Abstract
The nonspecific cross-reacting antigen-95 (NCA-95/CD66b), is a member of the human carcinoembryonic antigen (CEA) family encoded by the CGM6 gene that is exclusively expressed in neutrophils and eosinophils. No murine counterpart is known to exist. We have analyzed a cosmid containing the complete CGM6 gene. The coding sequence is contained within six exons spanning a 16.5 kb region. The main transcriptional start site was mapped to a tight cluster between nucleotides -95 and -101 relative to the translational start site. As with other members of the CEA gene family, no typical TATA or CAAT-box sequences were found in the CGM6 gene. Transgenic mice were established with the cosmid insert. CD66b expression is first seen in the fetal liver on day 12.5 of mouse embryonic development, and it first appears in the bone marrow at day 17.5. Northern blot analysis showed that CD66b transcripts are confined to the bone marrow of adult mice, whereas immunohistochemistry also showed CD66b-positive granulocytes in the spleen, thymus, and lungs. FACScan analyses of bone marrow and spleen cells showed CD66b expression to be exclusive to granulocytes. Thus, all the elements necessary for regulating granulocyte-specific expression are present within this cosmid clone. These mice could provide a model for transplantation and for inflammation studies using CD66b as a granulocyte-specific marker.
CD66b (FORMERLY CD67) HAS been characterized as a granulocyte-specific activation antigen.1 Using specific antibodies, CD66b has been located on the surface of neutrophilic and eosinophilic granulocytes at late stages of differentiation. To our knowledge, it is not known whether or not CD66b is expressed by basophils. In neutrophils, it is mainly found in the secondary granules within the cytoplasm, with lower amounts on the plasma membrane.2 On activation of granulocytes with various agents, the amount of CD66b on the plasma membrane is rapidly upregulated, presumably through release from the intracellular granules.3,4 By measuring stimulation of oxidative burst, granulocyte activation could be achieved through cross-linking CD66b surface molecules using F(ab′)2 fragments of CD66b-specific antibodies.5 Due to its granulocyte specificity, CD66b has clinical potential for locating regions of inflammation in patients with radiolabeled anti-CD66b antibodies.6 7
CD66b is synonymous with the nonspecific cross-reacting antigen-95 (NCA-95), a member of the carcinoembryonic antigen (CEA) family.8 The CEA gene family consists of 29 genes, of which 18 are transcriptionally active. The genes are divided into three main subgroups: the CEA subgroup, the pregnancy-specific glycoprotein (PSG) subgroup, and a subgroup of pseudogenes.9 Through molecular cloning, NCA-95 was found to be the product of the CGM6 gene, a member of the CEA subgroup.10,11 The derived protein structure of NCA-95 shows an immunoglobulin variable-like N-terminal domain, followed by two immunoglobulin constant-like domains. It is attached to the membrane through a glycosylphosphatidyl inositol (GPI) anchor. Some other members of the CEA family, ie, CGM1 (CD66d), biliary glycoprotein (BGP; CD66a), and NCA-50/90 (CD66c) are coexpressed with NCA-95 on granulocytes. Whereas CGM1 shows an identical expression pattern to NCA-95, BGP and NCA-50/90 are also present in other normal tissues, eg, in epithelial cells lining the gastrointestinal tract.12,13 Although its in vivo function is unknown, CD66b shows heterophilic cell adhesion properties with the closely-related CD66c molecule, which is coexpressed in granulocytes.14
Transcriptional regulation specific for the myeloid lineage has been well investigated,15-17 but less is known about specific transcriptional regulation in the granulocytic lineage, ie, after separation from the monocytic differentiation pathway. Due to its expression pattern, investigations on the regulation of expression of the CGM6 gene should help in a better understanding of gene regulation in later development of neutrophils and in eosinophils. For this reason, we are interested in locating the regions responsible for regulating the granulocyte-specific expression pattern of theCGM6 gene. As a basis for such investigations, we have characterized a cosmid clone containing the putative promoter region, as well as the complete coding sequence of the CGM6 gene, through restriction endonuclease and exon mapping. After determining the transcriptional start site, we have compared putative promoter sequences with corresponding regions from other members of the CEA gene family. Finally, we have established transgenic mice to determine whether all the cis-regulatory elements necessary for its tissue-specific expression pattern are present in this cosmid clone.
MATERIALS AND METHODS
Isolation and Characterization of a Cosmid Clone Containing the CGM6 Gene.
Cosmid clone F19632 was previously shown to contain the CGM6gene through sequencing a 1.3 kb BamHI fragment, which spans part of the first and second exons of this gene.18 A restriction endonuclease map of F19632 was constructed by first digesting it with the restriction endonuclease SfiI, which has two sites in the vector Lawrist 5 on either side of the multiple cloning site.19 The insert DNA was subjected to partial digestion with different restriction endonucleases. Partially digested fragments were hybridized on a Southern blot with oligonucleotides corresponding to sequences located to the right (5′-AACCTTATGTATCATACACATACG-3′) and left (5′-AATACGACTCACTATAG-3′) of the multiple cloning site, respectively.
The location of CGM6 gene exons within cosmid F19632 was established by hybridization of restriction endonuclease fragments with the following exon-specific oligonucleotides based on the sequence of a CGM6 cDNA clone.10 Oligonucleotide name, sequence: exon 1, 5′-GATGCGCCATCTGCAGGAAG-3′; exon 2a, 5′-GGCACAGCTTCAATAGTGAGCTGA-3′; exon 3a, 5′-ACTGAAGTTTGCACTCGCT-3′; exon 4a, 5′-ATGCGGCCTCTAATCCA-3′; exon 5a, 5′-GGAGTACTGGCCAGGGTG-3′; exon 6, 5′-ATCAGCCAATAGGGAGCAAT-3′.
Primers taken from the exons flanking the various introns were synthesized from published CGM6 cDNA sequences based on the knowledge that exon/domain structures are well conserved for all known members of the CEA gene family.8 10 The following primer combinations were used for polymerase chain reaction (PCR) amplification to determine intron sizes: intron 2, 5′-GGAATTCGCAGAGGGGAAGGAGGT-3′ (exon 2b) with the exon 3a primer; intron 3, 5′-AGGCTGCAGCTGTCCAA-3′ (exon 3b) with 5′-GTATTGCTGGAATGTGC-3′ (exon 4b); intron 4: exon 4a primer with 5′-ACAGTGGCTCTAGCTGAG-3′ (exon 5b).
It was not necessary to amplify intron 1 because of the known positions of the BamHI restriction endonuclease sites in exons 1 and 2. Various combinations of primers to amplify intron 5 were unsuccessful and the primers are therefore not listed. However, exon 6 could be mapped through an internal NarI restriction endonuclease site.
A 1.7-kb SstI fragment containing the putative promoter region, the first exon and part of the first intron was sequenced completely using a Pharmacia kit (Pharmacia, Freiburg, Germany).
Determination of the Transcriptional Start Site and Promoter Analyses of CGM6
To determine the transcriptional start site of the CGM6 gene, the exon 1-specific primer described above, which hybridizes with the CGM6 transcript at nucleotides +20 to +39 downstream from the translational start site in the leader region, was used for primer extension analyses. A total of 50 μg of total RNA, isolated from a chronic myeloid leukemia (CML) patient's leukocyte fraction as described previously, was used as a template.10The oligonucleotide was 5′-end labeled with [γ-32P] adenosine triphosphate (ATP) and the primer extension reaction was performed using avian myeloblastosis virus (AMV) reverse transcriptase in the presence of 2 U RNasin.20 To determine the exact location of the transcriptional start sites, a sequencing reaction was performed on the 1.7-kb SstI restriction endonuclease fragment from theCGM6 gene described above, applying the same primer as used for the primer extension analyses. Separation was on a 6% polyacrylamide sequencing gel. As negative controls, the primer extension reaction was performed with the same primer, but with 50 μg of total RNA from a human colon adenocarcinoma cell line (LoVo) and 50 μg of total RNA from a human cervix carcinoma cell line (HeLa), as templates. The extension products were analyzed on a 6% polyacrylamide sequencing gel and exposed for 96 hours to Kodak X-AR films (Siemens, Stuttgart, Germany). The promoter sequence was compared with other CEA gene family promoter sequences using the program Dot Matrix (Busch and Lucas, Freiburg, Germany).
Generation of CGM6 Transgenic Mice
Cosmid F19632 DNA was digested with SfiI and the 42-kb insert containing the complete CGM6 insert including 0.3 kb 5′- and 0.1 kb 3′-vector sequences was isolated from a 0.7% agarose gel by the freeze-squeeze method and used for microinjection into fertilized mouse oocytes derived from C57BL/6 × CB6 F1 mice (Ciba Animal Breeding Center, Basel, Switzerland) as described elsewhere.21,22 A mouse line was established from founder animals by backcross mating with C57BL/6 mice (Zentralinstitut für Versuchstierzucht, Hannover, Germany). Transgenic mice are routinely identified by PCR, using the CGM6-specific primers and PCR conditions essentially as described elsewhere, but with an annealing temperature of 58°C.23
Southern and Northern Blot Analyses
DNA was isolated from the thymuses of CGM6 transgenic and C57BL/6 mice. A total of 10 μg of each were digested withEcoRI, size-separated by gel electrophoresis, and blotted onto a positively charged nylon membrane. To estimate the number of theCGM6 transgene copies, C57BL/6 DNA was spiked with CGM6cosmid DNA, corresponding to 25 to 150 copies per haploid genome. After hybridization with [32P]-labeledEcoRI-digested F19632 cosmid DNA, the blot was washed twice in 0.1× SSPE (1× SSPE is 0.18 mol/L NaCl, 10 mmol/L sodium phosphate, pH 7.4, 1 mmol/L EDTA), 0.1% sodium dodecyl sulfate (SDS) for 30 minutes at 68°C. Autoradiography was performed for 48 hours.
Total RNA was isolated from mouse organs as described previously.24 Detection of CGM6 transcripts was achieved by standard Northern blot hybridization using a CGM6 cDNA probe.10,25 As an internal control of RNA intactness, a [32P]-labeled, mouse β-actin probe (1.2 kb PstI fragment from plasmid pAL41) was also hybridized.26 Final washing of the filters was performed at 65°C in 0.1× SSPE, 0.1% SDS.
Immunohistochemical Analyses
To determine tissue expression of CD66b by immunohistochemical methods, organs were isolated from adult transgenic mice, which had been anesthetized with Forene (Abbott GmbH, Wiesbaden, Germany) and killed by cervical dislocation. The organs were embedded and cryosectioned as described elsewhere.24 Rabbit anti-NCA antiserum was kindly provided by Dr Fritz Grunert (Institute of Immunobiology, Freiburg, Germany) and used at a 1:200 dilution in phosphate-buffered saline (PBS). This antiserum was developed against NCA-50/90 extracted from a breast tumor and was found to cross-react with other human CEA family members, including CD66b, but not with mouse CEA family members (F. Grunert, personal communication, May 1997). Binding of the primary antibody was followed by incubation with horseradish peroxidase-conjugated, goat antirabbit IgG at a dilution of 1:100 (Sigma-Aldrich, Deisenhofen, Germany). The staining reaction was performed using the substrate 3,3′-diaminobenzidine tetrahydrochloride. Sections were counterstained with hematoxylin. Staged nonfixed embryos were treated likewise. Earlier stage embryos (embryonic day 8.5) were sectioned in toto. Staged embryos were obtained from mating nontransgenic females overnight with transgenic males. Midday after mating was designated embryonic day 0.5.
FACScan Investigations
For identification of the cell population that express CD66b, three-color fluorescence FACScan analyses was performed on cells isolated from the bone marrow of mice transgenic for CGM6 as described.27 Erythrocytes were lysed by resuspending the isolated cell populations in ice-cold Gey's solution (130 mmol/L NH4Cl, 5 mmol/L KCl, 0.8 mmol/L Na2HPO4, 0.18 mmol/L KH2PO4, 5 mmol/L glucose, 10 mg/L phenol red, 0.1 mmol/L MgCl2, 0.03 mmol/L MgSO4, 0.1 mmol/L CaCl2, 13.5 mmol/L NaHCO3), which was immediately underlayered with fetal bovine serum and the intact cells were spun down and resuspended in fluorescence-activated cell sorting (FACS) buffer. The number of intact cells was determined in a hemacytometer after adding Trypan blue and equal cell numbers (2 to 5 × 105) were incubated with the labeled antibodies in concentrations ranging from 0.17 to 1.0 μg/mL as determined in titration experiments. The fluorescein isothiocyanate (FITC)-labeled monoclonal antibody (MoAb), antihuman CD66b (clone 80H3: Coulter-Immunotech, Hamburg, Germany) was used to identify theCGM6 gene product. The following phycoerythrin (PE)-conjugated MoAb was used to identify the main leukocyte populations: for B lymphocytes, antimouse B220, clone RA3-6B2; for T lymphocytes, antimouse Thy-1.2, clone 30-H12; for myeloid cells, antimouse GR-1, clone RBG-8CS. All were obtained from PharMingen (Hamburg, Germany). Single and double labelings were performed in FACS buffer (PBS containing 3% fetal bovine serum), whereby for the second incubation, FACS buffer without antibody was used for the single labelings to ensure identical conditions. Before FACScan analysis, 5 μg/mL propidium iodide (Fluka, Deisenhofen, Germany) was added to allow subtraction of dead cells.
RESULTS
Cosmid Clone F19632 Contains the Complete CGM6 Gene
Digestion of cosmid clone F19632 with SfiI resulted in a 5-kb vector fragment and a 42-kb fragment encompassing human genomic DNA flanked by 0.3 kb vector at the 5′ end and 0.1 kb vector 3′ to the CGM6 gene. A complete restriction endonuclease map, including the exon positions within this SfiI fragment is shown in Fig 1A. The CGM6 gene consists of six exons that span 16.5 kb. There is a strong correlation between the exon and derived protein domain borders for this gene.10 CGM6 is flanked by 13 kb of 5′- and approximately 12.5 kb of 3′-genomic sequences.
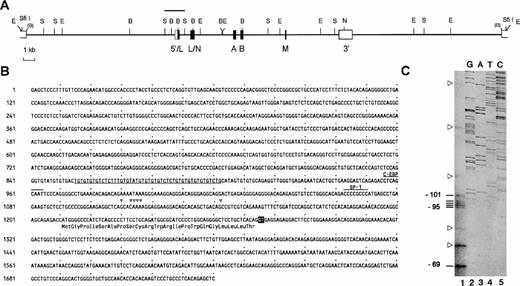
Exon structure of the human CGM6 gene, DNA sequence of the putative promoter and determination of the transcriptional start sites. (A) Restriction endonuclease map of the genomic insert from cosmid clone F19632, encompassing the CGM6gene, flanked by a pair of EcoRI/SfiI restriction endonuclease sites in the vector (S =SstI, E = EcoRI, B = BamHI, N =NarI). Boxes indicate the exons (L = leader, N = N-terminal domain, A and B = immunoglobulin constant-like domains, M = membrane domain). Open boxes correspond to the 5′- and 3′-untranslated regions. The bar above the first exon and flanking sequence designates the Sst I restriction endonuclease DNA fragment, whose sequence is shown in (B). The nucleotide sequence is numbered from the upstream SstI restriction endonuclease site (numbers at the left). The derived amino acid sequence from the leader exon is shown below the DNA sequence and the start of the intron 1 donor site (GT) is inversely shaded. Multiple transcriptional start sites are indicated as open triangles above the corresponding nucleotides. Further upstream, SP-1 and C-EBP concensus sequences are overlined and labeled. A simple repetitive DNA sequence is underlined. (C) Primer extension analysis to determine the transcriptional start site of the CGM6 gene (lane 1). Extension products only seen using CML leukocyte RNA are indicated by bars at the left and their nucleotide positions relative to the translational start site (+1) are included. Open triangles point out unspecific extension products that were also seen using RNA from a colon adenocarcinoma (LoVo) and a cervix carcinoma (HeLa) cell line as templates (data not shown). Lanes 2 to 5 show the DNA sequencing reaction with the 1.7-kbSstI restriction endonuclease DNA fragment as a template and using the same oligonucleotide primer as for the primer extension analyses. This allows direct identification of the transcriptional start sites.
Exon structure of the human CGM6 gene, DNA sequence of the putative promoter and determination of the transcriptional start sites. (A) Restriction endonuclease map of the genomic insert from cosmid clone F19632, encompassing the CGM6gene, flanked by a pair of EcoRI/SfiI restriction endonuclease sites in the vector (S =SstI, E = EcoRI, B = BamHI, N =NarI). Boxes indicate the exons (L = leader, N = N-terminal domain, A and B = immunoglobulin constant-like domains, M = membrane domain). Open boxes correspond to the 5′- and 3′-untranslated regions. The bar above the first exon and flanking sequence designates the Sst I restriction endonuclease DNA fragment, whose sequence is shown in (B). The nucleotide sequence is numbered from the upstream SstI restriction endonuclease site (numbers at the left). The derived amino acid sequence from the leader exon is shown below the DNA sequence and the start of the intron 1 donor site (GT) is inversely shaded. Multiple transcriptional start sites are indicated as open triangles above the corresponding nucleotides. Further upstream, SP-1 and C-EBP concensus sequences are overlined and labeled. A simple repetitive DNA sequence is underlined. (C) Primer extension analysis to determine the transcriptional start site of the CGM6 gene (lane 1). Extension products only seen using CML leukocyte RNA are indicated by bars at the left and their nucleotide positions relative to the translational start site (+1) are included. Open triangles point out unspecific extension products that were also seen using RNA from a colon adenocarcinoma (LoVo) and a cervix carcinoma (HeLa) cell line as templates (data not shown). Lanes 2 to 5 show the DNA sequencing reaction with the 1.7-kbSstI restriction endonuclease DNA fragment as a template and using the same oligonucleotide primer as for the primer extension analyses. This allows direct identification of the transcriptional start sites.
The CGM6 Gene Promoter Is Most Similar to PSG Gene Promoters
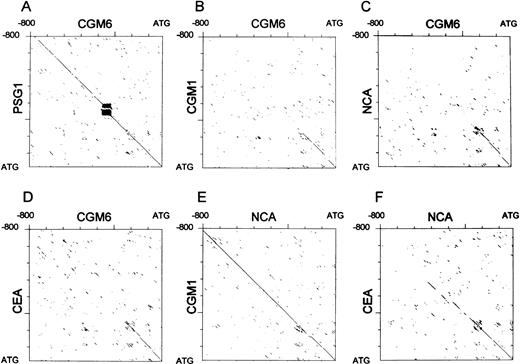
The transcriptional start site, as determined by primer extension experiments shows a tight cluster at nucleotide positions -95 to -101, with respect to the translational start (Fig 1B and C). An additional start site, not found in the control RNA samples was located at nucleotide -69, although this could also represent a strong stop site during the primer extension reaction, due to secondary structures in the template. Although the sequence reaction in Fig 1C is rather weak, additional experiments proved the exact positions of the transcriptional start sites. DNA sequencing of a 1,740-bp SstI fragment that contains the first exon and 1210 nucleotides upstream from the translational start site is shown in Fig 1B. Along with the transcriptional start cluster, a potential SP-1 binding element is marked at nucleotide positions -147 to -140 relative to the translational start site.28 A second element is indicated at nucleotide positions -255 to -247 relative to the translational start site, which shows a strong similarity to a C-EBP binding site.29 A repetitive simple sequence is found at nucleotides -356 to -309. As for other members of the CEA gene family,CGM6 lacks recognizable TATA or CAAT-box sequences at the expected positions. Sequence comparisons by dot matrix analyses have been performed between the promoter and 5′-upstream regions of various members of the CEA gene family. Surprisingly, the longest region of homology is found to the corresponding region in genes from the PSG subgroup (Fig 2A), which are mainly expressed in the syncitiotrophoblast of the placenta.13This region covers 800 nucleotides upstream from the translational start site. The repetitive simple sequence region found in theCGM6 gene promoter is also present in PSG gene promoters, which is visible in this alignment analysis (Fig 2A). In comparison with gene promoters from other members of the CEA subgroup, ie, CGM1 (Fig 2B), which like CGM6 is exclusively expressed in granulocytes, NCA (Fig 2C), which is expressed in granulocytes and epithelial cells, CEA (Fig 2D), whose expression is exclusive to certain epithelial cells and BGP(data not shown), that is expressed in granulocytes, epithelial cells and other tissues, this region of homology is limited in all cases to approximately 240 nucleotides upstream from the translational start site. Two of the CEA family members that are coexpressed in granulocytes (NCA and CGM1) are homologous to each other over the whole 800 nucleotide promoter region (Fig 2E), whereas the CEA and NCA gene promoters are less well conserved (Fig 2F).
Dot matrix analysis of the putative CGM6 gene promoter region and corresponding regions from other members of the human CEA gene family. Dot matrix sequence alignments were performed between the promoter regions of CGM6 and PSG1 (A), CGM1(B), NCA (C), and CEA (D); between NCA andCGM1 (E); and between NCA and CEA (F). In all cases, 800 nucleotides upstream from the translational start site (ATG) were compared and the stringency was set at 12 from 20 nucleotides conserved to register a dot.
Dot matrix analysis of the putative CGM6 gene promoter region and corresponding regions from other members of the human CEA gene family. Dot matrix sequence alignments were performed between the promoter regions of CGM6 and PSG1 (A), CGM1(B), NCA (C), and CEA (D); between NCA andCGM1 (E); and between NCA and CEA (F). In all cases, 800 nucleotides upstream from the translational start site (ATG) were compared and the stringency was set at 12 from 20 nucleotides conserved to register a dot.
Establishment of Mice Transgenic for the Human CGM6 Gene
To determine whether the CGM6 cosmid clone contains all the regulatory elements that are necessary for granulocyte-specific expression, the 42-kb genomic fragment was used to establish transgenic mice. From 16 founder mice that were born, only one was transgenic for the CGM6 gene, as determined by Southern blot analysis ofEcoRI-digested mouse tail DNA, hybridized with a radioactively-labeled EcoRI digest of the complete cosmid clone F19632. The CGM6 cosmid probe shows no cross-hybridization with C57BL/6 genomic mouse DNA (Fig 3, lane 1; see also Fig 1A). In the CGM6-positive strain, the transgene was found to contain all of the EcoRI fragments that were present in the original cosmid, apart from the 1.2-kb and the 3.75-kb vector fragments and the 3.2-kb and 4.3-kb end fragments (Fig 3, lane 2). However, a new 7.5-kb EcoRI fragment, resulting from the fusion of the 3.2-kb 5′-end EcoRI/SfiI fragment with the 4.3-kb, 3′-end EcoRI/SfiI fragment of the cosmid insert, is seen in the transgene, which comigrates with an internal EcoRI fragment (Fig 3, lane 2). This fusion fragment indicates the existence of multiple cosmid inserts in a tandem head-to-tail orientation. After inclusion of different cosmid copy numbers with C57BL/6 genomic DNA, we determined a total of 50 to 75 intact transgene copies to have become integrated in this mouse strain (Fig 3, lane 3 and titration data not shown).
Determination of the intactness and copy number of theCGM6 transgene. A total of 10 μg of thymus DNA each from a C57BL/6 mouse (lane 1) and from the CGM6-transgenic mouse 5322 (lane 2) was digested with EcoRI, size fractionated, and hybridized with [32P]-labeled F19632 cosmid DNA. To estimate the number of integrated transgene copies, F19632 DNA corresponding to 75 gene copies per haploid genome was added to 10 μg of C57BL/6 DNA before digestion with EcoRI (lane 3). Cosmid vector fragments and end fragments of the genomic insert of F19632 are indicated by arrowheads and arrows, respectively. The asterisk marks the band, which contains besides an internal EcoRI DNA fragment, the fusion fragment created by the head to tail insertion of the amplified transgene. The mobility and size in kb of the marker fragments is shown in the left margin.
Determination of the intactness and copy number of theCGM6 transgene. A total of 10 μg of thymus DNA each from a C57BL/6 mouse (lane 1) and from the CGM6-transgenic mouse 5322 (lane 2) was digested with EcoRI, size fractionated, and hybridized with [32P]-labeled F19632 cosmid DNA. To estimate the number of integrated transgene copies, F19632 DNA corresponding to 75 gene copies per haploid genome was added to 10 μg of C57BL/6 DNA before digestion with EcoRI (lane 3). Cosmid vector fragments and end fragments of the genomic insert of F19632 are indicated by arrowheads and arrows, respectively. The asterisk marks the band, which contains besides an internal EcoRI DNA fragment, the fusion fragment created by the head to tail insertion of the amplified transgene. The mobility and size in kb of the marker fragments is shown in the left margin.
CD66b Is Expressed Exclusively in Granulocytes of CGM6-Transgenic Mice
Northern blot analyses.
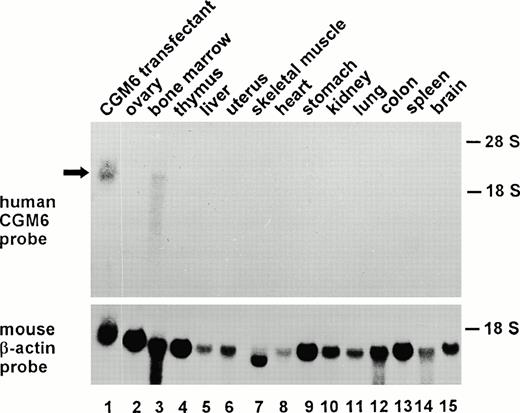
To investigate the presence of transcripts from the CGM6 gene, total RNA from 14 different organs of a CGM6 transgenic mouse was analyzed for the presence of CGM6 mRNA. A positive signal was only seen in RNA from bone marrow (Fig 4, lane 3). All other organs tested were negative for CGM6 mRNA. Although slightly degraded, the hybridizing mRNA in bone marrow shows a size that is similar to that reported elsewhere for human CGM6 transcripts.10 This mRNA was not present in the bone marrow of a nontransgenic sibling (data not shown). A positive signal was seen in all lanes after hybridization with a probe that detects mouse β-actin mRNA.
Northern blot analysis of human CGM6 mRNA in various organs from a CGM6-transgenic mouse. Total RNA from various organs of a CGM6-transgenic mouse and as a positive control RNA from a CGM6-HeLa transfectant10 were size-separated on a denaturing agarose gel. After transfer to a nylon membrane, they were hybridized with a CGM6 cDNA probe. The positions of the 28S and 18S rRNAs are indicated. In the lower part of the figure, the same blot was hybridized with a mouse β-actin cDNA probe.
Northern blot analysis of human CGM6 mRNA in various organs from a CGM6-transgenic mouse. Total RNA from various organs of a CGM6-transgenic mouse and as a positive control RNA from a CGM6-HeLa transfectant10 were size-separated on a denaturing agarose gel. After transfer to a nylon membrane, they were hybridized with a CGM6 cDNA probe. The positions of the 28S and 18S rRNAs are indicated. In the lower part of the figure, the same blot was hybridized with a mouse β-actin cDNA probe.
Immunohistochemical analyses.
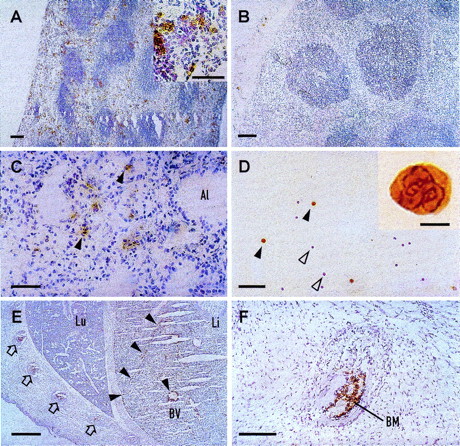
Immunohistochemical staining was performed on peripheral blood smears and 12 different organs (brain, tongue, stomach, small intestine, large intestine, spleen, thymus, lung, uterus, ovary, kidney, and liver) of adult CGM6-transgenic mice using a polyclonal rabbit anti-NCA serum that cross-reacts with CD66b. This antiserum was used because the CD66b-specific mouse MoAb 80H3, showed a high background staining on the mouse tissues (data not shown). It is obvious from Fig 5D that the cells staining positive in peripheral blood smears are polymorphonuclear and strongly resemble granulocytes, whereas the lymphocytes are negative. Positive staining was seen in the red pulp areas of the spleen (Fig 5A). No staining was seen in any tissues of nontransgenic sibling mice, shown in Fig 5B for the spleen, confirming that the anti-NCA serum used does not cross-react with any members of the mouse CEA family. The thymus (data not shown) and lungs (Fig 5C) of transgenic animals also showed labeled cells. All other organs, including the adult liver, were negative. The labeled cells in CGM6-transgenic mice often strongly resembled granulocytes in their morphology, although this could not be unequivocally determined in tissue sections.
Immunohistochemical detection of human CD66b expression in CGM6-transgenic mice. Cryosections from various organs were incubated with a polyclonal rabbit anti-NCA antiserum followed by a peroxidase-coupled, antirabbit IgG and stained for peroxidase activity (brown). Counterstaining of nuclei was with hematoxylin (blue). (A) The spleen from a CGM6-transgenic mouse shows staining of granulocytes (see inset) mainly in the red pulp areas. (B) The spleen from a nontransgenic sibling mouse is negative. The lung (C) and a blood smear (D) of a CGM6-transgenic mouse also show granulocyte staining (Al = alveolus). Black arrowheads indicate stained granulocytes (see inset in D) and white arrowheads are nonstaining lymphocytes. (E) Section through the lung (Lu) and liver (Li) of a day 17.5 CGM6-transgenic mouse fetus. Black arrowheads show granulocyte staining concentrated along the edge of a liver lobe and lining a blood vessel (BV). Open arrows show ribs containing stained granulocytes in the bone marrow. (F) Higher magnification showing stained granulocytes in the rib bone marrow (BM). Bars = 5 μm (D: inset), 50 μm (A: inset, C and D), 100 μm (A, B, and F), and 500 μm (E).
Immunohistochemical detection of human CD66b expression in CGM6-transgenic mice. Cryosections from various organs were incubated with a polyclonal rabbit anti-NCA antiserum followed by a peroxidase-coupled, antirabbit IgG and stained for peroxidase activity (brown). Counterstaining of nuclei was with hematoxylin (blue). (A) The spleen from a CGM6-transgenic mouse shows staining of granulocytes (see inset) mainly in the red pulp areas. (B) The spleen from a nontransgenic sibling mouse is negative. The lung (C) and a blood smear (D) of a CGM6-transgenic mouse also show granulocyte staining (Al = alveolus). Black arrowheads indicate stained granulocytes (see inset in D) and white arrowheads are nonstaining lymphocytes. (E) Section through the lung (Lu) and liver (Li) of a day 17.5 CGM6-transgenic mouse fetus. Black arrowheads show granulocyte staining concentrated along the edge of a liver lobe and lining a blood vessel (BV). Open arrows show ribs containing stained granulocytes in the bone marrow. (F) Higher magnification showing stained granulocytes in the rib bone marrow (BM). Bars = 5 μm (D: inset), 50 μm (A: inset, C and D), 100 μm (A, B, and F), and 500 μm (E).
To investigate the expression pattern of the CGM6 gene during embryogenesis, immunohistochemical analyses were also performed on mouse embryos from day 8.5 of embryonic development onwards. Transgenicity of the early embryos and fetuses was not determined, but at least 10 were analyzed to ensure the inclusion ofCGM6-positive animals, which should make up 50% of the offspring. Indeed, where determined postpartum, positive offspring do follow this expected Mendelian distribution pattern. Labeled cells are first seen at day 12.5 of development in cells within the embryonic liver, which are still visible at day 17.5 (Fig 5E) where staining of cells within the rib bone marrow (Fig 5F) is also apparent.
FACScan investigations.
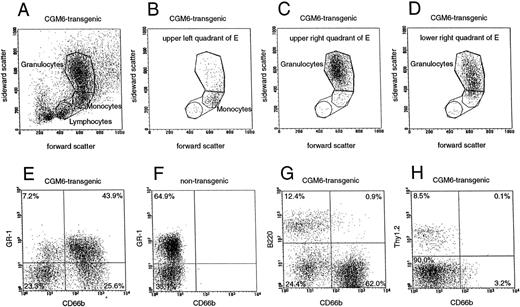
In an attempt to characterize the CD66b-expressing cells inCGM6-transgenic mice, FACScan analyses were performed on cells isolated from the bone marrow from mice in the fourth filial generation after backcrossing with C57BL/6 mice. The results of one experiment are shown in Fig 6. The forward/sideward scatter plot of the total cell population shows the three main leukocyte populations (Fig 6A), which were analyzed as a group for CGM6 expression. The remaining signals that represent debris and possible cell aggregates were excluded from the analyses. In the case of the T lymphocyte analysis (Fig 6H), only leukocytes within the lymphocyte gate (Fig 6A) were tested for antibody recognition so as to increase the number of T cells analyzed. Double-labeling experiments were performed using a FITC-labelled, CD66b-specific MoAb and PE-labelled rat antibodies specific for the mouse myeloid marker, GR-1 (Fig 6E and F), the B-lymphocyte marker, B-220 (Fig 6G) and the T-lymphocyte marker, Thy-1 (Fig 6H), respectively. As bone marrow cells from nontransgenic mice showed no signal with the CD66b-specific antibody (Fig 6F), cross-reactivity with murine CEA family members on mouse leukocytes, as well as possible binding by Fc-receptors, could be ruled out. These results showed that CD66b is not expressed on T, or B lymphocytes in the transgenic mice, but a large proportion of the GR-1–expressing myeloid cells also express CD66b (Fig 6E, upper right quadrant). Forward/sideward scatter analysis of these double-positive cells confirmed their identity as granulocytes (Fig 6C). Interestingly, some cells exist, which only express GR-1, but not CD66b (Fig 6E, upper left quadrant). Based on their forward/sideward scatter properties, these cells consist mainly of monocytes (Fig 6B). Cells also exist, which only express CD66b, but not GR-1 (Fig 6E, lower right quadrant), although these do not represent a separate population to the doubly-labeled cells. Analysis of this CD66b-positive, GR-1–negative group of cells using forward/sideward scatter confirmed that these cells are also granulocytes (Fig 6D), although there is a slight shift in the sideward scatter towards the monocyte population. These analyses suggest that the granulocytes in the transgenic animal are heterogenous for GR-1 expression.
FACScan analysis of CD66b expression in the bone marrow of a CGM6-transgenic and a nontransgenic mouse. (A) Forward/sideward scatter plot of bone marrow cells from aCGM6-transgenic mouse. The three main cell populations (lymphocytes, monocytes, and granulocytes) are gated separately or together for three-color fluorescence analyses, seen in the lower part of the figure (E-H) where four cell populations are shown and the percentages of events per quadrant are calculated. Double-labeling of bone marrow cells from a CGM6-transgenic mouse (E, G, and H) and from a nontransgenic mouse (F) with a FITC-labeled, CD66b-specific monoclonal antibody (x axis) and PE-labeled antibodies (y axis) for (E and F) a myeloid differentiation antigen (anti-GR-1), (G) a B-lymphocyte–specific marker (B220), and (H) a T-lymphocyte–specific marker (Thy-1). For the latter, only the lymphocyte-gated cell population was analyzed. (B) Forward/sideward scatter plot analysis of the GR-1 positive, CD66b-negative population from E; upper left quadrant. (C) Forward/sideward scatter plot analysis of the GR-1–positive, CD66b-positive population from E; upper right quadrant. (D) Forward/sideward scatter plot analysis of the GR-1–negative, CD66b-positive population from E; lower right quadrant.
FACScan analysis of CD66b expression in the bone marrow of a CGM6-transgenic and a nontransgenic mouse. (A) Forward/sideward scatter plot of bone marrow cells from aCGM6-transgenic mouse. The three main cell populations (lymphocytes, monocytes, and granulocytes) are gated separately or together for three-color fluorescence analyses, seen in the lower part of the figure (E-H) where four cell populations are shown and the percentages of events per quadrant are calculated. Double-labeling of bone marrow cells from a CGM6-transgenic mouse (E, G, and H) and from a nontransgenic mouse (F) with a FITC-labeled, CD66b-specific monoclonal antibody (x axis) and PE-labeled antibodies (y axis) for (E and F) a myeloid differentiation antigen (anti-GR-1), (G) a B-lymphocyte–specific marker (B220), and (H) a T-lymphocyte–specific marker (Thy-1). For the latter, only the lymphocyte-gated cell population was analyzed. (B) Forward/sideward scatter plot analysis of the GR-1 positive, CD66b-negative population from E; upper left quadrant. (C) Forward/sideward scatter plot analysis of the GR-1–positive, CD66b-positive population from E; upper right quadrant. (D) Forward/sideward scatter plot analysis of the GR-1–negative, CD66b-positive population from E; lower right quadrant.
DISCUSSION
As a first step towards investigating mechanisms controlling granulocyte-specific expression, we have analyzed a cosmid clone containing the complete CGM6 gene, which encodes the human granulocyte marker, CD66b/NCA-95.10 Furthermore, we have developed CGM6-transgenic mice with this cosmid that express CD66b exclusively in granulocytes. Experimental transgenic model systems using genomic fragments from YAC clones are currently being developed to analyze tissue-specific, cis-regulatory sequences in various genes.30 As seen from these investigations, similar studies are possible using cosmid inserts. We previously reported on mice that are transgenic for a cosmid insert containing the complete human CEA gene, which is exclusively expressed in epithelial cells. Those mice show a conserved spatiotemporal CEA expression pattern when compared with humans, despite the fact that mice do not have an endogenous CEA gene.24,31 These findings suggested the feasibility of determining the presence of tissue-specific cis-regulatory elements in the closely-relatedCGM6 gene. Although only one CGM6-transgenic line was developed, which contains 50 to 75 copies of the intact cosmid insert in a head-to-tail orientation, the granulocyte-specific expression pattern was found to be identical to that observed in humans.13 This indicates that all of the regulatory elements, which convey tissue-specificity, are present within this cosmid insert and that no additional effects due to the site of genomic integration can be registered. As mice apparently do not have counterparts to any of the GPI-linked members of the CEA family, including CD66b and CEA, it is rather surprising that the expression pattern is identical.32 However, the cis-regulatory elements present within these genes are obviously recognized and interact correctly with the murine trans-acting factors. Thus, the in vivo investigations on the CGM6 transgenic mice provide a basis for the identification of granulocyte-specific cis-regulatory elements and the interacting transcription factors.
Although myeloid-specific regulatory elements have been defined, including an initiator element that is essential for gene expression in myeloid cells, a myeloid-specific promoter that was characterized in transgenic mice and a myeloid-specific enhancer in the mouse lysozyme gene, less is known about granulocyte-specific gene regulation.15-17 In this context, a myeloid lineage-specific enhancer region has been characterized approximately 3 kb upstream of the gene encoding myeloperoxidase. This enzyme is activated at the promyelocytic stage of granulocyte development.33 The enhancer element was found to bind Pu.1 and members of the C-EBP family that are critical regulators of myeloperoxidase gene expression. Indeed, a model has been proposed whereby the temporal exchange of C-EBP isoforms mediates transcription from a primed state in multipotential cells to a transcriptionally active configuration in promyelocytes.34 Interestingly, a putative C-EBP element is also present in the CGM6gene promoter (Fig 1B). Two functional promoters have been characterized for genes that are specifically expressed in eosinophils, including the gene encoding the human interleukin (IL)-5 receptor alpha subunit and the human eosinophil peroxidase, but in both cases, the factors conveying tissue-specific expression remain to be determined.35 36 The DNA regions containing tissue-specific, cis-regulatory elements in the CGM6 gene must first be identified in vitro. For functional confirmation, however, new transgenic mice need to be generated to test enhancer-promoter/indicator gene constructs in vivo.
Attempts to analyze the CGM6 gene promoter in functional assays using promoter constructs of varying lengths to regulate the expression of the firefly luciferase gene in transient transfection assays have so far failed to show measurable activity. Various promyelocytic lines have been tested, with successful in vitro differentiation of HL60 cells and stimulation of CD66b expression using retinoic acid.37 However, only negligible luciferase activity was noted with all promoter constructs tested so far (constructs ranging from 600 up to 5,600 nucleotides from the 5′-flanking region), although internal controls using the cytomegalovirus (CMV) promoter to express the luciferase gene were successful (results not shown). It is possible that important enhancer sequences conveying cell-type specificity are located outside the regions tested. Alternatively, a strong silencer element could be present within the 600 nucleotide upstream region.
The transcriptional start site cluster of the CGM6 gene that is located between nucleotides -95 and -101 relative to the translational start site is homologous to the transcriptional start sites of theNCA and CEA genes, which we described previously.38 As with other members of the CEA gene family, no TATA, or CAAT-box sequences can be found upstream of this position. The main transcription initiation site cluster in the CGM6 gene (TCAG*CAC*A*A*A*AGGAGG [asterisk after a base indicates a transcriptional start site]) shows no homology to the Inr consensus sequence (YYA*NT/AYY) described for several other genes, including the gene encoding the platelet-activating factor receptor, whose expression is restricted to myeloid cells.15 However, the strong conservation of this initiation site within different CEA gene family members, including the CEA gene that is only expressed in epithelial cells and the CGM6 gene whose expression is restricted to granulocytes, indicates its importance for a general, but not for a tissue-specific transcriptional control of these genes.13,38 Interestingly, an upstream stimulatory factor (USF)-binding site that has been characterized in the promoters of the CEA and BGP gene, is only poorly conserved at the corresponding position starting at approximately 150 nucleotides upstream of the translational start site in theCGM6 gene.39-42 Although this region is also conserved in other CEA family genes, one section is deleted from those murine and human genes (including the CGM6 gene) that are not expressed in epithelial cells, indicating its importance for directing epithelial tissue-specific expression.31 The corresponding region in the CGM6 gene promoter does, however, contain a C/G-rich sequence (CCCCGCCC), that perfectly matches the Sp1-binding concensus sequence reported for other genes.29 This element is also conserved in the BGP gene and in two PSG genes (PSG6 and PSG11), where it was shown to be important for promoter activity.40,43 A second putative element was identified that shows similarity to a C-EBP binding element concensus sequence.29 The existence of this element in theCGM6 gene is interesting because it is known that C-EBP family members play a critical role in directing granulocyte development and it would thus not be surprising to find such elements in a gene whose expression is restricted to granulocytes.
The temporal expression pattern of CD66b was investigated immunohistochemically in CGM6 transgenic mice whereby it was first seen in cells located within the liver of day 12.5 fetuses. It has been reported that embryonic hematopoiesis is initiated in the blood islands of the yolk sac that appear at day 8.5 of mouse embryo development.44 These blood islands contain the majority of erythroid and myeloid progenitor cells at early stages of development.45 Our analyses failed to identify granulocytes expressing CD66b in the yolk sac islands of CGM6 transgenic mice. In this context, it has been reported that cells committed to the myeloid lineage are first seen at day 10 of mouse gestation.46 The liver is the leading hematopoietic organ of the 5 to 22 week human embryo and fetus, where proliferating granulomonocytic precursors have been found.47 In the developing mouse fetus, liver hematopoiesis has been reported to begin at day 13.46 Based on our studies in theCGM6-transgenic mouse, the first CD66b-expressing granulocytes appear in the fetal liver at day 12.5 of embryonic development. A shift to the bone marrow is apparent at day 17.5 of fetal development and this is maintained throughout adulthood.
The FACScan analyses indicate that CD66b in the transgenic mice represents a more specific granulocyte marker than the endogenous GR-1 epitope, which was also found to be present on monocytes (Fig 6). Indeed, the results presented here correlate well with the GR-1 expression pattern reported elsewhere, ie, GR-1 expression increases during granulocyte maturation and it is transiently expressed on cells in the monocytic lineage.48 Therefore, apart from their potential use for investigating fetal granulopoiesis, theCGM6-transgenic mice could also provide a model system, eg, for bone marrow transplantation studies using CD66b as a granulocyte-specific marker. After lethal irradiation of the recipient mouse bone marrow, pluripotent stem cells from CGM6-transgenic donor mice could be transplanted and reconstitution of the hematopoietic system monitored for the development of mature granulocytes. The use of specific markers for the various blood cell lineages can be used to test the ability of the transplanted stem cells to differentiate, which does not always occur.49 A comparable system to test pluripotent stem cell cohorts has been described, using genetic markers from transgenic bone marrow transplantates into nontransgenic mice.50 Finally, theCGM6 transgenic mice could provide a model system for testing the effectivity of antiinflammatory substances using CD66b, either as an endogenous granulocyte marker or to target these substances or derivatives thereof to granulocytes via CD66b-specific antibodies.51-53 Such in vivo studies could yield useful information on the efficacy and the potential side effects of this type of treatment before clinical trials.
ACKNOWLEDGMENT
We wish to thank Sabine von Kleist for her continuous support, as well as Rita Carsetti and Craig Stocks for their assistance and guidance with the FACScan analyses. We acknowledge the reliable technical assistance of Margarethe Ditter, Beate Fischer, Dirk Schillinger, and Joachim Grammel.
Supported by grants from the Deutsche Forschungsgemeinschaft (Bonn, Germany) and the Dr Mildred Scheel Stiftung für Krebsforschung (Bonn, Germany).
The nucleotide sequence presented in this report has been submitted to the EMBL Bioinformatics Institute and has been assigned the accession number Z95119.
Address reprint requests to John Thompson, PhD, Institut für Immunbiologie der Universität, Stefan-Meier-Str. 8, D-79104 Freiburg, Germany.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

![Fig. 3. Determination of the intactness and copy number of theCGM6 transgene. A total of 10 μg of thymus DNA each from a C57BL/6 mouse (lane 1) and from the CGM6-transgenic mouse 5322 (lane 2) was digested with EcoRI, size fractionated, and hybridized with [32P]-labeled F19632 cosmid DNA. To estimate the number of integrated transgene copies, F19632 DNA corresponding to 75 gene copies per haploid genome was added to 10 μg of C57BL/6 DNA before digestion with EcoRI (lane 3). Cosmid vector fragments and end fragments of the genomic insert of F19632 are indicated by arrowheads and arrows, respectively. The asterisk marks the band, which contains besides an internal EcoRI DNA fragment, the fusion fragment created by the head to tail insertion of the amplified transgene. The mobility and size in kb of the marker fragments is shown in the left margin.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/2/10.1182_blood.v91.2.663/3/m_blod4021003.jpeg?Expires=1763519863&Signature=M1k06Tb0T-oe70odWCDsTXuWXydTzz-YiU-0SDmO4~y1sF-CEtUYSTh8LANsJ83inhQiS6xxx0FM66mQ5lWsEAFqU10xJ7kDtzlpAx3wYtmiOn4bgQ7OHmrFJelcgYvcvPcRJfd4SOSYYxlYnkHWDfr4dZoD5ynPUZAooxWhPhe-FNJazxCOeIF3Z7TVV~DnBg71Pm~d8x6IS6bKyuQPxIRnKVBg0Xf3xUhmivhm1h5TG1D4BnDrzo~IoX8XfwwJxxIOzWPHgk9LPSz7mGMuOJycR1qfuToa0-d0Zg9xCxWqIvfEHiiUH5cwxzDqVxezNn~Itv6t6H6tU~Iu90pAUA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal