic DNA analysis of the RH50 locus.
Genomic DNA was extracted from peripheral whole blood collected onto EDTA.20 Allele-specific PCRs with wild-type and mutant specific primers were performed using primer combinations A and C or B and C (Table 1) to amplify the 836G wild-type allele or the 836A mutant allele, respectively (expected size, 89 bp). PCR mixtures were as already described using 200 ng genomic DNA. PCR cycles were as follows: denaturation for 5 minutes at 94°C, and 30 cycles of denaturation for 30 seconds at 94°C, annealing for 45 seconds at 63°C for primers A and C or 57°C for primers B and C, and extension for 30 seconds at 72°C. PCR products were subjected to electrophoresis on 4% Nusieve (FMC Bioproducts, Rockland, ME) and visualized by ethidium bromide staining.
PCR-RFLP assay.
PCR products amplified from genomic DNA or cDNA with D and C primers (101 bp) were digested for 1 hour at 37°C with 10 U MnlI in the buffer provided (New England Biolabs, Beverly, MA) made up to 20 μL and including 100 mg/mL BSA. After digestion, the enzyme was inactivated for 10 minutes at 65°C and the products were visualized on 4% Nusieve gel electrophoresis.
Sequencing of PCR products from genomic DNA and from cDNA.
PCR products were separated by electrophoresis on 2% Nusieve and excised and purified using the QIAEX II Agarose Gel Extraction Kit (Qiagen, Chatsworth, CA). Sequencing of PCR products was performed as suggested by the manufacturer. PCR extension products were purified by ethanol precipitation and sequenced on the Applied Biosystems (Foster City, CA) 373A DNA sequencer.
RESULTS
Rh antigen expression among family members of the Rhnullpropositus Y.T.
The Rh phenotype and genotype for the family members, deduced from serology and D gene dosage measurements,17,18 are shown in Table 2. The parents of the Rhnull propositus Y.T. were both DCe/DCe, and Y.T. therefore could only inherit DCe haplotypes. All three of Y.T.'s children were DCe/ce genotypes. The cehaplotype must have been transmitted from the father, E.T., as Y.T. could only transmit the DCe haplotype. The D andC antigens were expressed on red blood cells from all three children, indicating that the D and Ce genes carried by Y.T. were structurally normal. Interestingly, the flow cytometric studies on two children, M.P. and P.T., indicate that the level of expression of D antigen was below the normal range established from 39 donors with DCe/ce types (Table 2). Values for M.P. and P.T. were 71 and 91, compared with 174 ± 39 for the 39 controls, indicating a reduction in Rh antigen expression for the obligate heterozygote carrier of the regulator null trait. Additional typing analysis with specific monoclonal antibodies indicated that red blood cells from Y.T. lacked Rh50 glycoprotein and expressed only a low level of CD47 (3,000 v 30,000 to 50,000 mol/cell), as expected for Rhnull erythrocytes (data not shown). That the red blood cells from Y.T. lack the Rh50 protein was reported previously,16 and is confirmed.
Rh Phenotypes and D Genotypes for Family Members of the Rhnull Propositus
| Sample . | Rh Typing . | D Expression (mean ± SD)* . | |
|---|---|---|---|
| Serology . | D Genotype . | ||
| Family Y.T. | |||
| J.H. father | DCCee | DCe/DCe | Not tested |
| A.H. mother | DCCee | DCe/DCe | Not tested |
| Y.T. propositus | Null | DCe/DCe | 0 |
| E.T. husband | DCcee | DCe/ce | 184.4 |
| M.P. daughter | DCcee | DCe/ce | 70.6 |
| D.T. daughter | DCcee | DCe/ce | Not tested |
| P.T. son | DCcee | DCe/ce | 91.4 |
| Controls (n) | |||
| 10 | ccee | ce/ce | 2 ± 3 |
| 39 | DCcee | DCe/ce | 174 ± 39 |
| 22 | DCCee | DCe/DCe | 244 ± 55 |
| 16 | DCcEe | DCe/DcE | 369 ± 69 |
| Sample . | Rh Typing . | D Expression (mean ± SD)* . | |
|---|---|---|---|
| Serology . | D Genotype . | ||
| Family Y.T. | |||
| J.H. father | DCCee | DCe/DCe | Not tested |
| A.H. mother | DCCee | DCe/DCe | Not tested |
| Y.T. propositus | Null | DCe/DCe | 0 |
| E.T. husband | DCcee | DCe/ce | 184.4 |
| M.P. daughter | DCcee | DCe/ce | 70.6 |
| D.T. daughter | DCcee | DCe/ce | Not tested |
| P.T. son | DCcee | DCe/ce | 91.4 |
| Controls (n) | |||
| 10 | ccee | ce/ce | 2 ± 3 |
| 39 | DCcee | DCe/ce | 174 ± 39 |
| 22 | DCCee | DCe/DCe | 244 ± 55 |
| 16 | DCcEe | DCe/DcE | 369 ± 69 |
*Relative levels of D antigen expression are shown as the mean channel fluorescence intensity.
Analysis of the RH50 transcript in donor Y.T.
Reticulocyte RNAs isolated from Y.T. were reverse-transcribed, and theRH50 cDNA was amplified using the X and Y primer pair. Sequence analysis of the cloned PCR product (1.28 kb), corresponding to the complete coding region of the cDNA, revealed a G to A transition at nucleotide 836, creating a glycine to glutamic acid missense mutation at residue 279 of the Rh50 protein.
Genomic detection of the RH50 mutation.
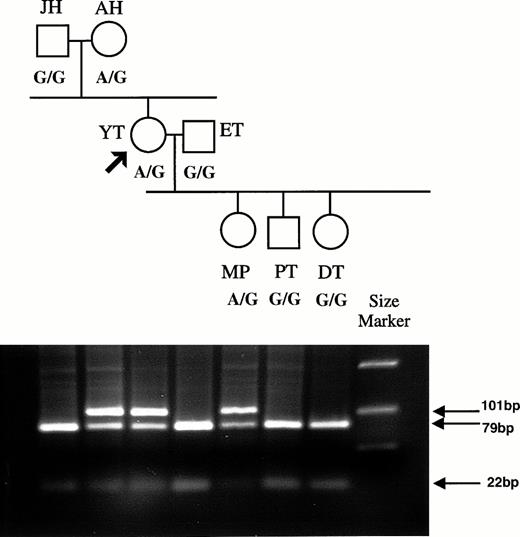
The G836A mutation abolishes an MnlI restriction site, and this polymorphism was used to determine the presence of the mutation in the genomic DNA of Y.T., the zygosity, and the inheritance of the mutation in Y.T.'s family. A 101-bp fragment encompassing nucleotide 836 was amplified from the genomic DNA of all family members using primer pair D and C. MnlI cleavage of the 101-bp resulted in two fragments of 79 and 22 bp. This pattern was observed for Y.T.'s father J.H., husband E.T., and children D.T. and P.T., consistent with their carrying two RH50 alleles with the G836 (Fig1). In contrast, Y.T.'s mother A.H., Y.T. herself, and her daughter M.P. showed the 101-bp fragment present after digestion in addition to the 79- and 22-bp cleaved products, consistent with the presence of two alleles with and without the G836 (Fig 1). This indicated that the A836 mutation was present in the genomic DNA of Y.T. at the heterozygous state. Sequencing the 101-bp products for A.H., Y.T., and M.P. showed mixed sequences at nucleotide 836, with both A and G present (data not shown).
Family tree for the Rhnull donor Y.T., showing inheritance of the RH50 allele containing the G836A mutation. PCR products were prepared from genomic DNA using primers D and C that span nt 820 to 908. Primer D has a 12-base tail added. The 101-bp PCR product obtained from each family member was digested withMnlI, and the fragments were analyzed on agarose gel. Products containing the G836 sequence are cut into 22- and 79-bp fragments, whereas those with A836 are uncut. The polymorphism at nt 836 (A or G) is indicated below each family member.
Family tree for the Rhnull donor Y.T., showing inheritance of the RH50 allele containing the G836A mutation. PCR products were prepared from genomic DNA using primers D and C that span nt 820 to 908. Primer D has a 12-base tail added. The 101-bp PCR product obtained from each family member was digested withMnlI, and the fragments were analyzed on agarose gel. Products containing the G836 sequence are cut into 22- and 79-bp fragments, whereas those with A836 are uncut. The polymorphism at nt 836 (A or G) is indicated below each family member.
Expression of the RH50 gene among Y.T.'s family members.
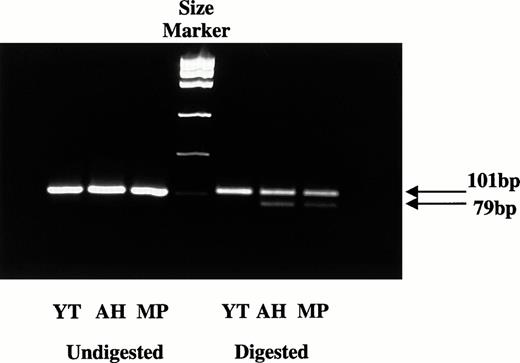
To determine whether the two RH50 alleles carried by Y.T. were both transcribed into mRNA, the 101-bp PCR product was amplified from the cDNA of Y.T. using D and C primers. A.H. and M.P. cDNAs were used as the controls, as both carry the A836 mutant and their secondRH50 allele (G836) must therefore be normal. The PCR products from A.H., M.P., and Y.T. cDNAs were subjected to restriction analysis with MnlI. After digestion, A.H. and M.P. cDNAs yielded two products at 101 and 79 bp, respectively, consistent with both carrying two alleles (G836 and A836) and both alleles being transcribed into mRNA (Fig 2). In contrast, the 101-bp fragment from Y.T. cDNA was not cut with MnlI. All PCR fragments were also directly sequenced to further check whether the mRNA could be detected from both RH50 genes. A.H. and M.P. were again used as controls. A.H. and M.P. showed mixed sequences at nucleotide 836, with both A and G present. In contrast, Y.T. showed only one sequence corresponding to A836, which was consistent again with the second RH50 gene (G836) not being transcribed. This silent allele must have been inherited from the father J.H.
Expression studies of the G836 or A836 alleles. cDNA was prepared from RNA extracts of blood samples and amplified by PCR using primers D and C. The 101-bp PCR products were digested withMnlI, and the fragments were analyzed on agarose gel. Products containing the G836 sequence are cut into 22- and 79-bp fragments, whereas those with A836 are uncut. After digestion, only the 101-bp fragment was detected with Y.T., indicating that only the allele carrying the A836 mutation is transcriptionally expressed. In contrast, both the 101- and 79-bp fragments were present in A.H. and M.P., indicating that both A836 and G836 are expressed.
Expression studies of the G836 or A836 alleles. cDNA was prepared from RNA extracts of blood samples and amplified by PCR using primers D and C. The 101-bp PCR products were digested withMnlI, and the fragments were analyzed on agarose gel. Products containing the G836 sequence are cut into 22- and 79-bp fragments, whereas those with A836 are uncut. After digestion, only the 101-bp fragment was detected with Y.T., indicating that only the allele carrying the A836 mutation is transcriptionally expressed. In contrast, both the 101- and 79-bp fragments were present in A.H. and M.P., indicating that both A836 and G836 are expressed.
Frequency of the G836A transition in the population.
Allele-specific PCR was used to test for the frequency of the G836A transition in blood donors from the Brisbane Center. Using the primer combination pair A and C specific for the wild-type 836G nucleotide, an 89-bp product was amplified for all 44 random individuals tested. In contrast, no product was obtained among these 44 donors using the primer pair B and C specific for the mutant 836A nucleotide (data not shown). E.T. and Y.T. were used as controls in these experiments. E.T. showed a product with the A and C primer pairs only, and as expected, Y.T. showed products with both primer pairs A and C and B and C. This result indicated that the G836A mutation did not correspond to a frequent polymorphism.
DISCUSSION
In this study, molecular alteration of the RH50 gene that encodes a major membrane component of the Rh membrane complex was characterized in a previously undescribed Rhnull family. The affected propositus Y.T. is a composite heterozygote, carrying two distinct RH50 alleles, one with the 836G nucleotide and one with the 836A nucleotide (Fig 3). The abnormality for the RH50 allele carrying the 836G nucleotide most likely resides at either the transcription or the posttranscription level, as no mRNA could be detected in the reticulocytes from Y.T. It follows that the nucleotide sequence for the coding region of this allele could not be deduced from the cDNA. The presence of transcriptionally silent RH50 genes has been reported for two other Rhnull of the regulator type, one homozygous and one heterozygous.12 The molecular basis for the abnormality has not been clarified and may require additional studies that analyze promoter and splice junction sites. The secondRH50 allele of Y.T. carries the A836 nucleotide and is successfully transcribed into mRNA, although no Rh50 protein could be detected at the red blood cell surface. The cDNA sequence of this mutant is identical to the previously published sequence, with the exception of the single nucleotide substitution G836A. Two other examples of structural abnormalities along the RH50 gene have been described previously for Rhnull types.12These involved, first, a two-nucleotide change and two-nucleotide deletion for two Rhnull individuals from South Africa, and second, a single nucleotide deletion observed for one Rhnull individual of Swiss origin. In both cases, the changes produced a frame shift of the coding sequence with a premature stop codon, and the Rh50 protein was not detected on red blood cells.12
RH and RH50 genotypes of family YT. RH50 alleles are as follows: +, wild-type carrying a G at nt 836; s, transcriptionally silent allele carrying a G at nt 836; *, mutant allele carrying an A at nt 836. The RH genotype of the Rhnull propositus Y.T. is shown in brackets since it is not phenotypically expressed.
RH and RH50 genotypes of family YT. RH50 alleles are as follows: +, wild-type carrying a G at nt 836; s, transcriptionally silent allele carrying a G at nt 836; *, mutant allele carrying an A at nt 836. The RH genotype of the Rhnull propositus Y.T. is shown in brackets since it is not phenotypically expressed.
The single nucleotide substitution described in the family of Y.T. results in a single amino acid change from Gly279Glu in the Rh50 protein. This change may be attributed to the Rhnull defect for three reasons. First, this change was not detected among a random selection of the community, indicating that it is not a frequently occurring polymorphism. Second, it is the only alteration along an otherwise normal RH50 gene that is transcribed successfully. Third, hypothetical models to explain the membrane topology of the Rh50 protein indicate that the Rh50 glycoprotein has 12 hydrophobic domains.11,21 The nonconservative change described in this report from a Gly to a Glu occurs within the ninth hydrophobic domain, and by introducing a charged residue, it has the potential to disrupt the conformation of the protein. This conformational change could potentially affect either the transport into the membrane and/or the insertion and interaction with other proteins within the membrane. This would provide an explanation for the previous studies, confirmed here, showing that neither Rh50 nor Rh30 proteins can be detected in the membrane of Y.T.14,15 In addition, this hypothesis is consistent with more recent studies showing that the Rh50 glycoprotein interacts with the Rh30 protein during biosynthesis, and this appears essential for transport of the Rh30 protein across the membrane compartments.15 21
While this report suggests that a single missense mutation in theRH50 gene for Y.T. prevents expression of the Rh50 glycoprotein in the membrane, it does not necessarily conflict with the possibility that a truncated Rh30 protein isoform, resulting from alternative splicing along the RH30 gene or from a posttranscriptional modification, may be inserted into the membrane.9Conceivably, the structural requirement permitting correct membrane insertion may be preserved in such a protein isoform, although it is lost in the Rh50 mutant protein of Y.T.
It is of interest that a mutant membrane protein, aquaporin-2, showed a single nonconservative change along the gene giving rise to a serine to proline transition in a hydrophobic membrane-spanning domain. This mutation was identified in a patient with nephrogenic diabetes insipidus, and the inability to facilitate water transport was attributed to an impaired routing to the plasma membrane.22
It is also of interest that one other example of a single nucleotide change in the RH50 gene has been described for an individual with reduced Rh antigen expression classified as Rhmod.12 The nucleotide change resulted in a serine to asparagine change at position 79 along the second hydrophilic segment of the Rh50 glycoprotein that protrudes within the cytosol of the red blood cell. Low amounts of Rh50 protein were detected in the membrane, consistent with a reduced transport and assembly process. It is likely therefore that a range of substitutions along theRH50 gene may be responsible for the varying degree of modulated Rh antigen activity observed.12 However, the change observed in Y.T. occurred in a critical position preventing any expression. Definitive proof that the missense mutation G836A is sufficient to account for the Rhnull phenotype of Y.T. will require mutagenesis and expression of the RH50 mutant in an expression system in which the RhD expression may also be examined. One such system is now available for D expression in the erythroleukemic K562 cells transduced with retroviral vectors; however, these have an endogenous Rh50 protein.8 There is currently no evidence that both RhD and RH50 (or mutants) may be expressed in a nonerythroid cell line. Studies of such systems are under investigation in Paris.
While the Rh50 glycoprotein appears essential for transport and/or expression of Rh30 polypeptides, the Rh50 glycoprotein does not have such a strict requirement for coexpression with the Rh30 polypeptides. Indeed, a low amount of the Rh50 glycoprotein is detectable with a murine monoclonal antibody on red blood cells from a subgroup of Rhnull individuals classified as U-positive (reacting weakly with anti-U antibodies), but not on those that are U-negative.23 Rhnull U-positive samples are less frequent than Rhnull U-negative samples, but occur for all amorph types and occasionally for regulator types of Rhnull.15,24 Red blood cells from both types exhibit a variable degree of suppression of Ss (and U) antigens, which has been correlated with a 60% to 70% decrease of glycophorin B.25 Although Rh30 proteins are undetectable on these cells, they still carry a small amount of Rh50 glycoprotein. One such example (patient D.A.A., Rhnull amorph type) with an intactRH50 transcript was reported in our previous report12 and showed that minor amounts of Rh50 glycoprotein may still be expressed at the red blood cell surface even in the absence of Rh30 proteins. Why reduced Rh50 expression is restricted to some Rhnull cells only, which apparently have a similar content of glycophorin B,24 is presently not understood. That some Rhnull cells may have a structurally altered glycophorin B is unlikely, but has not been formerly excluded. It is possible but not proven that Rh50 expression in certain Rhnull cells is permitted by some unidentified factor(s). This will result in the Rhnull U-positive phenotype, following protein interaction between glycophorin B and the residual amount of Rh50 glycoprotein, as previously postulated.21Clarification of the chemical nature of the U antigen may help to solve this issue.
The family study for Y.T. showed that the transcriptionally silentRH50 allele was inherited on the paternal side and was transmitted from the Rhnull mother (Y.T.) to one daughter, D.T., and one son, P.T. The second allele, carrying the hitherto unreported mutation G836A, was inherited from the mother (A.H.) and transmitted on to one daughter, M.P. The RH genes for Y.T. were normal, as the D and C antigens expressed by all three children could only have been inherited from the mother. However, the D antigen expression was reduced for both the daughter (M.P.) carrying the A836 gene and the son (P.T.) carrying the silent RH50 gene, as initially reported for obligate Rhnullheterozygotes.26 This again would be consistent with their carrying only a single dose of functional RH50 gene.
These findings definitively classify Y.T. as a regulator Rhnull with a DCe/DCe genotype. Most cases of Rhnull arise by consanguinity and result from the homozygosity for a defective gene.7 However, the donor reported here represents one of the rare cases in which two distinct alleles inherited from nonconsanguineous parents have produced the null trait. The family history supports these findings. The results from these studies provide further evidence that abnormalities along theRH50 gene are responsible for the Rhnull trait. They also provide some insight into the critical nature of the protein structure before successful assembly and expression of the Rh antigen complex can occur.
ACKNOWLEDGMENT
We warmly and gratefully acknowledge the support and interest that Y.T. and her family have continued to show throughout these studies.
Supported by the National Health and Medical Research Council of Australia, The Alexander Steele Young Memorial Lions Foundation, and Brisbane North Regional Health Authority Liver Transplant Unit.
Address reprint requests to C.A. Hyland, PhD, Australian Red Cross Blood Service, Queensland, PO 10325, Brisbane, 4000, Australia.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal