Abstract
The Fanconi anemia (FA) complementation group C (FAC) protein gene encodes a cytoplasmic protein with a predicted Mrof 63,000. The protein's function is unknown, but it has been hypothesized that it either mediates resistance to DNA cross-linking agents or facilitates repair after exposure to such factors. The protein also plays a permissive role in the growth of colony-forming unit–granulocyte/macrophage (CFU-GM), burst-forming unit–erythroid (BFU-E), and CFU-erythroid (CFU-E). Attributing a specific function to this protein requires an understanding of its intracellular location. Recognizing that prior study has established the functional importance of its cytoplasmic location, we tested the hypothesis that FAC protein can also be found in the nucleus. Purified recombinant Escherichia coli–derived FAC antigens were used to create antisera able to specifically identify an Mr = 58,000 protein in lysates from human Epstein-Barr virus (EBV)-transformed cell lines by immunoblot analysis. Subcellular fractionation of the cell lysates followed by immunoblot analysis revealed that the majority of the FAC protein was cytoplasmic, as reported previously; however, approximately 10% of FAC protein was reproducibly detected in nuclear fractions. These results were reproducible by two different fractionation methods, and included markers to control for contamination of nuclear fractions by cytoplasmic proteins. Moreover, confocal image analysis of human 293 cells engineered to express FAC clearly demonstrated that FAC protein is located in both cytoplasmic and nuclear compartments, consistent with data obtained from fractionation of the FA cell lines. Finally, complementation of the FAC defect using retroviral-mediated gene transfer resulted in a substantial increase in nuclear FAC protein. Therefore, while cytoplasmic localization of this protein appears to be functionally important, it may also exert some essential nuclear function.
FANCONI ANEMIA (FA) IS AN autosomal recessive disorder characterized by cell-cycle defects, cellular hypersensitivity to agents that damage DNA, bone marrow failure, diverse congenital anomalies, and a marked increase in the incidence of acute myelogenous leukemia.1,2 Diagnostically, the hallmark of this disorder is hypersensitivity of FA cells to the clastogenic effects of DNA cross-linking agents such as diepoxybutane (DEB) and mitomycin C (MMC).3,4 Based on these features, FA has been proposed to result from a disorder in DNA repair. FA is genetically heterogeneous, with at least five different complementation groups identified by somatic cell hybrid analysis.5,6Although the cDNA for FA complementation group C (FAC) has been cloned by functional complementation, sequenced, and mapped to chromosome 9q22.3,5,7 there is no substantial sequence homology with any known gene family, and the function of the gene product is unknown.7 Recent cloning of the FAA gene has likewise been unable to reveal any clear functional motifs or to shed light on the roles these genes play in the clinical disease.8,9 It is clear that the FAC gene product is necessary for optimal growth and differentiation of hematopoietic progenitor cells,9,10 at least in part by virtue of the capacity of the protein to modulate mitotic inhibitory signals induced in progenitor cells by gamma interferon.10 11
FAC cDNA contains an open reading frame of 558 amino acids and is predicted to encode an Mr = 63,000 protein that is predominantly hydrophobic with no obvious transmembrane domain, signal sequence, or other functional motifs. The first mutation identified in the gene was a point mutation that results in a leucine to proline substitution at position 554 (L554P).7 Other mutations have been discovered, including a splice mutation (termed IVS4+4A-T) that occurs in members of the Ashkenazi-Jewish population.12Both the L554P and IVS4+4A-T mutations are associated with a severe disease phenotype.
Although the cellular phenotype suggests that FA is a DNA repair disorder, recent reports indicate that FAC is principally, if not exclusively, cytoplasmic.13-15 Enforced expression of FAC protein in the nucleus by attachment of a nuclear localization signal renders the wild-type protein incapable of correcting the FAC phenotype in the MMC assay.15,16 Because the technical approach used in the latter studies effectively prohibited cytoplasmic FAC localization, it does not rule out a functional collaboration in the cytoplasm between other proteins and the FAC protein resulting in translocation to the nucleus. Consonant with the idea that other proteins collaborate with the FAC protein is the recent observation that MMC hypersensitivity occurs when L554P mutant FAC protein is overexpressed in normal cells17 and that other proteins coimmunoprecipitate or associate with FAC.13 15
In this report, we describe two new polyclonal antisera raised to highly purified FAC antigens, both of which are capable of detecting endogenous FAC protein in human peripheral blood cells and in cell lines by simple immunoblot and indirect immunofluorescence analyses. The antisera were used to determine the subcellular location of FAC protein in human cells. Based on evidence from two different subcellular fractionation procedures and confocal imaging experiments, we show that the FAC protein is located both in the cytoplasm and in the nucleus of human cells.
MATERIALS AND METHODS
Cell lines, viruses, plasmids, and chemicals.
Epstein-Barr virus (EBV)-transformed lymphoblast cell lines HSC536N (originally obtained from Dr Manuel Buchwald, Hospital for Sick Children, Toronto, Ontario, Canada) and PD4L were supplied by Dr Markus Grompe (Fanconi Anemia Cell Repository, Oregon Health Sciences University [OHSU]). The HSC536N cell line has mutations resulting in a leucine to proline substitution at amino acid position 554 on one allele and a deletion of the second FAC allele. The PD4L cell line has a deletion of a G residue in one allele resulting in a severe truncation, and a mutation that results in a stop codon at amino acid position 185 on the other allele.12 The HSC536N/FAC cell line is corrected for MMC hypersensitivity and was produced by infecting parental cell lines (HSC536N) with FAC-encoding retroviruses, LFACSN. The HSC536N/neo derivative used as a negative control in some experiments described was produced by infecting the HSC536N cell line with an amphotropic- or gibbon ape leukemia virus (GALV)-pseudotyped virus that encodes neomycin phosphotransferase (LXSN). The LXSN viruses were produced by PA31718 or PG1319 retroviral packaging cell lines transfected with the parental vector pLXSN.20 The EBV-transformed cell line JY, derived from a normal individual, was a gift from Dr Richard Maziarz (OHSU). All lymphoblast cell lines were grown in RPMI 1640 (GIBCO-BRL, Grand Island, NY) supplemented with 15% fetal bovine serum (FBS) (defined, heat-inactivated, low endotoxin; HyClone, Logan, UT), 1%l-glutamine (GIBCO-BRL), and 50 μg/mL gentamicin (GIBCO-BRL) at 37°C and 5% CO2 in a humidified atmosphere.
Constructs for expression of FAC protein in bacteria were made by subcloning an EcoRI-XbaI FAC fragment from plasmid pFAC3 (a gift from M. Buchwald, Toronto) into the vector pUC18 (Pharmacia, Piscataway, NJ). The resulting vector (pUCFAC3) was digested with EcoRI and SalI and then inserted into an EcoRI/Sal I-cut pGEX-4T-3 expression vector (Pharmacia) to produce pGEX-FAC1. To make pGEX-FAN2, the pUCFAC3 subclone was digested with EcoRI and ligated into a pGEX-4T-3 vector that was linearized with EcoRI. For COS-7 and 293 cell expression, full-length FAC was subcloned into vector pLXSN and a HpaI/XbaI fragment containing FAC from this subclone was inserted into an EcoRV/XbaI-cut pcDNA3 vector (Invitrogen, San Diego, CA). Cells were transfected with pcFAC or pcDNA3 using DEAE-dextran by standard methods. Chemicals were purchased from Sigma (St Louis, MO) unless indicated otherwise.
Purification of FAC antigens.
Glutathione S-transferase (GST) fusion proteins were produced in E coli strain HB101, which had been transformed with either pGEX-FAN2 or pGEX-FAC1 and induced with isopropyl β-d-galactoside (IPTG). The GST fusion proteins were purified by affinity chromatography on glutathione Sepharose 4B (Pharmacia) as described by the manufacturer and modified by Frangioni and Neel.21Briefly, E coli cell pellets were resuspended in 10 mmol/L Tris Cl (pH 7.4), 150 mmol/L NaCl, 1 mmol/L EDTA, 5 mmol/L dithiothreitol (DTT), and 1.5% (wt/vol) sarkosyl and disrupted by passage through a cold French pressure cell. The resulting crude cell extracts were centrifuged to remove debris, and Triton X-100 was added to the supernate to a final concentration of 2% (vol/vol). The extracts were incubated with glutathione Sepharose 4B and allowed to bind overnight at 4°C. The affinity matrices were first washed with phosphate-buffered saline (PBS), then with PBS plus 1% (vol/vol) Triton X-100, followed by PBS, and finally equilibrated in 50 mmol/L Tris Cl (pH 8.0). Cleavage of an estimated 9-mg GST fusion protein bound to the affinity matrix (10-mL bed volume) was performed in 50 mmol/L Tris Cl (pH 8.0), 2.5 mmol/L CaCl2, and 150 mmol/L NaCl containing approximately 24 to 30 NIH U thrombin for 4 hours at 37°C. The reaction was stopped by adding EDTA to a final concentration of 5 mmol/L. Preparative SDS-PAGE (model 491 Preparative Cell; Bio-Rad, Hercules, CA) was used to purify the cleaved proteins to homogeneity.
Polyclonal antibody production to FAC amino- and carboxy-terminal regions.
Antibodies were prepared by immunizing New Zealand white rabbits with purified FAN2 or FAC1 antigens as described previously.22Briefly, initial injections were given in complete Freund adjuvant (GIBCO-BRL), and injections of approximately 100 μg each in incomplete Freund adjuvant were given at monthly intervals thereafter. Antibody production was monitored from test bleeds by immunoblotting cell lysates from COS-7 cells that were transiently transfected with plasmid pcFAC compared with lysates from cells that had been transfected with the parental plasmid pcDNA3. Antisera with apparent specificity for FAC protein were further characterized against lymphoblast cell lines containing normal or mutant FAC protein. For confocal studies and for immunoblot analysis of the 293 and 293/FAC cell lysates, anti-FAC1 was affinity-purified against FAC1 antigen using the strip purification method.23
Immunoblotting.
Cells were washed twice with PBS, and the cell pellets were solubilized in RIPA (10 mmol/L Tris Cl [pH 7.6], 150 mmol/L NaCl, 1% [wt/vol] sodium deoxycholate, 1% [vol/vol] Triton X-100, 0.1% [wt/vol] SDS, 1% [vol/vol] aprotinin, 2 mmol/L Na3VO4, and freshly added leupeptin [1 μg/mL], pepstatin [1 μg/mL], and 1 mmol/L phenylmethylsulfonylfluoride [PMSF]). Lysates were centrifuged at 16,000g for 15 minutes at 4°C. Protein concentrations of the supernates were determined with a commercially available protein microassay (Bio-Rad) using BSA as a standard. Cell lysates were mixed with Laemmli sample buffer, heated at 94°C for 5 minutes, and separated by SDS-PAGE in 7.5% polyacrylamide gels. Proteins were electroblotted onto Bio-Blot nitrocellulose (Costar, Cambridge, MA) as previously described.24 Incubations and washes were performed at room temperature with gentle rocking. Nonspecific binding was blocked by incubating the blots in 5% (wt/vol) Carnation nonfat milk (Nestle Foods, Glendale, CA) for 1 hour. Each blot was incubated with one of the rabbit antisera at a 1:1,000 dilution in 5% nonfat milk for 1 hour. Primary antibody was followed by six (5-minute) washes of Tris-buffered saline (TBS) containing 0.005% Tween 20, and then incubation for 30 minutes with goat anti-rabbit IgG-HRP (Bio-Rad) at a 1:10,000 dilution in 5% nonfat milk. After incubation with secondary antibody, the blot was washed as described with TBS-Tween 20. Antibody-reactive proteins were detected using Amersham ECL reagents (Arlington Heights, IL) and visualized with x-ray film. Immunoblots reprobed with different primary antibodies were stripped according to the ECL manufacturer's recommendations (Amersham) as described by Kauffman et al.25
Subcellular fractionation.
Cellular fractionation was performed at 4°C as described previously by Lewis et al.26 Briefly, 5 × 108 JY cells were lysed in a hypotonic buffer (Tris Cl, pH 7.4, containing freshly added protease inhibitors: 10 mmol/L Tris Cl [pH 7.4], 1% [vol/vol] aprotinin, 2 mmol/L sodium vanadate, 1 μg/mL leupeptin, 1 μg/mL pepstatin, and 1 mmol/L PMSF) by 25 to 50 strokes of a Potter-Elvehjem homogenizer (Teflon pestle; clearance, 0.10 to 0.15 mm). Cells were observed microscopically to ensure that they were fully disrupted and that the nuclei were intact. The homogenate was made isotonic (final concentration: 10 mmol/L Tris Cl, pH 7.4, 0.1 mmol/L EDTA, 0.85% NaCl, and 0.1 mmol/L MgSO4) and then centrifuged at 1,000g to separate insoluble material from the cytoplasmic fraction. The pellet was resuspended in buffer C (10 mmol/L Tris Cl, pH 7.4, 0.1 mmol/L EDTA, 0.25 mmol/L sucrose, and protease inhibitors as described for RIPA) to a total vol of 5 mL. To separate plasma membranes, mitochondria, and nuclei, the solution was centrifuged at 1,000g for 30 minutes. The supernate was discarded, and the pellet was resuspended in buffer C, loaded onto a 32% to 52% (wt/vol) sucrose gradient cushion (containing 0.1 mmol/L EDTA, 10 mmol/L Tris Cl, pH 7.4, and protease inhibitors as described for RIPA), and centrifuged at 100,000g for 45 minutes. Distinct fractions representing plasma membranes, mitochondria, and nuclei were removed and prepared for SDS-PAGE analysis. An alternative method for subcellular fractionation was also performed as described by Tsuda and Alexander.27 After electrophoresis, the proteins were electroblotted onto nitrocellulose for immunoblotting as already described. Blots were reprobed with anti-β tubulin to enable estimation of the signal in various subcellular fractions by densitometry. Correction for cytoplasmic protein in nuclear fractions was calculated for each fractionated lysate. Densitometric results were obtained by analyzing immunoblots with a model 620 Video Densitometer (Bio-Rad).
Indirect immunofluorescence.
293 cell derivatives were grown on chamber slides, fixed with 3.7% paraformaldehyde, and permeabilized with 0.2% Triton X-100. The cells were blocked with a solution of 3% normal goat serum in PBS. Primary antibody was added (affinity-purified rabbit polyclonal anti-FAC1 and anti-β tubulin monoclonal [Boehringer Mannheim, Indianapolis, IN] or anti-p300 monoclonal [Upstate Biotechnology, Lake Placid, NY]) and allowed to bind overnight at room temperature with gentle rocking. After five 5-minute washes with PBS, secondary antibody was added (Texas Red–conjugated goat anti-rabbit and Oregon Green–conjugated goat anti-mouse; Molecular Probes). Primary and secondary antibodies were diluted in PBS containing 3% normal goat serum. Secondary antibody incubation was performed in the dark. The stained cells were mounted in SlowFade (Molecular Probes).
Stained cells were viewed with a Leica 900 confocal laser-scanning microscope (Leica Inc, Deerfield, IL) equipped with a krypton-argon laser, a 40× 1.3 NA oil objective lens, a simultaneous dual-channel detector, and a 24-bit imaging system including Leica's scanware software function. Appropriate filter sets were used to distinguish between Texas Red and Oregon Green emissions. For the optimal z series, slices were taken approximately every 1 μm over an 8-μm thickness with pinhole settings maintained at optimal levels for minimal slice thickness resolution. Settings were optimized using positively stained cells and maintained during scanning of control cells to maintain relative brightness. Collected images were imported into Adobe Photoshop 4.0 (San Jose, CA), pseudo-colored, and overlapped to produce merged images.
RESULTS AND DISCUSSION
Expression and purification of FAC proteins from bacteria.
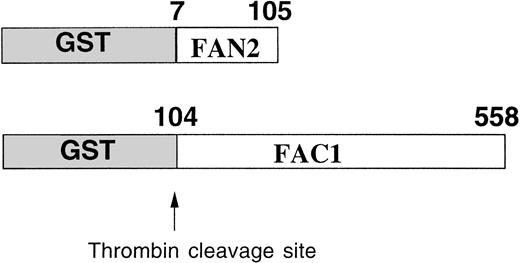
The GST system enables inducible high-level expression of fusion proteins and subsequent purification by glutathione-affinity chromatography, followed by thrombin cleavage to release the protein of interest. Two regions of FAC were expressed as GST-fusion proteins inE coli (Fig 1). GST-FAC1 is a fusion protein between GST and 453 amino acids of FAC, from amino acid position 104 to the last amino acid of the FAC protein at position 558. GST-FAN2 is the reciprocal GST-FAC fusion protein that contains residues 7 through 105 of amino-terminal FAC. Together, these two proteins encode all but the first six amino acids of the FAC coding region. GST-FAC1 and GST-FAN2 fusion proteins were expressed in E coli and purified on glutathione Sepharose 4B.
Expression and partial purification of GST-FAC fusion proteins. Diagram of the regions of the FAC protein used as antigen to stimulate antibody production. Numbers correspond to the amino acid position in the FAC peptide. Arrow indicates the thrombin cleavage site in the GST fusion proteins.
Expression and partial purification of GST-FAC fusion proteins. Diagram of the regions of the FAC protein used as antigen to stimulate antibody production. Numbers correspond to the amino acid position in the FAC peptide. Arrow indicates the thrombin cleavage site in the GST fusion proteins.
We observed that the purification strategy recommended by the manufacturer resulted in inadequate purification of FAC, which is a predominantly hydrophobic protein.7 Moreover, minor (but often very antigenic) contaminating proteins in E coli are difficult to purify away from the desired antigen using purification strategies that work well for predominantly hydrophilic proteins.22 In addition, although the antigenic response is difficult to predict with certainty, the hydrophobicity of FAC suggested that it might be less antigenic than the GST portion of the fusion protein. Based on all of these considerations, we designed a stringent purification strategy that included thrombin cleavage of the GST portion of the GST-FAC fusion proteins (the cleaved proteins are termed FAC1 and FAN2). After thrombin cleavage, FAC1 and FAN2 were completely solubilized and denatured before purification by preparative gel electrophoresis. Purity of the protein collected was estimated by subjecting a sample of the purified protein to SDS-PAGE, electroblotting to nitrocellulose, and detecting the proteins by sensitive colloidal gold staining. Purified antigens were injected into rabbits according to the procedure already described.
Detection of FAC in EBV-transformed lymphoblast cell lines by immunoblot analysis.
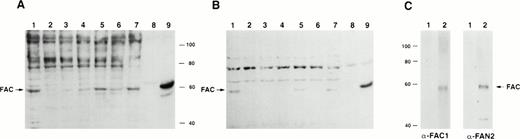
To determine if the rabbit polyclonal antibodies were specific for FAC, we analyzed lysates from several different cell lines. Using both carboxy- and amino-terminal antibodies, a protein of approximateMr = 58,000 was detected in lysates from the normal JY cell line, as well as those derived from the HSC536N cell line, which contains full-length FAC with the L554P mutation (Fig 2A and B). No obvious differences between mutant and wild-type FAC protein migration patterns were observed. The FAC EBV-transformed cell line PD4 has a C to T transition at nucleic acid position 808 that predicts a severely truncated protein of Mr = 20,000. As expected, protein comigrating with FAC protein in the PD4L was absent, as reported by Yamashita et al.13
Detection of FAC protein with anti-FAC antisera. One hundred micrograms of total protein from lymphoblast cell lines was immunoblotted with anti-FAC polyclonal antibodies as described. (A) Lane 1, JY; lane 2, PD4L; lane 3, HSC536N; lanes 4 and 6, HSC536N/neo; lanes 5 and 7, HSC536N/FAC transduced with amphotropic- and GALV-pseudotyped vectors, respectively; lane 8, COS-7/neo; lane 9, COS-7/FAC. FAC protein in A was detected with FAC1 antiserum. B shows the same blot as A, after stripping and reprobing with FAN2 antiserum. (C) Immunoblot of COS-7/FAC cell lysates (10 μg total cell protein per lane) demonstrating that the strong signal observed in immunoblots with FAC1 antisera and FAN2 antisera is completely blocked by addition of purified antigen. The first lane of each set represents blocking of FAC-reactive epitopes with 20 μg purified cognate antigen in a 1-hour preincubation step with 1 μL antiserum. The second lane of each set is the unblocked positive control. Complete blocking was also possible with 10 μg antigen (data not shown).
Detection of FAC protein with anti-FAC antisera. One hundred micrograms of total protein from lymphoblast cell lines was immunoblotted with anti-FAC polyclonal antibodies as described. (A) Lane 1, JY; lane 2, PD4L; lane 3, HSC536N; lanes 4 and 6, HSC536N/neo; lanes 5 and 7, HSC536N/FAC transduced with amphotropic- and GALV-pseudotyped vectors, respectively; lane 8, COS-7/neo; lane 9, COS-7/FAC. FAC protein in A was detected with FAC1 antiserum. B shows the same blot as A, after stripping and reprobing with FAN2 antiserum. (C) Immunoblot of COS-7/FAC cell lysates (10 μg total cell protein per lane) demonstrating that the strong signal observed in immunoblots with FAC1 antisera and FAN2 antisera is completely blocked by addition of purified antigen. The first lane of each set represents blocking of FAC-reactive epitopes with 20 μg purified cognate antigen in a 1-hour preincubation step with 1 μL antiserum. The second lane of each set is the unblocked positive control. Complete blocking was also possible with 10 μg antigen (data not shown).
For further verification of antibody specificity, we compared lysates from COS-7 cells that were transiently transfected with pcFAC (a vector that encodes full-length FAC) and lysates prepared from COS-7 cells transfected with the backbone vector pcDNA3. Both carboxy-terminal and amino-terminal antibodies recognized a strong band atMr = 58,000 in the COS-7/FAC lysates, but not in the pcDNA3-transfected COS-7 cell lysates or in experiments where purified cognate antigen was added to the antibody incubation step (Fig2C). Immunoblots of COS-7 cells transfected with the plasmid encoding FAC contained a signal that comigrated with the strong signal observed in the lysates derived from human hematopoietic cells.
FAC protein is located in the cytoplasm and nucleus.
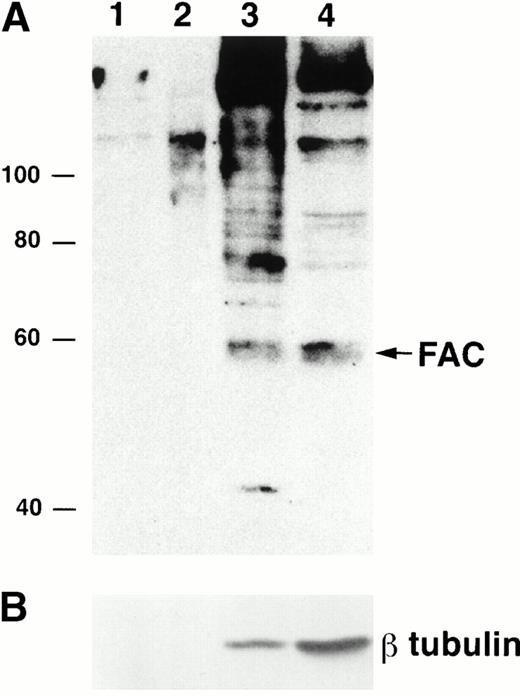
To examine FAC localization with anti-FAN2 and anti-FAC1 antibodies, lysates from JY (normal) EBV-transformed cell lines were fractionated and analyzed by immunoblotting. We found that the majority of FAC protein was located in the cytoplasm, as reported previously.13-15 However, FAC protein was reproducibly found in nuclear fractions in multiple separate experiments (Fig 3A; lanes 1 to 3 represent a fourfold increase in sample loading compared with the cytoplasmic fraction in lane 4). The quality of the fractionation was assessed by reprobing the blot with anti-β tubulin (Fig 3B). The amount of β tubulin present in the nuclear fractions and cytoplasmic fractions was used to estimate the percentage of cytoplasmic contamination of the nuclear fractions. We estimate that the amount of FAC present in the nuclear fraction after correction for cell number and cytoplasmic contamination was 10%. This observation was confirmed with anti-FAN2 and anti-FAC1 antisera, which recognize nonoverlapping epitopes in the FAC protein (data not shown). FAC protein was not detected in the membrane or mitochondrial fractions (Fig 3A, lanes 1 and 2).
FAC protein is located in the cytoplasm and nucleus of normal human lymphoblasts. JY cells were lysed by dounce homogenization and separated into subcellular fractions according to the method of Lewis et al.26 Individual fractions were separated by SDS-PAGE and immunoblotted with FAC1 (A) or β tubulin (B) antiserum. Lane 1, plasma membrane fraction; lane 2, mitochondrial fraction; lane 3, nuclear fraction; lane 4, cytoplasmic fraction. The cytoplasmic fraction contains one fourth the cell number contained in lanes 1 to 3. Similar results were obtained with FAN2 antiserum (data not shown). Values for quality of fractionation were calculated by desitometric analysis. The total amount of signal observed for β tubulin (a cytoplasmic marker) in all fractions was measured and then used to calculate the amount occurring in the nuclear fraction as an artifact.
FAC protein is located in the cytoplasm and nucleus of normal human lymphoblasts. JY cells were lysed by dounce homogenization and separated into subcellular fractions according to the method of Lewis et al.26 Individual fractions were separated by SDS-PAGE and immunoblotted with FAC1 (A) or β tubulin (B) antiserum. Lane 1, plasma membrane fraction; lane 2, mitochondrial fraction; lane 3, nuclear fraction; lane 4, cytoplasmic fraction. The cytoplasmic fraction contains one fourth the cell number contained in lanes 1 to 3. Similar results were obtained with FAN2 antiserum (data not shown). Values for quality of fractionation were calculated by desitometric analysis. The total amount of signal observed for β tubulin (a cytoplasmic marker) in all fractions was measured and then used to calculate the amount occurring in the nuclear fraction as an artifact.
To confirm and extend these observations, an alternate nuclear fractionation protocol was performed on normal and FA EBV-transformed lymphoblasts.27 We observed that FAC protein was in both the cytoplasmic (C) and nuclear (N) fractions of JY and HSC536N cells at an approximate C to N ratio of 3:2 and 9:1 for the two cell types, respectively, as estimated by densitometry and corrected for cell number and the amount of β tubulin, as already described (Fig4).
FAC protein is located in the cytoplasm and nucleus of normal and FA EBV-transformed human lymphoblasts. Cells were fractionated according to the method of Tsuda et al,27 and the separated nuclear and cytoplasmic lysates were analyzed by SDS-PAGE. FAC protein was detected by probing transferred proteins with FAN2 antisera. The blot was stripped and reprobed with anti-β tubulin as a control for artifactual presence of cytoplasmic protein in nuclear fractions as described in Fig 3. Reprobing with anti-FAC1 gave similar results (data not shown). Lane 1, JY cytoplasmic fraction; lane 2, HSC536N cytoplasmic fraction; lane 3, JY nuclear fraction; lane 4, HSC536N nuclear fraction. The cytoplasmic fractions (70 μg total protein loaded) contain approximately one fourth the cell number of the nuclear fractions (50 μg total protein loaded).
FAC protein is located in the cytoplasm and nucleus of normal and FA EBV-transformed human lymphoblasts. Cells were fractionated according to the method of Tsuda et al,27 and the separated nuclear and cytoplasmic lysates were analyzed by SDS-PAGE. FAC protein was detected by probing transferred proteins with FAN2 antisera. The blot was stripped and reprobed with anti-β tubulin as a control for artifactual presence of cytoplasmic protein in nuclear fractions as described in Fig 3. Reprobing with anti-FAC1 gave similar results (data not shown). Lane 1, JY cytoplasmic fraction; lane 2, HSC536N cytoplasmic fraction; lane 3, JY nuclear fraction; lane 4, HSC536N nuclear fraction. The cytoplasmic fractions (70 μg total protein loaded) contain approximately one fourth the cell number of the nuclear fractions (50 μg total protein loaded).
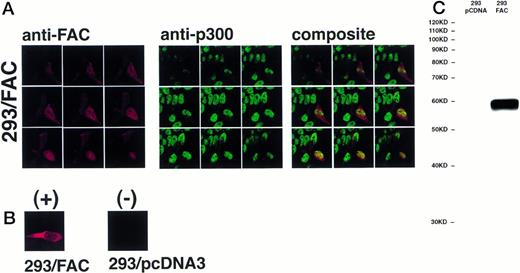
The definitive method for determining the exact location of a substance in a three-dimensional volume is to use cross-sectional imaging studies such as confocal image analysis. This strategy allows virtual examination of the internal structure of the object, free from overlapping and potentially confounding images occurring in different planes (eg, cytoplasm overlapping the nucleus in the cell). Accordingly, JY and HSC536N cells were examined by confocal laser-scanning microscopy after immunostaining with anti-FAC1 or anti-β tubulin. Although it is our impression that the pattern of dual cytoplasmic and nuclear FAC protein localization observed in fractionation procedures was also observed by this technique, the endogenous expression of FAC in these cells is at the detection limit for this assay (data not shown). Therefore, a second approach was taken to more definitively determine the subcellular distribution of the FAC protein by confocal analysis. Human 293 cells were transfected with pcFAC (termed 293/FAC) or the parental plasmid pcDNA3 (293/pcDNA3) as a control and selected for stable expression in G418. To ensure FAC expression, cell lysates were prepared from the cell lines and analyzed by SDS-PAGE followed by immunoblot with affinity-purified anti-FAC1. A strong unique band was observed at Mr = 58,000 in cells transfected with pcFAC (Fig 5C),indicating that the 293/FAC cells express FAC abundantly and that no cross-reacting signals were present in the control cell line by immunoblot analysis. For immunofluorescence studies, these cell lines were double-stained with affinity-purified FAC1 and nuclear (anti-p300) or cytoplasmic (anti-β tubulin) markers. The Texas Red and Oregon Green fluorophores conjugated to the secondary antibodies were selected to ensure the largest spread between emission wavelengths to maximize specificity of the signals (615 nm and 522 nm, respectively). As shown in the representative single optical sections (made parallel to the growing surface) in Fig 5B, the staining of FAC protein with affinity-purified anti-FAC1 is evident in 293 cells transfected with pcFAC as compared with an optical section through 293 cells transfected with pcDNA3 (parental vector). The 293/FAC cells examined were a population of stably transfected cells rather than a clonal population, enabling examination of cells expressing different levels of the FAC protein. The different levels of FAC protein observed in the transduced cells may also be due to involvement of FAC in a cell-cycle pathway or regulated FAC expression during the cell cycle, as suggested by recent reports by Kruyt et al28 and Kupfer et al.29This effect is demonstrated in Fig 5A, where cells were double-labeled with anti-p300 (green) and anti-FAC (red). In the first group (cells stained with anti-FAC), there is one cell (lower right) that stains brightly with the antibody, as well as a cell just above with weaker staining. Other cells in this field have weaker FAC staining, although all pcFAC-transfected cells that became G418-resistant were positive compared with the negative control. The nuclei of the stained cells are visible in the second group of images viewed for p300 staining. p300 staining is not observed in the cytoplasm, as expected. The confocal images resulting from anti-FAC and anti-p300 staining were merged to determine if the signals colocalized. The composite panel in Fig 5A demonstrates colocalization (yellow) of FAC protein and the nuclear marker p300. Colocalization of p300 and FAC protein can be observed even in the cell expressing a relatively low level of FAC protein.
FAC protein colocalizes with a nuclear protein marker in double-immunostained cells. Human 293 cells transfected with a FAC-encoding plasmid were analyzed by confocal laser-scanning microscopy after double labeling with affinity-purified anti-FAC1 and anti-p300 (a nuclear marker). The images shown contain a transfected cell population that express varying amounts of FAC protein. Nine optical sections taken from the apical (top left) to basal (bottom right) cell surface are shown. Composite images were created by merging corresponding images. The FAC protein is red, the p300 protein is green, and colocalization is yellow. (B) Optical section though a FAC-expressing cell (293/FAC) and 293 cells transfected with the parental pcDNA3 plasmid without the FAC gene insert (293/pcDNA3). (C) 293/pcDNA3 and 293/FAC cell lysates were analyzed by SDS-PAGE, transferred to nitrocellulose, and probed with the affinity-purified antibody used in the confocal analysis. Arrow indicates FAC protein. Molecular size markers are indicated at left.
FAC protein colocalizes with a nuclear protein marker in double-immunostained cells. Human 293 cells transfected with a FAC-encoding plasmid were analyzed by confocal laser-scanning microscopy after double labeling with affinity-purified anti-FAC1 and anti-p300 (a nuclear marker). The images shown contain a transfected cell population that express varying amounts of FAC protein. Nine optical sections taken from the apical (top left) to basal (bottom right) cell surface are shown. Composite images were created by merging corresponding images. The FAC protein is red, the p300 protein is green, and colocalization is yellow. (B) Optical section though a FAC-expressing cell (293/FAC) and 293 cells transfected with the parental pcDNA3 plasmid without the FAC gene insert (293/pcDNA3). (C) 293/pcDNA3 and 293/FAC cell lysates were analyzed by SDS-PAGE, transferred to nitrocellulose, and probed with the affinity-purified antibody used in the confocal analysis. Arrow indicates FAC protein. Molecular size markers are indicated at left.
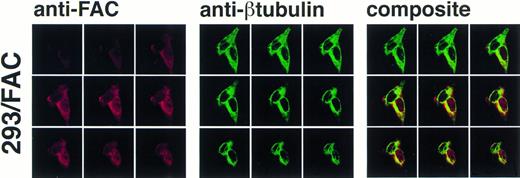
As permeabilization and fixation procedures can cause artifactual relocalization of protein,30 the cells were also stained with anti-β tubulin to determine if these procedures resulted in the appearance of β tubulin in the nucleus. The images in Fig 6 show that the nuclear margin is visible in the stained cells and β tubulin staining is not present in the nuclear compartment, although the cells stained with anti-FAC1 contain FAC protein in the nucleus (Fig6). The fractionation studies made in parallel using the EBV-transformed cell lines (JY and HSC536N) demonstrated that the nuclear localization of FAC protein is not merely a consequence of overexpression. Moreover, nuclear localization was decreased or absent in images of some 293 cells overexpressing FAC (data not shown), further suggesting that nuclear localization is not an artifact of overexpression and that translocation may be dependent on some unknown mechanism.
Confocal laser-scanning of double-stained cells reveals that FAC protein colocalizes with a cytoplasmic protein marker (β tubulin). The arrangement of images is as described in Fig 5. In the optical sections shown, FAC protein is stained red, β tubulin is green, and colocalization is yellow. Although the cytoplasmic marker does not appear in the nuclei of the cells, FAC protein staining is evident.
Confocal laser-scanning of double-stained cells reveals that FAC protein colocalizes with a cytoplasmic protein marker (β tubulin). The arrangement of images is as described in Fig 5. In the optical sections shown, FAC protein is stained red, β tubulin is green, and colocalization is yellow. Although the cytoplasmic marker does not appear in the nuclei of the cells, FAC protein staining is evident.
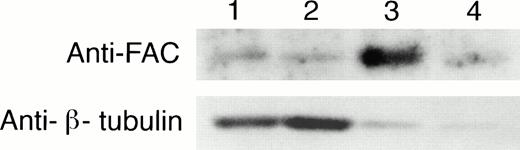
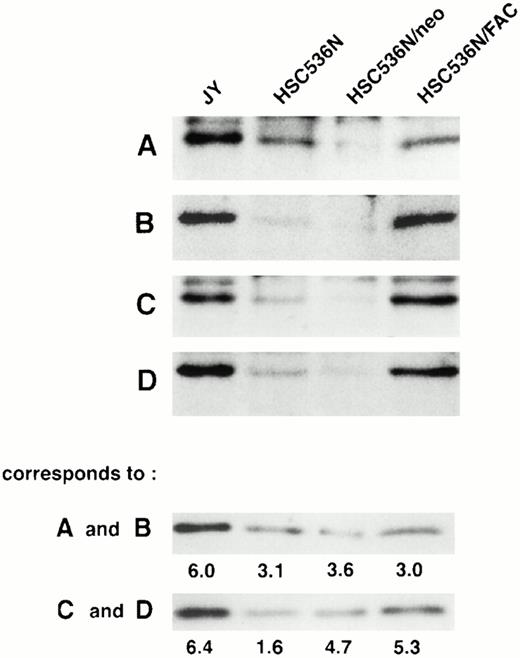
To determine if the amount of FAC protein in nuclear fractions increases after correction by FAC-encoding retroviral vectors, nuclear fractions from normal cells, FA group C cells, and FA cells that had been corrected by transduction with retroviral vectors encoding wild-type FAC were compared. The specific mutant cells we used for these experiments endogenously express a full-length mutant FAC protein in deceased amounts compared with wild-type cells. Figure 7 (column 1) shows FAC in nuclear fractions of JY cells, a normal EBV-transformed cell line. The FAC protein was detected by either anti-FAC1 (A and C) or anti-FAN2 (B and D). Figure 7 (column 2)shows lysates prepared from HSC536N probed with each antibody, as described for JY cells. The HSC536N cells have lower amounts of FAC protein detected in nuclear fractions compared with normal JY cells, as predicted by the loss of one of the FAC alleles in this cell line. Similarly, HSC536N cells transduced with retroviral vectors expressing neomycin phosphotransferase, but not wild-type FAC, have reduced amounts of FAC compared with the normal JY cells (Fig 7, column 3). In HSC536N cells transduced by retroviral vectors encoding wild-type FAC and corrected for MMC hypersensitivity, the relative amount of FAC in nuclear fractions is increased compared with the uncorrected HSC536N cells and is comparable to normal cells (Fig 7, column 4). In several separate fractionation experiments using these cell lines, we estimated cytoplasmic and nuclear fractions of FAC and observed that FAC consistently appeared in the nuclear fraction in increased amounts after the cells were corrected by FAC-encoding retroviral vectors (Fig4). Thus, in both of the corrected cell lines, using two nonoverlapping FAC antisera, we observed a reproducible increase in the amount of FAC in the nuclear fractions.
FAC protein is decreased in nuclear fractions of FA cells compared with normal and corrected FA cells. Cells were fractionated according to the method of Tsuda et al,27 and the nuclear lysates (50 μg each) were analyzed by SDS-PAGE. FAC protein (arrow) was detected by probing with FAC1 antisera (A and C) or stripping and reprobing with FAN2 antisera (B and D). A and B and C and D represent independent experiments where cells in columns 3 and 4 were transduced by pseudo-typed viruses encoding neo or FAC, respectively. Column 1, JY; column 2, HSC536N; column 3, HSC536N/neo; column 4, HSC536N/FAC. The blots were stripped and reprobed with β tubulin as shown in the bottom 2 panels, and the signals for nuclear and cytoplasmic β tubulin were estimated by densitometry. These fractionation quality controls are corrected for cell number, and the numbers shown are the percent of cytoplasmic protein contained in the nuclear fraction. Thereby, we estimate that the artifactual component of the signal detected for FAC in the nuclear fraction of JY cells (for A and B) is only 6%.
FAC protein is decreased in nuclear fractions of FA cells compared with normal and corrected FA cells. Cells were fractionated according to the method of Tsuda et al,27 and the nuclear lysates (50 μg each) were analyzed by SDS-PAGE. FAC protein (arrow) was detected by probing with FAC1 antisera (A and C) or stripping and reprobing with FAN2 antisera (B and D). A and B and C and D represent independent experiments where cells in columns 3 and 4 were transduced by pseudo-typed viruses encoding neo or FAC, respectively. Column 1, JY; column 2, HSC536N; column 3, HSC536N/neo; column 4, HSC536N/FAC. The blots were stripped and reprobed with β tubulin as shown in the bottom 2 panels, and the signals for nuclear and cytoplasmic β tubulin were estimated by densitometry. These fractionation quality controls are corrected for cell number, and the numbers shown are the percent of cytoplasmic protein contained in the nuclear fraction. Thereby, we estimate that the artifactual component of the signal detected for FAC in the nuclear fraction of JY cells (for A and B) is only 6%.
We attribute our capacity to detect FAC protein in the nuclei of human cells to technical differences between our methods and those used by other investigators. First, the subcellular fractionation procedures we used were unique to our study and may have allowed detection of the nuclear portion of the FAC protein. Second, one of our detection methods was based on a simple immunoblot, not a more lengthy immunoprecipitation procedure that might be associated with a loss of signal from the nuclear fraction. This is of particular importance because the FAC protein is labile and requires careful handling to maintain a signal. Third, individual antibodies often differ in the capacity to detect protein in different assays, such as immunoprecipitation, immunoblot, or immunofluorescence (IFA). Finally, it is possible that the FAC protein located in the nucleus cannot be immunoprecipitated, or it may be below the detection limit of that assay.
We have described two independent approaches that indicated that FAC protein is located not only in the cytoplasm of the cell but in the nucleus, as well. Interestingly, in studies using an immunofluorescence-based assay, a preliminary report using a FLAG epitope-tagged FAC described both nuclear and cytoplasmic localization.31 In addition, Youssoufian14observed an occasional cell with nuclear staining in cells overexpressing FAC using an IFA.
What is the implication for dual cytoplasmic and nuclear localization of FAC protein in terms of function? There are several possible explanations. First, precedent exists for proteins whose predominant location is distinct from their site of function. For example, the bulk of the env glycoprotein of the spleen focus-forming virus is located in the endoplasmic reticulum, yet only a very small portion (∼5%) transported to the cell surface has productive interaction with the erythropoietin receptor.32-34 Resolving the location of the interaction depended on a combination of genetic and biochemical evidence34,35; as is likely to be the case for the FAC protein. Second, since forced nuclear localization abrogates FAC protein function,17 it is possible that FAC protein collaborates with proteins in the cytoplasm that modify it for its function in the nucleus (eg, DNA repair) or that FAC-associated proteins15,17,36 have nuclear functions themselves that are activated by FAC protein in the cytoplasm. There are a variety of examples of this in mammalian cells. For example, the NF-kB protein is maintained in an inactive form in the cytoplasm by its inhibitor, IkB. In response to a variety of stimuli, IkB becomes phosphorylated and is quickly degraded, releasing NF-kB, which then translocates into the nucleus.37-39 Other examples of translocation in response to an extracellular signal include MAP kinases (p42mapk and p44mapk), Stat molecules,40 the Drosophila homeoproteinexd,41 and rsk-encoded protein kinases.42,43 A third possible explanation for the dual location of the FAC protein is that it may have different functions in the nucleus and the cytoplasm, similar to the hepatitis B virus HBx protein. The majority of HBx is in the cytoplasm, but a small amount is in the nucleus. The cytoplasmic HBx stimulates signal transduction pathways, while the nuclear form of HBx transactivates transcription factors in the nucleus.44
Our finding of nuclear localization is compatible with the recent speculation of Liebetrau et al,45 who examined the murine and human FAC protein for functional motifs. They reported that the FAC protein has two regions that may be leucine zippers, one of which is preceded by a basic region and a helix-turn-helix motif. They propose that FAC has a structure typical of a nuclear protein. Whether these proposed domains are essential for the observed nuclear localization of FAC is unknown. How and if the nuclear FAC differs from the cytoplasmic FAC protein and what factors dictate differential localization will be an important question for future study.
ACKNOWLEDGMENT
We thank Manuel Buchwald (Toronto, Ontario, Canada) for plasmid pFAC3 containing the full-length FAC cDNA and for cell line HSC536N. Other FA EBV-transformed cell lines were kindly provided by Marcus Grompe (Fanconi Anemia Registry, Portland, OR). We are grateful to Lorene Langeberg (Portland, OR) for advice and help in preparation of the confocal data for publication. We thank the members of the Fanconi anemia research community at large for stimulating discussions, particularly the members of the OHSU Fanconi Anemia Program Project team.
Supported by grants from the National Institutes of Health (HL48546, G.C.B.), the Department of Veterans Affairs (G.C.B. and M.C.H.), the Medical Research Foundation of Oregon (M.E.H. and M.C.H.), and the Fanconi Anemia Research Fund (M.E.H.).
Address reprint requests to Grover C. Bagby Jr, MD, Division of Hematology and Medical Oncology, Oregon Health Sciences University, 3181 SW Sam Jackson Park Rd, Portland, OR 97201.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal