Abstract
Mammalian Polycomb group (Pc-G) genes, constituting some 5 subfamilies based on their identity to the Drosophila genesPc, Psc, ph, esc, and E(z), appear to play critical roles in maintaining the transcriptional repression state ofHox/HOM-C genes during development. Despite increasing evidence of the important role of Hox genes in both normal hematopoiesis and leukemic transformation, little is known about the expression and possible function played by Pc-G genes in hematopoietic cells. To address this, we first examined the expression of Pc genes in purified CD34+ human bone marrow cells by reverse transcriptase-polymerase chain reaction (RT-PCR), using degenerate primers that specifically amplify the majority of Pcgenes. This analysis showed the expression of 8 different Pcgenes in CD34+ bone marrow cells, includingHP1Hsα, HP1Hsγ, the heterochromatin p25 protein, the human homologue of the murine M32 gene, and 4 novel members of this family. To assess whether Pc-G mRNA levels change during differentiation of bone marrow cells, a quantitative RT-PCR method was used to amplify the total cDNA originating from three purified subpopulations of CD34+bone marrow cells known to differ in their ability to grow in long-term or semisolid cultures. In sharp contrast to Hox gene expression, which is highest in the most primitive bone marrow cells, these studies show that the expression level of 8 of the 9 Pc-Ggenes studied (ie, HP1Hsα, HP1Hsγ, M31, M32, M33, Mel-18, Mph1/Rae-28, and ENX-1) markedly increases with differentiation of bone marrow cells. Interestingly,BMI-1 exhibits a strikingly different pattern of expression, with high expression levels in primitive cells and very little expression in mature CD34− cells. Together, these results document for the first time that differentiation of human bone marrow cells is accompanied by profound changes in Pc-G gene expression levels.
IN DROSOPHILA, the homeotic(HOM-C) genes of the ANT-C and BX-C complexes encode highly conserved transcription factors involved in cell fate determination.1 During embryogenesis, the spatial and temporal expression of these genes is collinear relative to their 3′ → 5′ position on the chromosome. Although this unique expression pattern is established by segmentation genes of the pair-rule (activators) and gap (repressors) families, the maintenance of HOM-C gene expression during later stages of development is dependent on the trithorax(trx-G) and Polycomb group (Pc-G) gene products (reviewed in Simon et al2). For the Pc-G genes, this repressive function appears to be achieved initially by direct interaction with the transiently expressed gap proteins and, later, by contributing to the formation and stable transmission of heterochromatin.
The DrosophilaPolycomb(Pc) gene was one of the first members of the Pc-G family to be identified based on its ability to maintain segment-specific expression of theHOM-C genes.3 At least 12 other mutations leading to a phenotype similar to Pc (or to its enhancement) have been described. These include: Posterior sex combs(Psc)4; polyhomeotic(ph)5; Polycomblike(Pcl)6; extra sex combs(esc)7; Additional sex combs(Asx)4; Enhancer of zeste[E(z)],8 also known as polycombeotic(pco)9; l(4)102EFc,10 recently renamed pleihomeotic11; Sex combs extra(Sce)12; Sex combs on midleg(Scm)4; super sex combs(sxc)13; multi sex combs(mxc)14; and Enhancer of Polycomb[E(Pc)].15 These various Pc-G names originate from the ectopic expression of organs called sex combs on the second and third legs of male mutants.16
Recent studies have provided evidence for the existence of mammalianPc-G genes and their involvement in repressing the transcription of homeotic (Hox) genes.17M31, M32, M33 (murine), the heterochromatin p25 protein (the human homologue of M31),HP1Hsγ, and HP1Hsα (human) are all homologues of the DrosophilaPcgene18; bmi-1 and mel-18 are both murine and human homologues of the DrosophilaPosterior sex combs(Psc) gene19; Enx-1 (human and mouse), eed and Mph1/Rae-28 (mouse), and HPH1and HPH2 (human) likely represent the mammalian counterparts of the DrosophilaE(z), esc, and ph genes, respectively.20 21
An important clue to Pc-G gene function in contributing to the formation of heterochromatin came from the finding that a conserved region called the chromodomain (for chromatin organizer domain) is shared between Pc and the heterochromatin-associated protein HP1.22 Furthermore, it has been recently shown that bothDrosophila and mammalian Pc-G proteins interact with chromatin as heterogeneous multimeric complexes.20,23,24 The exact mechanisms by which these complexes repress transcription of their target genes are not yet fully understood. The chromatin accessibility model predicts that the cooperative interaction of Pc-G protein complexes interacting with Pc-G response elements (PREs) regionally compacts the chromatin structure, thus eliminating the accessibility of DNA to transcriptional regulators.25-29However, several pieces of evidence suggest that the mechanism of Pc-G gene-mediated silencing may not be solely achieved by the general inaccessibility of regulatory sequences. For example, it was shown that Pc proteins can inhibit Gal-4– but not T7-dependent transcription in Drosophila embryos.30Thus, other models by which Pc-G genes might be acting have been proposed. For example, Pc-G genes could act as transcriptional repressors,6,31-35 they could interact with specific molecules required for pol II transcription, or, alternatively, they could block looping interactions between promoters and enhancers.36
The pleiotropic phenotypes observed in many Pc-G mutants indicate their participation in numerous cellular processes such as anterio-posterior segmentation, dorso-ventral patterning, neural development, oogenesis, and hematopoiesis.11-13,37-40 In support of the importance of Pc-G genes in hematopoiesis, severe hypomorphic alleles of the multi sex comb(mxc)gene in Drosophila were shown to result in premature hemocyte differentiation and tumorous overgrowth of the larval hematopoietic organs.14,41 42
Recent studies in mice also support a critical role for Pc-Ggenes in hematopoiesis. B and T cell populations in M33−/− mice exhibit a decreased proliferative response to plant agglutinin.23 Mice lacking bmi-1display a progressive aplastic disease characterized by replacement of bone marrow space by adipocytes, as well as a smaller spleen and thymus than control littermates. Although all thymocyte populations are initially normal in newborn bmi-1−/−mice, a progressive loss of CD4+CD8+ cells is observed such that adult thymi contain more than 90% CD4−CD8− cells. B-cell development is also abnormal, with bone marrow pro-B and pre-B cells being most affected. In addition, bone marrow macrophage colony-stimulating factor (M-CSF) and interleukin-7 (IL-7), but not IL-3, responsive clonogenic progenitors are decreased in numbers. Interestingly, erythropoiesis does not appear to be altered in these mice. Together, these data suggest that bmi-1 function in hematopoietic cells is lineage- and stage-specific, displaying a redundant role during embryogenesis but being essential for proliferation of certain adult hematopoietic lineages.43 Similarly, targeted disruption of the other mammalian Psc gene, mel-18, leads to B- and T-cell developmental defects caused by an insufficient response to IL-7 stimulation of the lymphoid precursors.20 An additional line of evidence further suggesting a role for Pc-G proteins in hematopoietic cell function is provided by the finding of a direct interaction between the Pc-G protein Enx-1 and Vav, a proto-oncogene expressed predominantly in hematopoietic cells.44
Interestingly, perturbation in expression levels of certainPc-G and Hox genes produce comparable phenotypes in hematopoietic cells. Similarly to what has been observed in bmi-1and mel-18−/− mice, retroviral overexpression of HOXA10 or HOXB3 in murine bone marrow cells causes a block in differentiation of early B and T cells, respectively.45,46 These observations underscore the importance of preserving the downregulation of Hox gene expression that occurs during normal differentiation of primitive hematopoietic cells47 and suggest that Pc-G gene products play a key role in assuming this function. Furthermore, inactivation ofPc-G genes may lead to leukemic growth as a consequence ofHox gene overexpression.45 48-51
In this work, we have investigated the expression of mammalianPc-G genes in purified subpopulations of human bone marrow cells and in leukemic cell lines. We document the existence of a highly regulated program of Pc-G gene expression with mature bone marrow subpopulations showing much higher Pc-G gene mRNA levels relative to less differentiated precursors. These results contrast with previously documented Hox gene expression profiles and thus suggest a role for Pc-G proteins in regulating differentiation and/or proliferation of human hematopoietic cells by silencingHox gene expression.
MATERIALS AND METHODS
PCR amplification of the chromodomains of the Polycomb (Pc) subfamily of Pc-G genes.
A set of degenerate oligonucleotides was designed to match all the different sequences of the conserved 5′-end [5′-CAT-GAA-TTC-(GATC)GA-(GA)AA-(GA)AA-(GA)G-T(GATC) (TC)-T(GATC)GA-(TC)(AC)G-3′] and 3′-end [5′-TCT-AGA-TCT-(TC)T-C(GATC)G-G(TC)T-CCC-A(GATC)GT-(GA)T-T-3′] of the chromodomains of most members (from Drosophila to mammals; Table 1) of the Polycomb(Pc) subfamily of Pc-G genes. These primers were used to polymerase chain reaction (PCR)-amplify the conserved chromodomain of Pc genes from a phage cDNA library made from purified CD34+ human bone marrow cells originating from a single donor.46 Briefly, phage DNA was obtained by 2 successive phenol-chloroform extractions and approximately 0.1 μg of this DNA introduced in a PCR mixture containing 200 pmol of each degenerate primer (see above), 250 μmol of each four deoxyribonucleotides (dNTP; Pharmacia, Uppsala, Sweden), 1.5 mmol of MgCl2, 10 mmol of Tris-HCl, pH 8.9, 50 mmol of KCl, 5 U of Taq polymerase (Life Technology, Burlington, Ontario, Canada), and water to 50 μL. Parameters for PCR amplification were 30 seconds at 94°C, 2 minutes at 50°C, and 2 minutes at 72°C for 35 cycles. A unique 115-bp fragment was obtained and subcloned into theEcoRV site of Bluescript KS (Stratagene, La Jolla, CA) as described.47
Alignment of the Chromodomains of thePc Genes and Generation of a Consensus Sequence to Design the Degenerate Oligonucleotides Used in These Studies
| Pc Genes . | Chromodomain Conserved Sequences . |
|---|---|
| HP1(D) | YVVEKVLDRRVRKGKVEYYLKWKGYPETENTWEPENN-150 |
| HP1Hsα(H) | YAVEKIIDRRVVKGQVEYLLKWKGFSEEHNTWEPEKN |
| HP1HSγ(H) | FVVEKVLDRRVVNGKVEYFLKWKGFTDADNTWEPEEN |
| M31(M) | YVVEKVLDRRVVKGKVEYLLKWKGFSDEDNTWEPEEN |
| M32(M) | FVVEKVLDRRVVNGKVEYFLKWKGFTDADNTWEPEEN |
| p25(H)-151 | YVVEKYLDRRVVKGKVEYLLKWKGFSDEDNTWEPEEN |
| Consensus | YVVEKVLDRRVVKGKVEYYLKWKGFPETDNTWEPENN |
| FA II RN Q F YSDEH K | |
| L T AE E |
| Pc Genes . | Chromodomain Conserved Sequences . |
|---|---|
| HP1(D) | YVVEKVLDRRVRKGKVEYYLKWKGYPETENTWEPENN-150 |
| HP1Hsα(H) | YAVEKIIDRRVVKGQVEYLLKWKGFSEEHNTWEPEKN |
| HP1HSγ(H) | FVVEKVLDRRVVNGKVEYFLKWKGFTDADNTWEPEEN |
| M31(M) | YVVEKVLDRRVVKGKVEYLLKWKGFSDEDNTWEPEEN |
| M32(M) | FVVEKVLDRRVVNGKVEYFLKWKGFTDADNTWEPEEN |
| p25(H)-151 | YVVEKYLDRRVVKGKVEYLLKWKGFSDEDNTWEPEEN |
| Consensus | YVVEKVLDRRVVKGKVEYYLKWKGFPETDNTWEPENN |
| FA II RN Q F YSDEH K | |
| L T AE E |
Abbreviation: D, Drosophila; H, human; M, mouse.
Letters in bold represent the amino acid sequence used to construct the 2 degenerate primers used in these studies.
Human heterochromatin p25 protein is also referred to asHSM1 orHP1Hsβ and is the human homologue of the mouse M31 gene.
DNA sequencing and sequence analysis.
DNA sequencing was performed by the dideoxy chain termination method using [35S] dATP and a T7 sequencing kit (Pharmacia) according to the manufacturer's recommendations. Either the universal, reverse, T3, or T7 primers complementary to sequences within the cloning vector were used. Nonredundant nucleotide sequence databases (GenBank, EMBL) were screened for homologous sequences using the search algorithms BLAST and FastA of the GCG program (Genetic Computer Group, Madison, WI). The BestFit program was used to obtain the similarities and identities of the Pc-G genes of the Pcsubfamily.
Purification of CD34+ human bone marrow cell subpopulations.
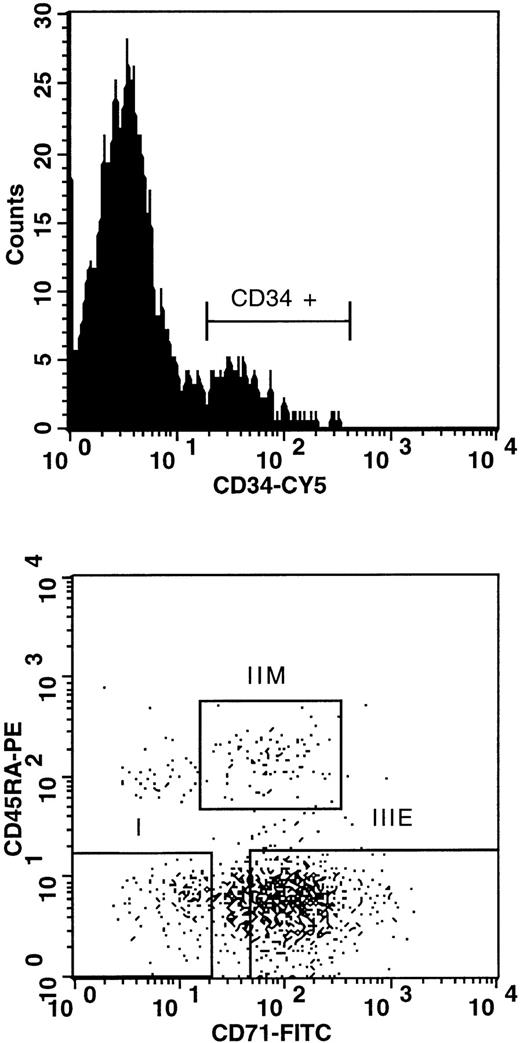
Low-density cells (<1.077 g/mL) obtained from 3 different healthy bone marrow (BM no. 1, 2, and 3) donors were isolated by centrifugation on Ficoll-Paque (Pharmacia LKB, Uppsala, Sweden) and kept frozen in Iscove's medium containing 10% fetal calf serum (FCS) and 7.5% dimethyl sulfoxide. For fluorescence-activated cell sorting (FACS) experiments, cells were thawed in the presence of DNase I (Sigma, St Louis, MO) to avoid clogging; stained with a series of directly conjugated fluorescent antibodies to CD34 (8G12-Cy5), CD45RA (8d2-R-phycoerythrin), and CD71 (OKT9-fluorescein isothiocyanate); and washed twice (propidium iodide at 1 μg/mL was included in the last wash [Sigma]) before sorting on a FACStarplus (Becton Dickinson Immunocytometry, San Jose, CA) as described.47 Cells were sorted in three phenotypically and functionally distinct subpopulations: subpopulation I (CD34+CD45RA−CD71−), IIM (CD34+CD45+CD71lo), and IIIE (CD34+CD45RA-CD71hi), as characterized before.47 Cells from bone marrows were also stained with 8G12-Cy5 alone and separated into total CD34+and CD34− subpopulations. Aliquots from each subpopulation were analyzed and found to be greater than 98% pure.
cDNA generation and amplification.
A previously described method for generating representative amplified total cDNA from small numbers of hematopoietic cells using an oligodT-based primer and polyA tailing strategy was used, with modifications designed to improve cDNA yield of even rare transcripts and to provide amplified sequences extending up to 2 kb 5′ of the polyadenylation site.47 In brief, 1,500 to 10,000 cells of each subpopulation were pelleted and then lysed in a 5 mol/L guanidium isothiocyanate solution containing 20 mmol/L dithiothreitol. Nucleic acids were precipitated by adding 25 μL of 7.5 mol/L ammonium acetate, 20 μg of glycogen as a carrier, and 2 vol of 95% ethanol. The washed pellets were dried at room temperature and resuspended in 9.5 μL of a solution containing 6.1 μL of diethyl pyrocarbonate-treated water, 2 μL of 5× RT buffer (Life Technology), 1 μL of 0.1 mol/L dithiothreitol, 0.2 μL of 25 mmol/L dNTPs (Pharmacia), 0.2 μL of a special 60-mer oligo(dT) primer [1 μg/μL; 5′-CAT-GTC-GTC-CAG-GCC-GCT-CTG-GAC-AAA-ATA-TGA-ATT-CT(24)-3′], and 0.5 μL of Moloney murine leukemia virus (MMLV) SuperScript II reverse transcriptase (200 U/μL; Life Technology). The samples were left at 40°C for 1 hour, heated to 75°C for 10 minutes, and ethanol-precipitated with ammonium acetate and a linear polyacrylamide carrier, as described.47 The pellet was washed once; resuspended in 5 μL of a tailing solution containing 1 μL of 5× terminal deoxynucleotidyl transferase (TdT) buffer (Life Technology), 0.5 μL of 100 mmol/L dATP (Pharmacia), 3.5 μL of water, and 0.5 μL of TdT enzyme (15 U/μL; Life Technology), and incubated for 15 minutes at 37°C. After heat inactivation (75°C for 10 minutes), this solution was directly added to a PCR amplification mixture consisting of 25 μL of a 2× buffer (20 mmol/L Tris, pH 8.8, 100 mmol/L KCl, 9 mmol/L MgCl2), 4 μL of the 60-mer primer (same as described above), 0.5 μL of nuclease-free bovine serum albumin (10 mg/mL; Sigma), 5.25 μL of water, and 2 μL of d(GCT) deoxynucleotides adjusted at 25 mmol/L each. Four micrograms of gene 32 protein (Pharmacia) and 5 U of Taq polymerase (Life Technology) were added to each tube and total cDNA was amplified with an Ericomp thermal cycler (Ericomp, San Diego, CA) using the following parameters: 94°C for 1 minute; 55°C for 2 minutes except for the first cycle, which was performed at 37°C; and 72°C for 10 minutes for 44 cycles.
Southern blot analysis of total amplified cDNA.
One-fifth of the total amplified cDNA prepared from each purified subpopulation or cell line was electrophoresed in a 1% agarose gel and transferred to an ionic nylon membrane (Zeta-probe; Bio-Rad, Hercules, CA). Probes were labeled with 32P-dCTP by random priming and purified on Sephadex-G50 columns (Pharmacia). Blots were prehybridized and hybridized at 65°C in 4.4× SSC, 7.4% formamide, 0.74% sodium dodecyl sulfate (SDS), 1.5 mmol/L EDTA, 0.74% skim milk, 370 μg/mL salmon sperm DNA, and 7.5% dextran sulphate. Membranes were washed three times for 30 minutes at 65°C in 0.3% SSC, 0.1% SDS, and 1 mg/mL sodium pyrophosphate and exposed to Kodak BioMax MS films (Interscience Inc, Markham, Ontario, Canada). Blots were stripped in a 1% SDS solution at 95°C for 30 minutes and tested for the absence of signals by overnight exposure to Kodak BioMax MS films (Eastman Kodak, Rochester, NY) using appropriate intensifying screens (Interscience Inc).
Most probes used in these studies were generated by PCR amplification of cDNA or genomic DNA obtained from various sources and were all sequenced as described above. These probes included (1) a 264-bp fragment of the human HP1Hsα gene (nucleotides 416-679; accession no. S62077; 5′ primer CTCAAACAGTGCCGATGACA and 3′ primer TCCGCATCCTCAGGATATGC) located downstream of the chromodomain; (2) a 222-bp fragment of the humanHP1Hsβ gene (or p25heterochromatingene) also located downstream of the conserved chromodomain (nucleotides 558-779; accession no. U35451; 5′ primer GAAAGCTGGCGGGCACTAT and 3′ primer GAGCGTTAGTTCTTGTCATC); (3) a 326-bp fragment corresponding to the untranslated exon 7 of the 3′ UTR of the murine M31 gene (nucleotides 68-393; accession no. X95397; 5′ primer TGTCTTGACACCATAGAGGT and 3′ primer CTACACACATGCTAGGCTGT); (4) a 268-bp fragment of the murineM32 gene located downstream of the chromodomain (nucleotides 252-519; accession no. X56683; 5′ primer ATCTGACAGTGAATCTGAT and 3′ primer TTGTGCTTCATCT TCAGGAC); (5) a 322-bp fragment of the murine M33 gene located downstream of the chromodomain (nucleotides 1346-1667; accession no. X62537; 5′ primer AGCTGACTTGCAAGGCAACG and 3′ primer GACTCCTTCACGGTGACAGT); (6) a 329-bp fragment of the human BMI-1 gene located in the 3′ UTR (nucleotides 1938-2248; accession no. L13689; 5′ primer GATGAATTCGTCACTGTGAATAACGATTT and 3′ primer TCTAGATCTACAATCATTTCTGAATGCAT); (7) a 287-bp fragment of the humanMel-18 gene located downstream of the RING finger domain (nucleotides 876-1168; accession no. D13969; 5′ primer CAAGTACCGTGTCCAGCCAG and 3′ primer TCTGCAGGCAGTTCAAGCTA); (8) a 228-bp fragment of the human ENX-1 gene located downstream to the SET domain (nucleotides 2338-2565; accession no. U52965; 5′ primer CTGAAGTATGTCGGCATCGA and 3′ primer ACACTTTGCAGCTGGTGAGA); and (9) a 251-bp fragment of the murine Mph1/Rae-28 gene located in the 3′ UTR (nucleotides 3190-3440; accession no.U63386; 5′ primer GTGCTACATGGTGACAGCTT and 3′ primer AGCTAGGAAAGCTGACCTCT). Probes for HP1Hsα,HP1Hsβ, and ENX-1 were isolated from cDNA obtained from CD34+ human bone marrow cells; M31and M32 were obtained from a pool of total RNA extracted from the Ba/F3, 32D, FEL, and FDC-P1 murine hematopoietic cell lines;M33 and Mph1/Rae-28 were obtained from murine genomic DNA; and BMI-1 and Mel-18 were obtained from human genomic DNA isolated from K562 cells. Probe for the full-length humanHP1Hsγcoding cDNA (519 bp) was isolated as aBamHI/EcoRI fragment of pGEX-2T (kindly provided by Dr H.J. Worman, College of Physicians & Surgeons of Columbia University, New York, NY). Probes for β-actin, humanCD34, Multi-Drug Resistance(MDR), andβ-globin were isolated as described.47
After database searches, probes were designed to minimize any cross-hybridization between the various Pc-G genes and other related sequences. To determine whether these probes recognized single copy gene, Southern blot analysis was performed on genomic DNA isolated from mouse thymus and human leukemic cell lines (K562, HL-60) using the same hybridization conditions as described above. A single band was detected with the following probes: M31, M33,Mel-18, BMI-1, Mph1/Rae-28, and ENX-1. However,M32 and HP1Hsγ hybridized to 5 to 10 different DNA fragments digested with either of the following restriction enzymes: EcoRI, HindIII, Bgl II,Kpn I, BamHI, and Xho I. The probe used to detect HP1Hsα also showed a single band. However, by using a different probe, it has been shown that this gene is part of a larger family18 and highly similar sequences toHP1Hsα are found in different EST databases.
Cell lines.
Hematopoietic cell lines used in this study were obtained from the American Type Culture Collection (ATCC; Rockville, MD), unless specified otherwise. They included the HL-60 cells derived from a patient suffering from acute myeloid leukemia, the K562 cells obtained from pleural effusion of a patient suffering from blast phase chronic myelogeneous leukemia, the MOLT-4 cells established from the peripheral blood of a patient suffering from acute T-cell lymphoblastic leukemia, the KG-1a cells obtained from a patient suffering from acute myeloid leukemia, the TF-1 human erythroleukemic cell line, the 32D murine mast cell line, the FDC-P1 myeloid cell line derived from long-term bone marrow cultures of DBA-2 mice, the Ba/F3 murine cell line (a gift from A. Miyajima, DNAX Research Institute, Palo Alto, CA), the FEL-745 Friend murine erythroleukemic cell line, and the murine Rat-1 fibroblasts. All cell lines were maintained in RPMI 10% FCS (GIBCO/BRL) except for FDC-P1, 32D, and Ba/F3, which grow in the presence of 5 ng/mL of mIL-3 and TF-1 cells that were maintained in the presence of 5 ng/mL of human granulocyte-macrophage colony-stimulating factor (hGM-CSF). All growth factors were used as diluted COS-cell supernatants produced at Institut de Recherches Cliniques de Montréal (IRCM).
Northern blot analysis.
Total cellular RNA from 1 × 107 cells of the murine FEL, FDC-P1, 32D, and Ba/F3 and human K562, HL-60, TF-1, MOLT-4, and KG-1a hematopoietic cell lines was isolated with TRIzol (Life Technology). Approximately 5 μg of each sample was size fractionated by electrophoresis on a 1% agarose gel containing 1× Na-MOPS and 5% deionized formaldehyde and transferred to Zeta-probe nylon membranes. Blots were prehybridized and hybridized at 45°C in 48% deionized formamide, 4.8% SDS, 480 mmol/L phosphate buffer (Na2HPO4/NaH2PO4, pH 7), 960 μg/mL of nuclease-free bovine serum albumin, and 400 μg/mL of salmon sperm DNA. Membranes were washed twice in 0.2× SSC, 0.1% SDS and once in 0.1× SSC, 0.1% SDS for 30 minutes at 55°C. Total RNA loading was verified by hybridization of membranes with radiolabeled oligonucleotides (ACG-GTA-TCT-GAT-CGT-CTT-CGA-ACC) specific to 18S ribosomal RNA.
RESULTS
Expression of known and novel Pc genes in CD34+ human bone marrow cells.
To obtain an initial indication of the range and expression pattern of known and potentially novel Pc genes that might be expressed in human bone marrow cells, a set of degenerate oligonucleotides spanning a region of the conserved chromodomain of most Pc genes was synthesized (Table 1) and used to PCR-amplify cDNA isolated from a CD34+ human bone marrow cDNA library. A PCR fragment of 115 bp presumably containing several different chromobox sequences was subcloned and the nucleotide sequences of 73 independent clones was compared with known Pc-G genes.
The results from this analysis showed that at least 8 different Pcgenes are expressed in human bone marrow cells (Table 2). The HP1Hsαand HP1Hsγ genes were most represented, with 31 (42%) and 15 (21%) of the 73 clones corresponding to these 2 genes. Three Pc subfamily members having a chromobox highly similar to that of HP1Hsα were identified in 2 independent PCR reactions, suggesting that they represent novelPc genes. They were named HP1Hsα-like A, B, and C. DNA and protein sequence comparison of these clones toHP1Hsα is shown in Table 2.
Pc Gene Sequences Obtained From cDNA Isolated From Purified CD34+ Human Bone Marrow Cells With Degenerate Oligonucleotides Designed Based on Conserved Sequences ofPcGenes
| Pc Genes . | cDNA Sequences Between Degenerate Oligos (and Translation) . | No. of Clones* Sequenced . | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R | V | V | K | G | Q | V | E | Y | L | L | K | W | K | G | F | S | E | E | H | |||
| 1. HP1Hsα | G | CGC | GTG | GTT | AAG | GGA | CAA | GTG | GAA | TAT | CTA | CTG | AAG | TGG | AAA | GGC | TTT | TCT | GAG | GAG | CAC | 31† |
| H | ||||||||||||||||||||||
| 2. HP1Hsα-like A | . | .AT | ... | ... | ... | ..G | ... | ... | ..G | ... | ..G | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | 3 |
| E | ||||||||||||||||||||||
| 3. HP1Hsα-like B | . | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | G.. | ... | ... | ... | ... | ... | ... | ... | ... | 4† |
| F | ||||||||||||||||||||||
| 4. HP1Hsα-like C | . | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | .T. | ... | ... | ... | 3† |
| R | V | V | N | G | K | V | E | Y | F | L | K | W | K | G | F | T | D | A | D | |||
| 5. HP1Hsγ | A | CGT | GTA | GTG | AAT | GGG | AAA | GTG | GAA | TAT | TTC | CTG | AAG | TGG | AAG | GGA | TTT | ACA | GAT | GCT | GAC | 15† |
| R | V | V | K | G | K | V | E | Y | L | L | K | W | K | G | F | S | D | E | D | |||
| 6. p25 | T | CGA | GTG | GTA | AAG | GGC | AAA | GTG | GAG | TAC | CTC | CTA | AAG | TGG | AAG | GGA | TTC | TCA | GAT | GAG | GAC | 3 |
| D | E | |||||||||||||||||||||
| 7. p25-like | . | ... | ... | ... | ... | .A. | ... | ... | ... | ... | ... | ... | ... | ... | ... | .A. | ... | ... | ... | ... | ..T | 1 |
| R | V | V | N | G | K | V | E | Y | F | L | K | W | K | G | F | T | D | A | D | |||
| 8. M32 (homologue) | T | CAT | GTA | GTG | AAT | GGG | AAG | GTG | GAG | TAT | TTC | CTG | AAG | TGG | AAG | GGG | TTC | ACA | GAT | GCT | GAT | 2† |
| 9. Several HP1Hsα-like with a single mutation (considered as potential PCR-induced mutations) | 6 | |||||||||||||||||||||
| 10. One HP1Hsγ-like with a single mutation | 1 | |||||||||||||||||||||
| Pc Genes . | cDNA Sequences Between Degenerate Oligos (and Translation) . | No. of Clones* Sequenced . | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R | V | V | K | G | Q | V | E | Y | L | L | K | W | K | G | F | S | E | E | H | |||
| 1. HP1Hsα | G | CGC | GTG | GTT | AAG | GGA | CAA | GTG | GAA | TAT | CTA | CTG | AAG | TGG | AAA | GGC | TTT | TCT | GAG | GAG | CAC | 31† |
| H | ||||||||||||||||||||||
| 2. HP1Hsα-like A | . | .AT | ... | ... | ... | ..G | ... | ... | ..G | ... | ..G | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | 3 |
| E | ||||||||||||||||||||||
| 3. HP1Hsα-like B | . | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | G.. | ... | ... | ... | ... | ... | ... | ... | ... | 4† |
| F | ||||||||||||||||||||||
| 4. HP1Hsα-like C | . | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | .T. | ... | ... | ... | 3† |
| R | V | V | N | G | K | V | E | Y | F | L | K | W | K | G | F | T | D | A | D | |||
| 5. HP1Hsγ | A | CGT | GTA | GTG | AAT | GGG | AAA | GTG | GAA | TAT | TTC | CTG | AAG | TGG | AAG | GGA | TTT | ACA | GAT | GCT | GAC | 15† |
| R | V | V | K | G | K | V | E | Y | L | L | K | W | K | G | F | S | D | E | D | |||
| 6. p25 | T | CGA | GTG | GTA | AAG | GGC | AAA | GTG | GAG | TAC | CTC | CTA | AAG | TGG | AAG | GGA | TTC | TCA | GAT | GAG | GAC | 3 |
| D | E | |||||||||||||||||||||
| 7. p25-like | . | ... | ... | ... | ... | .A. | ... | ... | ... | ... | ... | ... | ... | ... | ... | .A. | ... | ... | ... | ... | ..T | 1 |
| R | V | V | N | G | K | V | E | Y | F | L | K | W | K | G | F | T | D | A | D | |||
| 8. M32 (homologue) | T | CAT | GTA | GTG | AAT | GGG | AAG | GTG | GAG | TAT | TTC | CTG | AAG | TGG | AAG | GGG | TTC | ACA | GAT | GCT | GAT | 2† |
| 9. Several HP1Hsα-like with a single mutation (considered as potential PCR-induced mutations) | 6 | |||||||||||||||||||||
| 10. One HP1Hsγ-like with a single mutation | 1 | |||||||||||||||||||||
*Four additional clones were unrelated to Pc-G genes.
Sequences obtained from 2 independent PCR reactions.
Three additional sequences were identical to the humanheterochromatin p25 protein and one clone, referred to asp25-like, was most similar to p25 but contained 3 mismatches (Table 2). Two clones, each obtained from independent PCR reactions, had a single mismatch to the murine M32 gene and likely represent its human homologue. Finally, seven additional sequences containing a single nucleotide mismatch to eitherHP1Hsα or HP1Hsγ were identified. It is impossible at this point to ascertain whether any of these represent distinct genes or reflect PCR artifacts.
Together, the data obtained with this approach show the presence of at least 8 different Pc genes in human bone marrow cells, 4 of which are potentially novel genes, and one is the human homologue of the murine M32 gene.
Quantitative analysis of Pc-G gene expression in functionally distinct subpopulations of human bone marrow cells.
We next assessed the variation in expression levels of Pc-Ggenes during differentiation of bone marrow cells. This was performed by generating PCR-amplified total cDNA from functionally and phenotypically distinct FACS-purified human bone marrow subpopulations, using antibodies directed against CD34, CD45RA, and CD71 surface antigens (Fig 1). In total, 5 different subpopulations were purified from 3 healthy bone marrow donors: (1) total CD34+ cells, which phenotypically represent 1% to 5% of bone marrow cells and contain all types of progenitors; (2) CD34+CD45−CD71− cells (subpopulation I), which are highly enriched for very primitive long-term culture-initiating cells (LTC-IC); (3) CD34+CD45−CD71hi, which are enriched in erythroid (ie, burst-forming unit-erythroid [BFU-E]) clonogenic progenitors (subpopulation IIIE); (4) CD34+CD45+CD71lo cells, which are highly enriched in granulocyte-macrophage progenitors (colony-forming unit–granulocyte-macrophage [CFU-GM]; subpopulation IIM); and (5) total CD34− subpopulation, which contains mature bone marrow cells with no progenitor activity. Detailed functional characterization of each of these purified populations has been described elsewhere.47 52
FACS profiles of the CD34+ subpopulations isolated from donor no. 1: subpopulation I, CD34+CD45RA−CD71− (highly enriched in LTC-IC); subpopulation IIM, CD34+CD45RA+CD71lo (highly enriched in CFU-GM); subpopulation IIIE, CD34+CD45RA−CD71hi (highly enriched in BFU-E ). FITC, fluorescein isothiocyanate; PE, phycoerythrin.
FACS profiles of the CD34+ subpopulations isolated from donor no. 1: subpopulation I, CD34+CD45RA−CD71− (highly enriched in LTC-IC); subpopulation IIM, CD34+CD45RA+CD71lo (highly enriched in CFU-GM); subpopulation IIIE, CD34+CD45RA−CD71hi (highly enriched in BFU-E ). FITC, fluorescein isothiocyanate; PE, phycoerythrin.
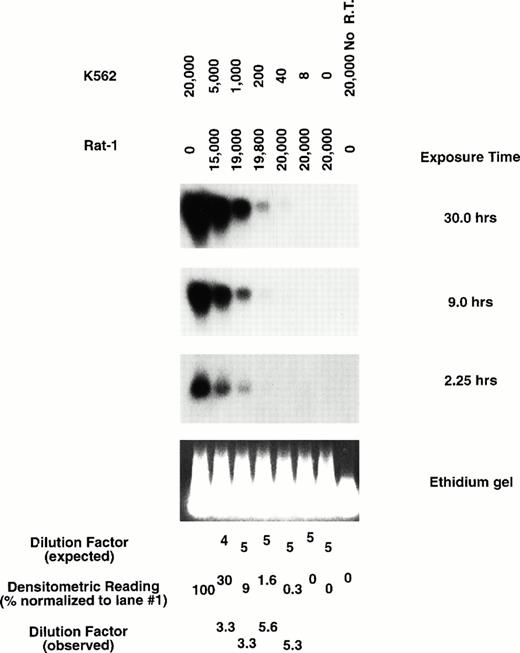
Total cDNA isolated from each purified subpopulation was PCR-amplified using a method previously shown to preserve quantitative differences in mRNA abundance using limited cell numbers.47 53 To assess both the sensitivity and the ability of this procedure to measure quantitative differences of Pc-G messages in hematopoietic cells, various numbers of the human K562 cells, which express HP1Hsγ, were mixed with Rat-1 fibroblasts that do not express this gene. Total cDNA amplified from each cellular preparation was blotted and hybridized with a probe specific for HP1Hsγ and showed linearity of expression in a range between 40 and 20,000 K562 cells (Fig2).
Representative amplification of mRNA by quantitative RT-PCR. Expression of HP1Hsγ in total amplified cDNA obtained from various cellular preparations of K562 (which express HP1Hsγ) and Rat-1 cells (HP1Hsγ negative) by Southern blot analysis. Exposure times are as indicated.
Representative amplification of mRNA by quantitative RT-PCR. Expression of HP1Hsγ in total amplified cDNA obtained from various cellular preparations of K562 (which express HP1Hsγ) and Rat-1 cells (HP1Hsγ negative) by Southern blot analysis. Exposure times are as indicated.
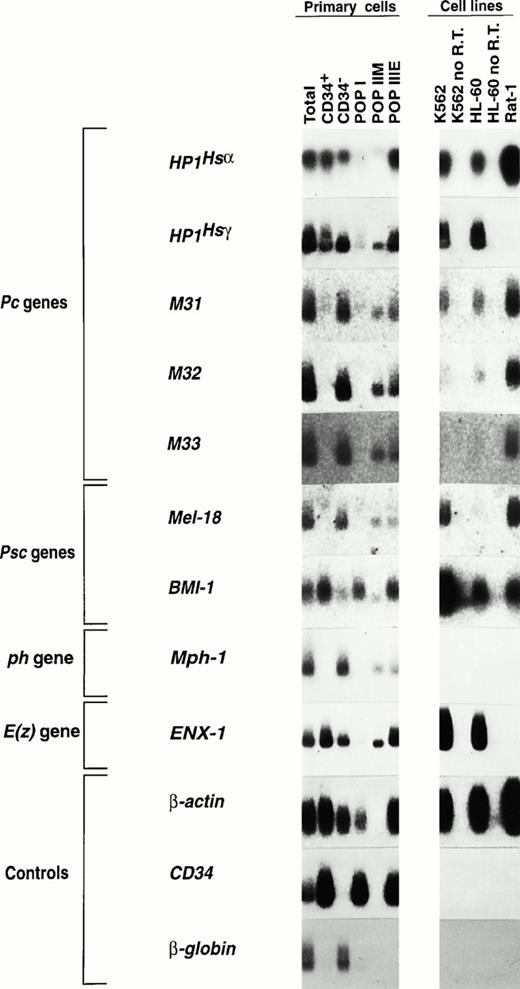
Total amplified cDNA from each of the 5 purified subpopulations obtained from one bone marrow donor was analyzed for the expression of the majority of mammalian Pc, Psc, ph, and E(z) genes characterized to date (Fig 3). Probes were carefully designed to minimize the presence of conserved or repetitive domains within the various Pc-G genes. All Pc-G genes analyzed to date are expressed at some levels in at least one of the purified subpopulations shown in Fig 3. Interestingly, and in contrast to Hox gene expression, which is predominantly observed in primitive subpopulations of CD34+ human bone marrow cells,47 the expression of most Pc-G genes is higher in cells lacking the CD34 surface antigen (compare the signals shown in lane 3 [CD34− cells] with those in lane 2 [CD34+]; Fig 3). The detailed analysis of these results suggests a progressive increase in Pc-G gene expression with bone marrow cell differentiation (Fig 3). Some important variations in expression levels were observed within each subfamily. For thePc subfamily, M31, M32, and M33 had a similar pattern of expression, with very high expression levels in CD34− cells, moderate levels in subpopulations IIM and IIIE, and little (M31) or no detectable expression (M32, M33) in the most primitive subpopulation I. The expression pattern of HP1Hsα differed significantly from the other Pc genes with similar abundance in CD34+ and CD34− cells (Fig 3). Expression also differed between the 2 known members of the Psc familyBMI-1 and Mel-18. Whereas Mel-18 expression was most prominent in mature cells (CD34− and subpopulation IIM), that of BMI-1 was highest in the most primitive subpopulation and minimal in CD34− cells. The expression of ENX-1 [a mammalian E(z) gene] was similar to that of HP1Hsα and the expression ofMph1/Rae28 (a ph gene) paralleled that ofMel-18. Interestingly, Mph1/Rae28 expression was not observed in the 3 cell lines shown in Fig 3, whereas Mel-18expression was detected only in human K562 and murine Rat-1 cells.
Expression of mammalian Pc-G genes in purified bone marrow CD34+ subpopulations. Five to ten thousand cells were isolated from each subpopulation (>98% purity upon reanalysis) and their total RNA was reverse-transcribed and PCR-amplified as described in the Materials and Methods. From primitive to mature subpopulations: subpopulation I, CD34+CD45RA−CD71−; subpopulation IIM, CD34+CD45RA+CD71lo; subpopulation IIIE, CD34+CD45RA−CD71hi and CD34− cells. Exposure times (all at −70°C unless specified) were as follows: HP1Hsα, 6.5 hours at room temperature; HP1Hsγ, 1.2 hours; M31, 6 days; M32, 27 hours; M33, 26 hours; Mel-18, 24 hours; BMI-1, 8 days; Mph-1/Rae-28, 3 days;ENX-1, 4.5 hours; β-globin, 5 minutes; and β-actin andCD34, 5 hours each.
Expression of mammalian Pc-G genes in purified bone marrow CD34+ subpopulations. Five to ten thousand cells were isolated from each subpopulation (>98% purity upon reanalysis) and their total RNA was reverse-transcribed and PCR-amplified as described in the Materials and Methods. From primitive to mature subpopulations: subpopulation I, CD34+CD45RA−CD71−; subpopulation IIM, CD34+CD45RA+CD71lo; subpopulation IIIE, CD34+CD45RA−CD71hi and CD34− cells. Exposure times (all at −70°C unless specified) were as follows: HP1Hsα, 6.5 hours at room temperature; HP1Hsγ, 1.2 hours; M31, 6 days; M32, 27 hours; M33, 26 hours; Mel-18, 24 hours; BMI-1, 8 days; Mph-1/Rae-28, 3 days;ENX-1, 4.5 hours; β-globin, 5 minutes; and β-actin andCD34, 5 hours each.
Several control probes were hybridized to the membranes containing the amplified cDNA obtained from each subpopulation. Actin showed comparable loading in each of the 9 lanes, except for lane 4, which was moderately underloaded, and lane 5 (subpopulation IIM), in which a clear distinct signal could be detected only upon prolonged exposure (not shown). Expression of CD34 in these subpopulations correlated with the expression of this antigen as determined by FACS analysis, with a progressive decrease in expression levels from subpopulation I to IIM and IIIE (data not shown and Sauvageau et al47). Expression of CD34 could not be detected in the CD34− cDNA even upon prolonged exposure (Fig 3). As might be expected, β-globin expression was only detected in populations containing mature red blood cell precursors (total unseparated bone marrow and CD34− cells). Finally, consistent with the findings of our previous studies,47 the expression of the multidrug resistance gene (MDR-1) was highest in subpopulation I and was also abundant in K562 cells (data not shown).
The reproducibility of these data was examined in identical bone marrow subpopulations isolated from 2 additional donors. Probes forHP1Hsα, HP1Hsγ, andENX-1 were hybridized to membranes containing total amplified cDNA, and the results from these experiments were superimposable to those shown in Fig 3 (data not shown). Together, these data highlight the complexity of the regulation of Pc-G gene expression during differentiation of human bone marrow cells and suggest the existence of diverse Pc-G protein complexes during hematopoietic cell differentiation.
Several Pc-G genes are expressed as multiple alternative transcripts in primary cells and in leukemic cell lines.
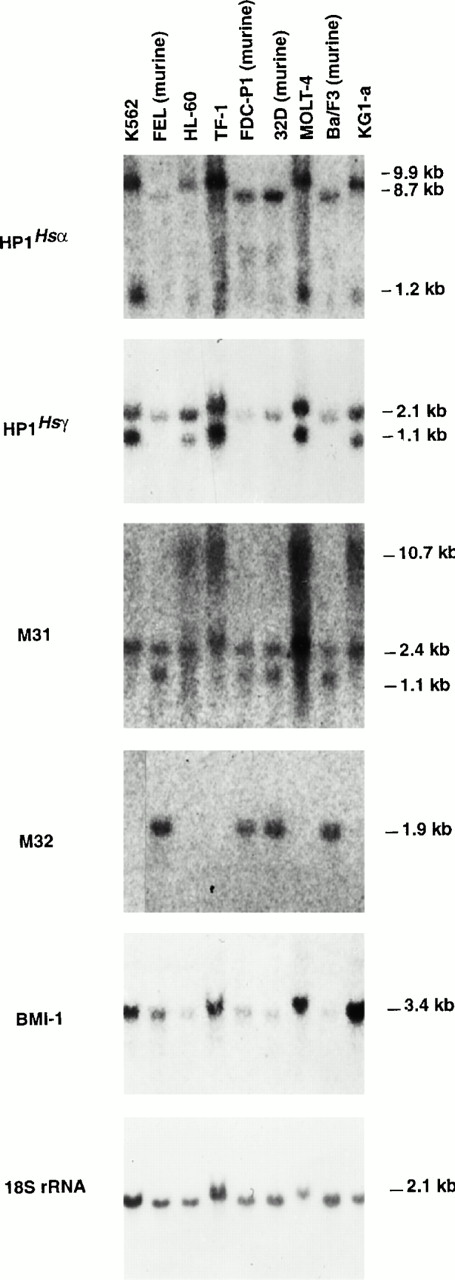
Previous studies have shown that one of the hallmarks of Pc-Ggenes is their expression as several alternative transcripts. To evaluate whether hematopoietic cells also express multiple Pc-Ggene transcripts, total RNA isolated from 5 different human (K562, HL-60, MOLT-4, TF-1, and KG1-a) and 4 murine (FEL, FDC-P1, 32D, and Ba/F3) hematopoietic cell lines, representing different lineages and stages of differentiation, was assessed for Pc-G gene expression by Northern blot (Fig 4).
Northern blot analysis showing the expression of selected members of the Pc-G family in human and murine hematopoietic cell lines. Five micrograms of total RNA isolated from each cell line was hybridized to probes specific to HP1Hsα (14 hours of exposure), HP1Hsγ (5.5 hours),M31 (96 hours), M32 (6 days), BMI-1 (14 hours), and 18S rRNA (4 minutes).
Northern blot analysis showing the expression of selected members of the Pc-G family in human and murine hematopoietic cell lines. Five micrograms of total RNA isolated from each cell line was hybridized to probes specific to HP1Hsα (14 hours of exposure), HP1Hsγ (5.5 hours),M31 (96 hours), M32 (6 days), BMI-1 (14 hours), and 18S rRNA (4 minutes).
This analysis showed the presence of multiple transcripts for 3 of the 5 Pc-G genes examined. These includeHP1Hsα, which had a human and a mouse-specific transcript of 9.9 and 8.7 kb, respectively, and shared a transcript of 1.2 kb in all cell lines examined (Fig 4). Different transcripts of theM31 gene were detected in human versus murine cell lines, the former expressing transcripts of 10.7 and 2.4 kb and the latter expressing transcripts of 2.4 and 1.1 kb. The probe forHP1Hsγ also detected 2 distinct species (2.1 and 1.1 kb) in human cell lines, whereas murine cells only expressed a transcript of 2.1 kb.
Two of the 5 genes examined expressed a single transcript. These include the M32 gene, whose expression could only be detected in cell lines of murine origin (Fig 4). The lack of expression ofM32 in human cell lines contrasts with its relative abundance in primary bone marrow cells (Fig 3), suggesting either the presence of different transcripts specifically recognized by our probe in primary cells or its complete absence in the immortalized lines examined here. Similarly, M33 and Mph1/Rae-28 expression could not be detected in any hematopoietic cell line examined, although both were easily detectable in the purified subpopulations shown in Fig 3. Finally, although Mel-18 expression could not be detected by Northern blot analysis of total RNA (data not shown), RT-PCR analysis showed its presence at low levels in K562 but not in HL-60 cells (Fig3). These results were confirmed using polyA mRNA isolated from the same cell lines. Two different transcripts for Mel-18 (1.8 and 3.4 kb) were detected in all the cell lines examined, except in HL-60 cells (data not shown).
To test whether the transcripts identified in the human cell lines reflect those present in primary isolates, mononuclear cells were isolated from 2 different human bone marrow specimens and total RNA hybridized to probes specific for the HP1Hsα andHP1Hsγ genes. In both cases, the transcripts observed in primary cells corresponded to those found in human cell lines, except for the 1.2-kb transcript of HP1Hsαwhose presence in primary cells remains unclear (data not shown). Together, these data suggest that alternative transcription patterns for the majority of Pc-G genes represent an additional level for regulation of the Pc-G gene action during hematopoietic cell differentiation.
DISCUSSION
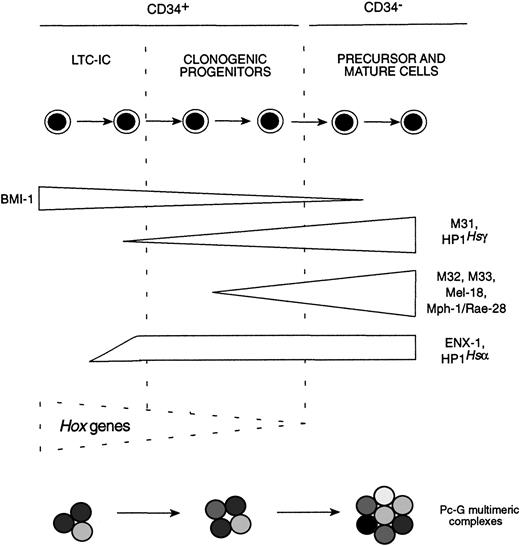
These studies report the expression of at least 13 differentPc-G genes in human bone marrow cells, including 4 potentially novel homologues of the DrosophilaPc gene. Moreover, our data suggest the existence of a highly defined program of Pc-Ggene expression in phenotypically distinct subpopulations of human bone marrow cells representing various stages of differentiation. In contrast to the preferential expression of Hox genes in the early hematopoietic cells,47 our study showed that the expression levels of 8 of the 9 Pc-G genes studied is much higher in the more mature bone marrow cells than in the primitive subpopulations. For some of the Pc-G genes, such asENX-1, M31, HP1Hsα, andHP1Hsγ, this upregulation seems to appear in the earliest stages of hematopoietic differentiation, whereas for others (ie, M32, M33, Mel-18, and Mph1/Rae-28), increase in their expression levels coincides with later stages of differentiation (summarized in Fig 5). This suggests that Pc-G protein complexes present in primitive hematopoietic cells (ie, population I) differ from those found in mature bone marrow cells. These results document, for the first time, changes in Pc-Ggene expression levels with cellular differentiation. This contrasts with their ubiquitous expression in Drosophilasyncytial blastoderms24 and thus point to an additional level for regulating Pc-G gene functions in mammalian cells.
Summary of Pc-G gene expression patterns observed in different purified subpopulations of human bone marrow cells.
Summary of Pc-G gene expression patterns observed in different purified subpopulations of human bone marrow cells.
Using degenerate primers, 4 novel chromobox sequences similar to that of the previously described Pc genes (referred to asHP1Hsα-like A, B, and C andp25-like) were identified. In most cases, these sequences were obtained in 2 independent PCR reactions, suggesting that they may represent novel human Pc members. Because the bone marrow used for this part of the work originated from a single donor, it is impossible to rule out that one of these sequences (eg, clone no. 4, Table 2) represents polymorphism. However, this is unlikely, because each novel sequence contains at least one mutation that affects the primary sequence of the highly conserved chromodomain. In support of the existence of several uncharacterized HP1Hsαmembers, several HP1Hsα sequences have been found in EST databases. Hybridization of mouse and human genomic DNA with a probe for HP1Hsα have also shown several bands.18 Importantly, the otherHP1Hsα-related sequences identified by Saunders et al18 were not detected by the probe used in our studies. Together, these data indicate that our knowledge of the full complement of Pc-G genes expressed in hematopoietic cells has not been resolved yet.
The ability of each probe to hybridize to specific sequences is shown in Fig 3 by the absence of a signal in at least one of the cell line controls (HP1Hsγ, M32, M33,Mel-18, Mph1/Rae-28, and ENX-1) or one of the purified subpopulations analyzed (HP1Hsαand M31). Except for M32 andHP1Hsγ, all Pc-G gene probes used in these studies only detected one DNA fragment, as shown by Southern blot analysis of human and mouse genomic DNA (data not shown). Therefore, M32 and HP1Hsγ probes possibly cross-hybridized with other Pc genes whose expression would also be prominent in mature bone marrow cells (Fig 3), further supporting our observation that Pc gene expression is higher in more mature bone marrow cells.
Further insight into Pc-G gene expression was provided by Northern blot analysis, which showed multiple Pc-G gene signals in several human and murine hematopoietic cells. Because our probe for M32 and HP1Hsγ may recognize more than one gene, some of the signals detected by Northern blot analysis may be derived from related genes. However, the signals observed with the probes specific for HP1Hsα, M31, andMel-18 represent alternative transcripts. It would be interesting to investigate whether some of these transcripts are hematopoietic-specific, lineage-specific, or encode proteins with altered function (eg, dominant negative, etc). Interestingly, the low to undetectable expression levels of the M32, M33, andMph1/Rae-28 genes in all cell lines (Figs 3 and 4) contrast with their relatively high expression levels in differentiated primary hematopoietic cells (Fig 3), raising the possibility that these proteins normally perform antiproliferative functions in mature bone marrow cells. Indeed, a correlation can be made between the expression pattern of BMI-1 and Mel-18 (the 2 known mammalianPsc genes) and their ability to control cellular proliferation.BMI-1, a known proto-oncogene, is preferentially expressed in bone marrow cells displaying a high proliferative potential (ie, subpopulation I; Fig 3), whereas Mel-18, a gene recently shown to exhibit tumor suppressive activity,54 is only expressed in mature and nonproliferating CD34− cells.
Together, our results show a progressive upregulation of mostPc-G genes concomitant with differentiation of human bone marrow cells and support a complex-constitution model. In this model, newly expressed Pc-G gene products would progressively interact with existing Pc-G protein complexes, favoring novel interactions with target sequences. This, in turn, would allow a progressive packaging of DNA into an heterochromatin-like structure and, for the Hox genes, a progressive 3′ to 5′ closure of the clusters, allowing proper differentiation of the hematopoietic stem cells. This is most interesting in the view thatHox gene expression decreases 3′ to 5′ during differentiation of hematopoietic cells47 and overexpression of Hox genes in hematopoietic cells profoundly alters their differentiation and proliferation.45,46 55
ACKNOWLEDGMENT
The authors gratefully acknowledge the expert technical assistance of Nathalie Tessier, head of the Flow Cytometry Service of IRCM for cell sorting, and of Dr Peter Lansdorp and Vishia Dragowska for providing the CD34, CD45RA, and CD71 conjugated antibodies used in this study. Dr Trang Hoang and Nathalie Bouchard are also acknowledged for providing frozen bone marrow cells. We also thank Dr David Lohnes, Dr Keith Humphries, and Dr Terry Magnuson for critically reviewing this manuscript.
Supported by grants from the National Cancer Institute, the Medical Research Council (MRC), and the Leukemia Research Fund of Canada. J.L. is a recipient of an MRC scholarship and G.S. is a Clinician-Scientist scholar of the MRC.
Address reprint requests to Guy Sauvageau, MD, PhD, Institut de Recherches Cliniques de Montréal, 110 Pine Ave W, Montréal, Québec, Canada H2W 1R7.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal