Abstract
The hybrid gene BCR-ABL that typifies chronic myeloid leukemia (CML) represents an attractive target for therapy with antisense oligodeoxyribonucleotides (ODN). A central obstacle in the therapeutic application of ODN is their poor cellular uptake. Adding various lipophilic conjugates to the ODN backbone has been reported to improve uptake, and electroporation of target cells has also been shown to enhance intracellular ODN delivery. We have shown that (1)BCR-ABL–directed ODN will specifically decrease the level ofBCR-ABL mRNA, provided that cells are first permeabilized with Streptolysin-O (SL-O), and (2) chimeric methylphosphonodiester:phosphodiester ODN directed against 9 bases either side of the BCR-ABL junction are more efficient ODN effectors than structures composed solely of phosphodiester or phosphorothioate linkages. In this study, we compared the efficacy of lipophilic conjugation, SL-O permeabilization and electroporation on the intracellular delivery and molecular effect ofBCR-ABL–directed ODN. b2a2- and b3a2-directed chimeric ODN were synthesized either unmodified or with one of the following groups at the 5′ end: cholesterol, vitamin E, polyethylene glycol of average molecular weight 2,000 or 5,000, N-octyl-oligo-oxyethylene, or dodecanol. ODN associated with Lipofectin was also studied. Comparison was made in untreated, electroporated, and SL-O permeabilized KYO1 cells. Uptake was examined by fluorescence microscopy and flow cytometry, using ODN structures that were 3′ labeled with fluorescein. The effect on target BCR-ABL mRNA expression was analyzed by Northern blotting. Several conjugated structures associated avidly with the cell membrane without achieving significant intracellular uptake or molecular effect. Similarly, ODN:Lipofectin complexes moderately increased cell association, without enhancing intracellular levels of ODN or inducing detectable molecular effect. In SL-O permeabilized or electroporated cells, uptake was approximately 1 to 2 logs greater than in untreated cells, and rapid nuclear localization was seen, especially with unmodified chimeric ODN. In SL-O permeabilized cells treated with ODN directed to the b2a2 and b3a2 junctions respectively, b2a2 BCR-ABL mRNA levels at 4 hours were reduced to 2.6% ± 2.1% and 38.4% ± 1.3% of control values. In cells permeabilized by electroporation, BCR-ABL mRNA levels were decreased to 4.0% ± 1.4% of control levels by b2a2 directed ODN, although very little nontargeted suppression was seen with b3a2-targeted ODN (93.4% ± 4.2% of control). Greater cell to cell variation in ODN uptake was seen for SL-O permeabilized cells when compared with electroporated cells, suggesting that, after SL-O permeabilization, relatively unpermeabilized and overpermeabilized populations may coexist. No structure had any effect on the level of irrelevant (p53, MYC, and GADPH) mRNA levels. We conclude that the conjugation of chimeric ODN with one of the above-mentioned lipophilic groups or the complexing of ODN with Liopfectin does not improve either intracellular delivery of ODN or the molecular effect. In contrast, both electroporation and SL-O permeabilization (1) considerably enhanced uptake of chimeric ODN (even for structures without a conjugate group) and (2) achieved significant suppression of target mRNA levels.
IN CHRONIC MYELOID leukemia (CML), the characteristic Philadelphia abnormality translocates part of theABL gene on chromosome 9q34 to the major breakpoint cluster region BCR on chromosome 22q11.1 The resultant hybrid gene BCR-ABL is leukemia-specific, and its protein product (p210) is likely to be central in the maintenance of the leukemic phenotype. BCR-ABL mRNA therefore represents an attractive target for antisense oligodeoxyribonucleotides (ODN).
Several groups have reported their experience with short (18-26mer) ODN on CML cells and cell lines.2-5 Most groups report that ODN decrease p210 expression, BCR-ABL transcript level, or CML cell growth in in vitro culture, although the effects may be nonspecific.3,4,6 In these studies, all the constituent nucleosides have been either wholly phosphodiester (PO)-linked or wholly phosphorothioate (PS)-linked. However, PO-linked ODN are highly sensitive to nucleases present both intracellularly and in serum.7 Furthermore, PS-linked analogues, although significantly more resistant to nuclease degradation, may bind nonspecifically to a variety of intracellular molecules, such as nucleic acid polymerases,8 and PS-linked ODN may produce nonantisense aptameric effects in CML cells.9,10 Apparent positive results with PS-linked ODN may therefore be due to nonspecific nonantisense effects,3 which may not be leukemia-specific when mixed populations of leukemic and normal cells are studied.
ODN in which all the bases are linked by methylphosphonodiester (MP) linkages are totally resistant to nucleases. All MP-linked ODN have been used to target BCR-ABL mRNA,11 with reported selective decrease in BCR-ABL p210 protein levels and an antiproliferative effect. However, all MP-linked ODN are poorly soluble and exhibit low affinity for their target mRNA. All MP-linked ODN are also unable to direct destruction of target mRNA by RNase H, a ubiquitous cellular enzyme that cleaves the RNA component of DNA:RNA heteroduplexes.12 In the hope of combining useful properties of both MP- and PO-linked ODN structures, we have therefore designed novel MP:PO:MP chimeric ODN, consisting of terminal MP-linked sections flanking a central PO-linked region (Fig 1). The flanking MP linkages confer protection against exonucleases, whereas the central PO-linked section will retain highly specific cleavage of BCR-ABL mRNA by targeting RNase H. We have shown in previous work that such chimeric ODN have superior specific antisense properties and are more stable in cell extracts when compared with conventionally linked ODN.13
Sequence and internucleoside linkages for the ODN studied. A dash (-) between bases indicates a PO linkage, and a slash (/) indicates an MP linkage. Fluorescein attachment to the 5′ and 3′ termini, via a 6-amino-1-hexanol linker, is indicated by ∼F and F∼, respectively. CH, cholesterol; DD, dodecanol; PEG2 and PEG5, polyethylene glycol of average molecular weight 2,000 and 5,000, respectively; POE, N-octyl-oligo-oxyethylene; VITE, vitamin E. In the ODN code, 2B and 3B denote sequences targeting the b2a2 and b3a2 junctional sequences, respectively.
Sequence and internucleoside linkages for the ODN studied. A dash (-) between bases indicates a PO linkage, and a slash (/) indicates an MP linkage. Fluorescein attachment to the 5′ and 3′ termini, via a 6-amino-1-hexanol linker, is indicated by ∼F and F∼, respectively. CH, cholesterol; DD, dodecanol; PEG2 and PEG5, polyethylene glycol of average molecular weight 2,000 and 5,000, respectively; POE, N-octyl-oligo-oxyethylene; VITE, vitamin E. In the ODN code, 2B and 3B denote sequences targeting the b2a2 and b3a2 junctional sequences, respectively.
A central problem in the application of ODN to living cells is their poor cytosolic uptake.14,15 Reported antiproliferative effects from experiments in which ODN are simply added to the culture medium may result from nonantisense mechanisms,16,17 and this is especially true for wholly PS-linked ODN structures. Several reports have suggested that introduction of lipophilic groups into ODN structure will enhance intracellular delivery of ODN, with concomitant increase in antisense efficacy. Introduction of a lipophilic dodecanol chain18 or cholesterol19 at the 3′ end increased cellular uptake by up to 2 orders of magnitude. Entrapping ODN in liposomes11 or complexing them with cationic lipids20,21 has also been reported to improve their biological action. Although these modifications were reported to increase ODN association to cells and in some cases to improve intracellular delivery, in no case was there unequivocal evidence of a specific antisense effect. None of these studies has used chimeric ODN, and few have targeted genes of relevance to hematological malignancy. Ex vivo electroporation has also been reported to enhance intracellular ODN delivery, apparently killing neoplastic cells while sparing normal marrow cells in a murine model.22 We have previously reported that human leukemic cells may be temporarily permeabilized by Streptolysin-O (SL-O) to load them with ODN.13,23 24
In the present work, we set out to compare these three different strategies of improving BCR-ABL–directed ODN delivery to CML cells: (1) SL-O permeabilization of target cells, (2) electroporation of target cells, and (3) conjugation of ODN with various lipophilic structures. We have examined their effect on ODN uptake and intracellular localization and have investigated ODN delivered by these techniques on inhibiting target mRNA expression.
MATERIALS AND METHODS
ODN structure and synthesis.
All ODN structures studied were 18-mers, complementary to an 18-base mRNA sequence spanning either the b3a2 or the b2a2 BCR-ABLfusion breakpoint (Fig 1). The overall structure was chimeric in design, composed of 4MP:9PO:4MP internucleoside linkages. To assess cellular uptake and intracellular localization, all ODN were labeled with fluorescein. As well as unconjugated ODN, structures were studied that were conjugated to one of the following lipophilic groups at the 5′ end: cholesterol, dodecanol, polyethylene glycol of average molecular weight 2,000 (PEG2), polyethylene glycol of average molecular weight 5,000 (PEG5), N-octyl-oligo-oxyethylene (POE), and vitamin E.
We have previously reported in detail the synthesis of chimeric MP:PO:MP ODN directed against BCR-ABL.13 Briefly, standard cyanoethyl phosphoramidites and methylphosphonamidites (Glen Research; supplied by Cambio Ltd, Cambridge, UK) were used on an Applied Biosystems 381A DNA Synthesiser (Applied Biosystems, Warrington, Cheshire, UK). For the lipophilic conjugates, syntheses were initiated throughout using fluorescein-derivatized CPG25 to give 3′ fluorescein-labeled ODN. The nonlipophilic unconjugated ODN were synthesized with 5′-Amino-Modifier C6 TFA (Glen Research) at their 5′ end and tagged with fluorescein, postsynthesis, using Fluos reagent (Boehringer Mannheim, Lewes, East Sussex, UK).
Phosphoramidite derivatives of lipophilic molecules were synthesized and conjugated to ODN at the 5′ termini during automated synthesis as follows. Cholesterol (BDH, Lutterworth, Leicestershire, UK), dodecanol (Aldrich, Gillingham, Dorset, UK), vitamin E (Fluka, Gillingham, Dorset, UK), and POE (Alexis) were phosphitylated without prior modification, and the resultant phosphoramidites were purified using a general procedure for the preparation of DNA synthesis monomers.26 As diols, PEG2 and PEG5 (Aldrich) required conversion into mono-(4,4′-dimethoxytrityl) derivatives before phosphitylation.27 Vitamin E containing ODN were synthesized according to the method of Will and Brown.28All lipophilic phosphoramidite monomers were used as 1.5 mol/L solutions in anhydrous dichloromethane. Deprotection and purification of the lipophilic oligonucleotides was performed as previously described for unconjugated chimeric MP:PO:MP ODN.13 All ODN were associated with sodium as the cation.
ODN were purified by C18 Sep Pak cartridges (Waters Chromatography Division of Millipore Ltd, Watford, UK) and subsequently by gel electrophoresis to remove fluoresceinated impurities. All ODN, once synthesized and purified, were stored at −20°C in the dark until use.
Cells and cell lines.
The human b2a2 BCR-ABL–containing CML cell line KYO1 (generous gift of Dr S. O'Brien, LRF Leukaemia Unit, Hammersmith Hospital, London, UK) was used as a target in which to study ODN uptake. KYO1 cells were cultured in RPMI 1640 medium supplemented with L-glutamine (GIBCO Ltd, Paisley, UK) and 10% heat-inactivated fetal bovine serum (FBS; Sera Lab, JRH Biosciences, West Sussex, UK). Cells were maintained in logarithmic phase of growth by subculturing thrice weekly.
Uptake experiments and flow cytometry.
All uptake studies were performed using purified ODN to ensure that only intact oligonucleotide was studied. A total of 106cells in 50 μL of RPMI/FBS were incubated with 20 μmol/L ODN at 37°C for up to 6 hours. Ten-microliter aliquots were removed at intervals into 1 mL of RPMI/FBS containing 10 μg/mL of propidium iodide (PI) at 4°C for 10 minutes and washed three times in RPMI/FBS. All experiments were performed in duplicate.
An Ortho Cytoron Absolute flow cytometer (Ortho, High Wycombe, Buckinghamshire, UK) was used to measure uptake, using a 15-mW argon ion laser set to measure fluorescein and PI (dead cells) in separate channels. The cytometer was calibrated using glutaraldehyde-fixed chick red cells (Sigma, Poole, UK). Standard curves for quantitation of ODN were derived by using APS-Hyperspheres (Shandon Southern Products Ltd, Runcorn, UK), to which we had irreversibly bound known quantities of fluorescein-tagged ODN. From these calibration curves, the mean fluorescence was converted into attomoles of ODN per cell as previously described.14 Uptake was recorded for cells that excluded PI.
SL-O permeabilization.
The technique for SL-O permeabilization was essentially that of Barry et al,29 modified as previously described.24,25 30
Briefly, after washing in serum-free RPMI medium, 5 × 106 KYO1 cells were exposed to dithiothreitol-activated SL-O at optimal concentration and 20 μmol/L ODN in 200 μL of serum free RPMI in a Falcon 3047 multiwell tissue culture plate (Becton Dickinson Labware, Lincoln Park, NJ). After incubation for 10 minutes at 37°C, 1 mL of RPMI/FBS medium was used to reseal the cells. Ten minutes later, the cells were transferred to tissue culture flasks containing 10 mL prewarmed gassed RPMI. After sampling, PI counterstaining, and washing, the efficiency of permeabilization and the amount of ODN introduced into viable cells was assessed by calibrated dual fluorescence flow cytometry.
Electroporation.
After washing once in RPMI, 5 × 106 KYO1 cells were resuspended in 800 μL RPMI at 4°C. ODN was added where required to give a final concentration of 20 μmol/L. The cell suspension was transferred to ice-cold 0.4-mm gap electroporation cuvettes (BioRad, Hemel Hempstead, Hertfordshire, UK), and cells permeabilized with a single pulse from a Gene Pulser attached to optional capacitance extender (BioRad) set to 960 μF, 250 V. These conditions were defined as producing optimal permeabilization with minimal toxicity in preliminary experiments and resulted in an electroporation time constant of approximately 20 milliseconds. The cells were incubated for 60 minutes at 4°C in the electroporation cuvettes and then transferred to flasks containing 10 mL of prewarmed gassed RPMI as for SL-O permeabilization.
Transfection by Lipofectin.
ODN and 30 μL Lipofectin (GIBCO) were allowed to associate for 15 minutes at 37°C, using sufficient ODN to achieve a final concentration of 2 μmol/L after addition to the cell suspensions. This ratio of ODN:Lipofectin was defined in preliminary experiments as that maximizing cell-associated ODN. KYO1 cells (5 × 106) were suspended in 200 μL RPMI, incubated with ODN/Lipofectin mixture at 37°C for 15 minutes, and then transferred to flasks containing 10 mL warmed gassed RPMI. No attempt was made to remove the Lipofectin. The dose of ODN/Lipofectin used was the maximum dose that did not induce excessive acute toxicity (cell death <30% at 4 hours, as measured by PI counterstaining).
RNA purification and analysis.
Total cellular RNA was prepared from samples of experimental cells by the guanidinium thiocyanate-acid phenol method.31 RNA precipitates were resuspended in deionized diethylpyrocarbonate-treated formamide,32 and their relative concentrations were estimated by gel electrophoresis and ethidium bromide staining. Denaturing agarose gel electrophoresis, transfer of RNA to Nytran nylon membrane (0.2-μm pore; Schleicher and Schuell; UK distributor Anderman and Co Ltd, Kingston-upon-Thames, Surrey, UK), hybridization to digoxigenin-labeled probe, and subsequent chromogenic visualization procedures were all performed as previously described,33except that digoxigenin-labeled antisense RNA was used as probe34 rather than random-primed DNA. The blots were quantified using a Shimadzu CS9000 flying spot densitometer (Shimadzu; UK distributor Howe and Co Ltd, Banbury, Oxfordshire, UK), as previously described.35 The expression of BCR-ABLand irrelevant control (C-MYC, p53, and glyceraldehyde-3-phosphate dehydrogenase [GAPDH]) mRNAs were corrected for the flow cytometrically determined number of cells from which the RNA was extracted. Results are expressed as relative to an untreated control.
RESULTS
ODN uptake.
Figure 2 gives the ODN uptake into KYO1 cells exposed continuously to b2a2-directed ODN of various structures for 6 hours. All ODN were used at a concentration of 20 μmol/L, except for ODN conjugated to cholesterol or vitamin E, which were used at a concentration of 2 μmol/L, because higher ODN concentrations caused lysis of target cells because of membrane damage. Uptake is expressed as the log of attomoles of fluorescein-labeled ODN per cell. No difference in ODN uptake is seen for conjugated structures in comparison with unconjugated ODN. Similarly, no difference was seen for ODN associated with Lipofectin (data not shown). For ODN directed against the b3a2 junction, the same pattern and level of uptake was seen (data not shown).
ODN uptake over a 6-hour incubation period. (•) 2 μmol/L 5′CH-2B494AS-3′F; (⧫) 20 μmol/L 5′F-2B494AS; (▪) 20 μmol/L 5′DD-2B494AS-3′F; (*) 20 μmol/L 5′PEG5-2B494AS-3′F; (▴) 20 μmol/L 5′PEG2-2B494AS-3′F; (▵) 20 μmol/L 5′POE-2B494AS-3′F; (+) 2 μmol/L 5′VITE-2B494AS-3′F; (○) 20 μmol/L 5′F-2B494AS with SL-O.
ODN uptake over a 6-hour incubation period. (•) 2 μmol/L 5′CH-2B494AS-3′F; (⧫) 20 μmol/L 5′F-2B494AS; (▪) 20 μmol/L 5′DD-2B494AS-3′F; (*) 20 μmol/L 5′PEG5-2B494AS-3′F; (▴) 20 μmol/L 5′PEG2-2B494AS-3′F; (▵) 20 μmol/L 5′POE-2B494AS-3′F; (+) 2 μmol/L 5′VITE-2B494AS-3′F; (○) 20 μmol/L 5′F-2B494AS with SL-O.
Figure 2 also shows the uptake of unconjugated ODN into KYO1 cells that were first permeabilized with SL-O and then resealed after ODN loading. In this case, ODN uptake is seen to be more than 1 log greater than that into nonpermeabilized cells. The uptake of conjugated ODN structures into SL-O permeabilized KYO1 cells was also studied (data not shown). In no case was any advantage obtained over unmodified ODN.
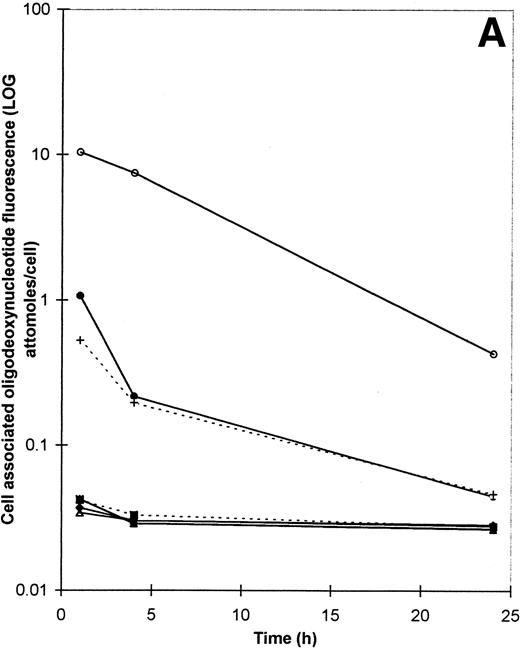
Figure 3A shows how cell-associated ODN fluorescence decays over the 24 hours after a pulse of ODN treatment, also expressed as the log of attomoles of fluorescein-associated ODN per cell. All ODN concentrations are 20 μmol/L; because the exposure to ODN is pulsed and brief, it was now possible to use this concentration also for cholesterol- or vitamin E-conjugated structures. Although there is a gradual decrease in the amount of cell-associated ODN, at any given time there is significantly greater amounts of ODN present in SL-O permeabilized cells than in nonpermeabilized cells. Comparable data were again obtained for b3a2-directed ODN (data not shown).
(A) The decay of cell-associated ODN fluorescence after treatment of KYO1 cells with ODN conjugates or loading with SL-O. Symbols are the same as for Fig 2, except that the 5′CH-2B494AS-3′F and 5′VITE-2B494AS-3′F concentrations are 20 μmol/L. (B) Comparison of the decay of cell-associated ODN fluorescence in SL-O permeabilized and electroporated KYO1 cells. Details of the experimental conditions are given in the text. (▴) 20 μmol/L 5′F-2B494AS with SL-O; (+) 20 μmol/L 5′F-3B494AS with SL-O; (⧫) 20 μmol/L 5′F-2B494AS with electroporation; (▪) 20 μmol/L 5′F-3B494AS with electroporation.
(A) The decay of cell-associated ODN fluorescence after treatment of KYO1 cells with ODN conjugates or loading with SL-O. Symbols are the same as for Fig 2, except that the 5′CH-2B494AS-3′F and 5′VITE-2B494AS-3′F concentrations are 20 μmol/L. (B) Comparison of the decay of cell-associated ODN fluorescence in SL-O permeabilized and electroporated KYO1 cells. Details of the experimental conditions are given in the text. (▴) 20 μmol/L 5′F-2B494AS with SL-O; (+) 20 μmol/L 5′F-3B494AS with SL-O; (⧫) 20 μmol/L 5′F-2B494AS with electroporation; (▪) 20 μmol/L 5′F-3B494AS with electroporation.
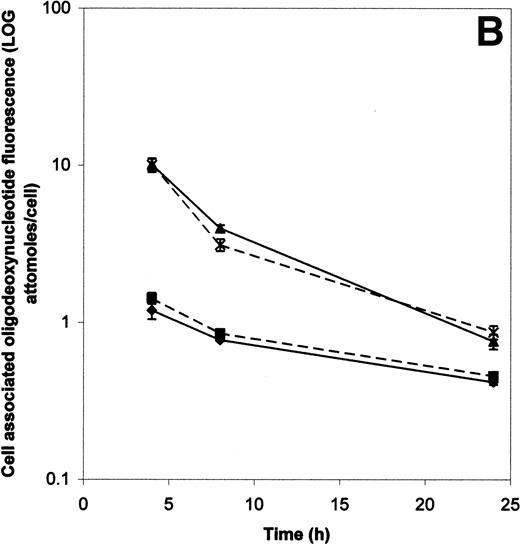
Figure 3B compares the decay of cell-associated ODN fluorescence in cells permeabilized by SL-O and by electroporation. Using the electroporation conditions described in the methods section, 14.2% ± 1.1% of cells were killed (as evidenced by PI uptake) and 6.6% ± 1.8% of cells were unpermeabilized. SL-O permeabilized cells therefore received a supraoptimal dose of SL-O, designed to give comparable cell death to electroporated cells, and this killed 12.3% ± 3.2% of cells, leaving 2.9% ± 0.4% of the cells unpermeabilized. Under these conditions, SL-O permeabilization achieves a mean apparent intracellular concentration of ODN of approximately 1 log greater than that achieved by electroporation.
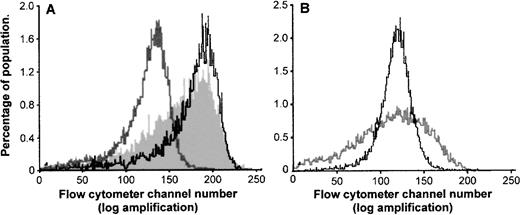
Figure 4 gives the mode and variation of intracellular fluorescence 4 hours after SL-O or electroporation facilitated delivery of fluorescent ODN (20 μmol/L extracellular concentration) into KYO1 cells. In Fig 4A, the frequency distribution of fluorescence after electroporation (grey line) is compared with that after optimal (<5% of cells killed) and supraoptimal (cell death equivalent to that of electroporation, as described in the preceding paragraph) doses of SL-O. At each dose of SL-O, significantly higher levels of ODN uptake are seen in the majority of cells (mean intracellular ODN levels of 5.2 and 11.2 attomoles per cell, respectively, compared with 1.2 attomoles per cell for electroporated cells). However, Fig 4B compares electroporation with a suboptimal dose of SL-O designed to give similar mean ODN loading (mean intracellular ODN level of 1.2 attomoles per cell). At this SL-O concentration, in comparison with electroporation, substantial numbers of cells have low levels of ODN uptake, yet some cells remain highly loaded.
Frequency histograms showing the mean and variation of intracellular fluorescence 4 hours after SL-O and electroporation delivery of fluorescent ODN (20 μmol/L extracellular concentration) into KYO1 cells. (A) The filled grey area represents cells permeabilized with an optimal concentration of SL-O (data from Fig 3A). The solid line represents cells permeabilized with a supraoptimal concentration of SL-O (data taken from the experiment described in Fig3B). The grey line represents electroporated cells (data from Fig 3B). (B) The solid line represents electroporated cells. The grey line represents cells permeabilized with a suboptimal dose of SL-O, designed to give equivalent ODN loading (mean of 1.2 attomoles per cell).
Frequency histograms showing the mean and variation of intracellular fluorescence 4 hours after SL-O and electroporation delivery of fluorescent ODN (20 μmol/L extracellular concentration) into KYO1 cells. (A) The filled grey area represents cells permeabilized with an optimal concentration of SL-O (data from Fig 3A). The solid line represents cells permeabilized with a supraoptimal concentration of SL-O (data taken from the experiment described in Fig3B). The grey line represents electroporated cells (data from Fig 3B). (B) The solid line represents electroporated cells. The grey line represents cells permeabilized with a suboptimal dose of SL-O, designed to give equivalent ODN loading (mean of 1.2 attomoles per cell).
Effect on mRNA expression.
Figure 5A shows the effect of various ODN structures on the expression of BCR-ABL mRNA in KYO1 cells, 4 hours after a pulsed exposure to ODN. No structure decreased mRNA expression to less than 80% of that of an untreated control. ODN associated with Lipofectin had no effect (data not shown). No consistent difference was seen between the effect of b2a2- and b3a2-directed ODN. No ODN (whether b2a2- or b3a2-directed) decreased the expression of p53, C-MYC, or GAPDH mRNA (data not shown).
(A) Effect of various ODN structures on BCR-ABLmRNA expression in KYO1 cells 4 hours after treatment with 20 μmol/L ODN. (B) BCR-ABL mRNA expression in electroporated (EP) and SL-O permeabilized cells. ODN nomenclature is as for Fig 1.
(A) Effect of various ODN structures on BCR-ABLmRNA expression in KYO1 cells 4 hours after treatment with 20 μmol/L ODN. (B) BCR-ABL mRNA expression in electroporated (EP) and SL-O permeabilized cells. ODN nomenclature is as for Fig 1.
In cells permeabilized by an optimal concentration of SL-O (<5% cell death), unmodified b2a2-directed ODN decreased target mRNA levels to 11% ± 3% of control. b3a2-directed ODN showed some cross-reactivity in reducing the b2a2 target to 54% ± 17% of control.
Figure 5B gives the BCR-ABL mRNA expression in KYO1 cells that were first permeabilized by either electroporation or by SL-O at a supraoptimal dose designed to give equivalent toxicity to electroporation (12% to 14% cell death). For each technique, the relevant control was treated cells not exposed to ODN. In SL-O permeabilized cells, unmodified b2a2-directed ODN decreased target mRNA levels to 2.6% ± 2.1% of control. b3a2-directed ODN showed some cross-reactivity in reducing the b2a2 target to 38.4% ± 1.3% of control. In cells permeabilized by electroporation, BCR-ABLmRNA levels were decreased to comparable levels by b2a2-directed ODN (4.0% ± 1.4% of control), although very little nontargeted suppression was seen using a b3a2-targeted ODN (93.4% ± 4.2% of control).
Cellular localization of ODN.
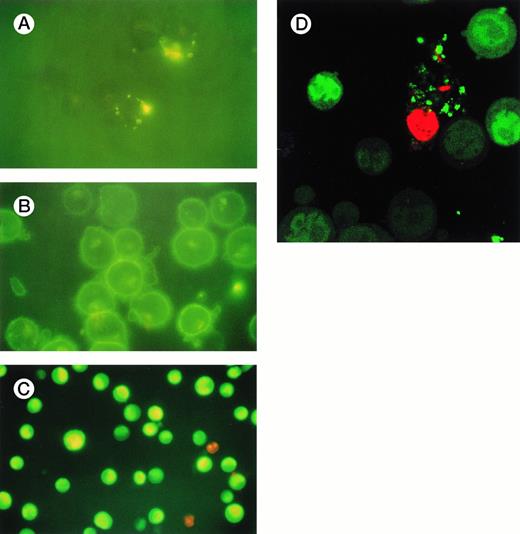
Figure 6 shows a series of fluorescence microscopic appearances of KYO1 cells after exposure to ODN. Figure 6A shows the appearances with 100 μmol/L 4-9-4 chimeric ODN, without SL-O permeabilization. There is a vesicular intracellular distribution, compatible with uptake by fluid phase pinocytosis. Similar appearances were seen with ODN conjugated with POE, Dodecanol, and PEG2 or PEG5, with no evidence of improved intracellular delivery.
Fluorescence microscopic appearances of KYO1 cells 6 hours after exposure to (A) 100 μmol/L unconjugated ODN without SL-O permeabilization; (B) 20 μmol/L cholesterol conjugated ODN without SL-O permeabilization; (C) 20 μmol/L unconjugated ODN after initial cellular permeabilization with SL-O; and (D) 20 μmol/L unconjugated ODN after electroporation.
Fluorescence microscopic appearances of KYO1 cells 6 hours after exposure to (A) 100 μmol/L unconjugated ODN without SL-O permeabilization; (B) 20 μmol/L cholesterol conjugated ODN without SL-O permeabilization; (C) 20 μmol/L unconjugated ODN after initial cellular permeabilization with SL-O; and (D) 20 μmol/L unconjugated ODN after electroporation.
Figure 6B shows appearances with 20 μmol/L cholesterol-conjugated ODN, again without SL-O permeabilization. There is avid association with the outer cell membrane, but little evidence of intracellular uptake. The areas of denser uptake are consistent with endosomal localization, in which ODN is still separated from the cytosol by an endosomal membrane. Similar appearances were seen for vitamin E-conjugated ODN (data not shown).
Figure 6C shows the uptake appearances of 20 μmol/L unconjugated ODN with KYO1 cells that had first been permeabilized with SL-O. There is bright and avid nuclear association, with no appreciable association with the outer cell membrane or with endosomes.
Figure 6D shows the cellular localization after incubation of electroporated cells with 20 μmol/L ODN. The appearances are virtually identical to those after SL-O permeabilization.
These appearances are therefore consistent with improved delivery of ODN to the site of target mRNA and pre-mRNA.
DISCUSSION
Although leukemic cells may take up ODN more readily than normal cells,36 for ODN to have a worthwhile biological effect, some means of enhancing cellular uptake is essential. As a prelude to clinical studies of ODN for purging hemopoietic cell harvests from patients with CML, we have examined three different strategies for improving the effectiveness of ODN in CML cells. The present data again demonstrate the lack of significant intracellular ODN uptake and molecular effect if ODN (of whatever structure) are simply added to cells. We report that SL-O permeabilization or electroporation of target cells is superior to lipophilic conjugation or Lipofectin association for improving intracellular ODN delivery, achieving levels of approximately 5 to 10 and 1 attomole respectively per cell. The approximately 0.1 to 1 attomole per cell achieved with a variety of conjugated structures without prior permeabilization of target cells is comparable with that observed by Smetsers et al4 using 10 μmol/L PS-linked 26-mer ODN without uptake enhancement in BV173 cells, also a b2a2 BCR-ABL–containing cell line. The fluorescence micrographs demonstrate that the cellular distribution of ODN is predominantly intranuclear after SL-O permeabilization or electroporation, in contrast to the surface membrane and vesicular localization seen for conjugated structures. The superior uptake and cellular localization seen with SL-O permeabilization and electroporation translates into increased biological effectiveness, as evidenced by the decreased target mRNA levels, not seen in unmanipulated target cells treated by conjugated or Lipofectin-associated ODN structures.
Although SL-O permeabilization may achieve a greater mean ODN concentration than electroporation, each technique suppressesBCR-ABL mRNA to a comparable degree. The data of Fig 4A show that most cells permeabilized with SL-O contained much higher concentrations of ODN than the electroporated cells. This may explain the apparent nontargeted suppression of b2a2 BCR-ABL mRNA by b3a2-targeted ODN in SL-O permeabilized cells (Fig 5) induced through its complementarity to 9 ABL-derived bases at the b2a2 breakpoint.
In parallel with the present data on BCR-ABL mRNA suppression, we have recently investigated the expression of p210 after ODN treatment of SL-O permeabilized cells. No change in p210 expression was seen, despite impressive reductions in transcript level.37p210 may be a particularly difficult oncoprotein to target by ODN, because its half-life is in excess of 24 hours,38 yet ODN-induced suppression of BCR-ABL mRNA is wearing off after 8 hours, and levels have returned to control values by 24 hours. In contrast, MYC protein may be significantly and durably suppressed by ODN targeting a sequence in the coding region of c-MYC; the half-life of MYC is of the order of 10 minutes in KYO1 cells.37 The half-life of target protein may therefore be of particular importance in determining the biological effect of ODN. More biologically stable ODN targeting BCR-ABL may produce more durable mRNA suppression and thus result in suppression of p210.
The nature of the chemical linkages between each nucleoside is critical in determining the fate of ODN in biological systems. All PO-linked ODN are highly sensitive to nucleases, and all PS-linked molecules may bind nonspecifically. We have shown previously that, in the presence of human RNase H, chimeric MP:PO:MP ODN directed against b3a2 or b2a2BCR-ABL mRNA will produce sequence-specific mRNA cleavage in a cell-free system, in contrast to the lower specificity of all PO- or all PS-linked structures.13 No effect was seen using ODN targeting irrelevant sequences or for ODN recognizing partially complementary sequences.13,23 We have also previously shown that, when incubated with cell extracts to simulate intracellular conditions, all PO-linked ODN are entirely degraded within 5 minutes, whereas chimeric ODN are largely unaltered at 30 minutes, as assessed by high-performance liquid chromatography (HPLC).13 Our previous findings therefore suggest that chimeric ODN have a better chance of producing a sequence-specific (and therefore leukemia-specific) antisense effect in intact cells than conventionally linked ODN. We have therefore concentrated on studying the effect of chimeric MP:PO:MP ODN. We have focused on the b2a2 junctional sequence of BCR-ABL, because this may be more readily targetable than the the b3a2 junction.13 39
ODN may, in certain situations, act by nonantisense but sequence-specific mechanisms. Examples of inhibitory sequences include four contiguous Guanosine nucleosides40,41 and also the dinucleoside motif CpG, especially if present in the hexamer palindromic sequence AACGTT, which will induce interferon production and augment natural killer cell activity.42 Neither of these motifs are present in either of the sequences used in the present work. Vaerman et al5 have described a TAT sequence at the 3′end of BCR-ABL–directed ODN that may be responsible for nonspecific antiproliferative effects, which may result from its degradation by nucleases and release of nucleoside and nucleotide products. They have argued that such effects strongly question the use of anti–BCR-ABL ODN as purging agents in CML. However, these observations are solely on ODN that are wholly PO-linked and may not apply well to ODN held together by other combinations of internucleoside linkages.10 Although the ODN sequence for the b2a2 target in the present work contains a TAT sequence at the 3′ end, this is linked solely by MP linkages. MP linkages are very resistant to nuclease attack,7 and the current structures are unlikely to induce this nonspecific effect.13
We recently reported the effects of SL-O permeabilization on ODN uptake into marrow and peripheral blood stem cell harvests from CML patients.30 SL-O significantly enhanced ODN uptake in samples which were first selected for CD34+ cells. This effect was achieved without either direct toxicity or inhibition of colony-forming unit–granulocyte-macrophage (CFU-GM) growth. When taken together with the present data, our findings suggest that SL-O permeabilization can be used to enhance the intracellular uptake and molecular effect of BCR-ABL–directed ODN without significant toxicity.
ODN-purged marrow has been used for transplantation in CML. Eight patients have been transplanted with marrow purged with ODN directed against the c-MYB oncogene.43 A single patient has received marrow purged with a 26-mer directed against the b2a2 junction, without evidence of hematological toxicity.44Both these studies used wholly PS-linked ODN and did not use any means of enhancing cellular ODN uptake. We have recently reported the clinical use of SL-O facilitated ODN targeting BCR-ABL in purging harvests from 3 CML patients, with evidence of molecular benefit in the single b2a2-positive patient. All 3 patients underwent transplantation uneventfully, with acceptable engraftment times,45 and 1 remains in hematologic although not cytogenetic remission 11 months after the procedure.
The data of Fig 4B suggest that, at low concentrations of SL-O, some cells may be resistant to SL-O permeabilization and that a more homogeneous population of permeabilized cells may be generated by electroporation. Increasing the concentration of SL-O results in permeabilization of the bulk of the cell population. However, this is achieved at the expense of a certain degree of loss of specificity, because some cells then receive sufficiently high concentrations of ODN for significant antisense effects to occur at sites of partial complementarity in nontargeted mRNA. It is not known whether a degree of loss of specificity might be acceptable in a therapeutic setting to ensure maximum cell permeabilization using SL-O. Alternatively, although electroporation is less efficient in terms of amount of ODN delivered under optimal conditions, it might prove to be the superior technique when all things are considered. Further studies are indicated to examine the application of ODN permeabilization strategies for purging CML cells before autografting.
Supported by the Leukaemia Research Fund and the North West Cancer Research Fund of Great Britain.
Address reprint requests to Richard E. Clark, MD, University Department of Haematology, Royal Liverpool University Hospital, Prescot St, Liverpool, L7 8XP UK.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal