Abstract
NPM-ALK chimeric transcripts, encoded by the t(2;5), lead to an aberrant expression of ALK by CD30+ systemic lymphomas. To determine if t(2;5) is involved in cutaneous lymphoproliferative disorders, we studied 37 CD30+ cutaneous lymphoproliferations, 27 mycosis fungoides (MF), and 16 benign inflammatory disorders (BID). NPM-ALK transcripts were detected by nested reverse transcription-polymerase chain reaction (RT-PCR) in 1 of 11 lymphomatoid papulosis (LyP), 7 of 15 CD30+ primary cutaneous T-cell lymphoma (CTCL), 3 of 11 CD30+ secondary cutaneous lymphoma, 6 of 27 MF, and 1 of 16 BID. However, the expression of NPM-ALK transcripts was not associated with ALK1 immunoreactivity in MF, LyP, or BID cases. Only 1 CD30+ primary CTCL and 3 CD30+ secondary cutaneous lymphoma were ALK1 immunoreactive. The ALK1+cases were also characterized by amplification of tumor-specific genomic breakpoints on derivative chromosome 5. These cases, except for 1 secondary cutaneous lymphoma, were also characterized by reciprocal breakpoints on derivative chromosome 2, leading to the expression of reciprocal ALK-NPM transcripts. Amplification of chromosomal breakpoints on both derivative chromosomes could represent an alternative to conventional cytogenetics for the diagnosis of t(2;5) and seems to be more reliable than the detection of cryptic NPM-ALK transcripts by nested RT-PCR.
UNLIKE SYSTEMIC CD30+lymphomas with cutaneous involvement, CD30+ primary cutaneous T-cell lymphomas (CTCL) are characterized by an indolent course with spontaneous remission and good prognosis.1-4Other CD30+ cutaneous disorders, such as lymphomatoid papulosis (LyP), also belong to the spectrum of CD30+lymphoproliferations and may be associated in some instances with CD30+ CTCL or mycosis fungoides (MF).4-7 No single morphological or biological feature can be used as a gold standard to differentiate CD30+ primary cutaneous lymphoma from CD30+ systemic lymphoma at the time of diagnosis and therapeutic choice.3 Moreover, descriptions of a common clonal cell origin between LyP and CD30+ cutaneous and systemic lymphoma in the same patients8 9 have suggested that these entities may be biologically related in some instances.
The t(2;5)(p23;q35) translocation fuses the NPM (nucleophosmin) gene at 5q35 with the newly identified ALK (anaplastic lymphoma kinase) gene at 2p23.10 This results in the expression of a chimeric fusion protein NPM-ALK/p80 containing the entire intracellular portion of ALK, including its tyrosine kinase domain but lacking its extracellular and transmembrane domains.10,11 Whereas ALK protein is not expressed by normal lymphocytes, the ubiquitously activated NPM promoter drives the expression of a chimeric NPM-ALK protein with oncogenic properties that may contribute to lymphomagenesis.10,12-14 NPM-ALK transcripts have been detected in a significant proportion (ranging from 16% to 66%) of systemic CD30+ T-cell lymphoma.15-20 We and others18 have shown the presence of NPM-ALK transcripts in a subset of CD30+ cutaneous lymphoproliferations, including LyP cases.21 However, several studies have suggested that the absence of t(2;5) may be a common feature of CD30+cutaneous lymphoproliferations, as opposed to its presence in CD30+ systemic lymphomas.17,20,22-24 Such discrepancies have been observed for Hodgkin's disease,25-31 suggesting either polymerase chain reaction (PCR) artifacts or the presence of normal cells expressing NPM-ALK transcripts within the above lymphoproliferations.32 33This led us to further characterize NPM-ALK breakpoints and transcripts in CD30+ cutaneous lymphoproliferations. Therefore, we designed DNA-PCR assays for the detection of the t(2;5) breakpoints on both derivative chromosomes 5 and 2. We also developed a reverse transcription-PCR (RT-PCR) assay for the detection of the ALK-NPM reciprocal transcript. The findings were analyzed in view of the results of immunodetection of the chimeric protein with either the monoclonal ALK1 antibody or the polyclonal anti-p80 antibody. Furthermore, we studied a larger series of CD30+ cutaneous lymphomas, LyP, and a group of MF and benign inflammatory skin disorders (BID) to determine whether NPM-ALK transcripts or protein could be detected in epidermotropic T-cell lymphoma or in benign cutaneous disorders.
MATERIALS AND METHODS
Patient samples.
This multicentric study included 37 patients with a CD30+cutaneous lymphoproliferation. Twenty-three of them were included in a previous study.21 The diagnosis was revised by the French Study Group of Cutaneous Lymphoma both for clinical and histopathological data, especially for LyP cases. Expression of CD30 antigen by more than 75% of lymphomatous cells was required for the diagnosis of CD30+ lymphoma. After a complete initial staging procedure and a minimum 6 months of clinical follow-up, these 37 cases were divided into three anatomo-clinical groups: (1) LyP (n = 11), (2) CD30+ primary CTCL without extracutaneous involvement for at least 6 months after diagnosis (n = 15, including 3 cases with a past record of LyP), and (3) CD30+ secondary cutaneous lymphoma arising either in the course of a CD30+systemic lymphoma (n = 4) or de novo with concomitant cutaneous and systemic involvement (n = 7). In addition, 27 MF and 16 BID such as eczemas (n =12) and psoriasis (n = 4) were studied by RT-PCR, DNA-PCR, and immunohistochemistry.
Immunohistochemistry.
After a high-pressure cooking antigen-retrieval procedure, a three-stage streptavidin-peroxidase assay (Dako, Les Ullis, France) was used for the detection of the CD30, CD3, and L26 antigens (Dako). The LSAB kit was used with the monoclonal ALK1 antibody34 and a StreptABC HRPkit (Dako) was used with the polyclonal anti-p8035 (Nichirei Co, Tokyo, Japan). These antibodies were both applied at a 1:50 dilution for 16 hours at 4°C.
Primers and probes.
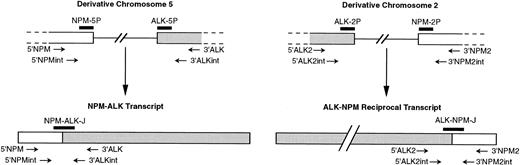
All primers were purchased from Eurogentec (Seraing, Belgium). Standard procedures of RT-PCR for NPM-ALK transcript and genomic PCR (DNA-PCR) on derivative chromosome 5 used the primers 5′NPM (5′-TCCCTTGGGGGCTTTGAAATAACACC-3′) and 3′ALK (5′-CGAGGTGCGGAGCTTGCTCAGC-3′), whereas nested procedures used the internal primers 5′NPMint (5′CCAGTGGTCTTAAGGTTGAAG-3′) and 3′ALKint (5′-TTGTACTCAGGGCTCTGCAGC-3′), as previously described (Fig 1).21 To amplify the reciprocal ALK-NPM breakpoints and cDNA, for the standard PCR we used the primers 5′ALK2 (5′-ATCCTCTCTGTGGTGACCTC-3′) and 3′NPM2 (5′-TGGAACCTTGCTACCACCTC-3′).36Internal primers 5′ALK2int (5′-CCCTCGTGGCCGCCCTGGTC-3′) and 3′NPM2int (5′-GGGGCAGACCGCTTTCCAGA-3′) were designed for nested rounds. Amplifications were performed on a Hybaid OmniGene automated thermal cycler (Hybaid, Teddington, UK). A junction-specific oligoprobe NPM-ALK-J (5′GCTCCTGGTGCTTCCGGCGGTACACTACTAAGTGCTGTCCACT-3′) was used for Southern hybridization of NPM-ALK amplified cDNA, as previously described.21 Southern analysis of the reciprocal ALK-NPM amplified cDNA used the junction-specific oligoprobe ALK-NPM-J (5′-TCCGGCATCATGATTGCTGTGGAGGAAGAT-3′). Southern hybridization of DNA-PCR products was performed with two exonic probes flanking both sides of the breakpoint either on derivative chromosome 5 (NPM-5P, 5′-GTGGTTCAGGGCCAGTGCATATT-3′, and ALK-5P, 5′-ATCTGCATGGCTTGCAGCTCCTG-3′) or on derivative chromosome 2 (NPM-2P, 5′-TATACTTAAGAGTTTCACATCC-3′, and ALK-2P, 5′-GTCCTGGCTTTCTCCGGCAT-3′).
Schematic diagram of NPM and ALK regions rearranged by t(2;5) (according to Ladanyi and Cavalchire55) and mRNA. NPM and ALK exons, whose normal sizes are unknown, are shown as open boxes, and single lines represent introns. The relative approximate positions of PCR primers and probes used in our study are indicated.
Schematic diagram of NPM and ALK regions rearranged by t(2;5) (according to Ladanyi and Cavalchire55) and mRNA. NPM and ALK exons, whose normal sizes are unknown, are shown as open boxes, and single lines represent introns. The relative approximate positions of PCR primers and probes used in our study are indicated.
RT-PCR detection of chimeric t(2;5)-encoded NPM-ALK transcript and ALK-NPM reciprocal transcript.
RT-PCR was performed on total RNA extracted with Trizol reagent (GIBCO-BRL, Gaithersburg, MD) from a 500-μm thick section of frozen skin biopsy.21 The final reaction mix of reverse transcription contained 1.5 μg of total RNA, 500 pmol of random hexamers [pd(N)6; Boehringer Mannheim, Mannheim, Germany], 50 mmol/L Tris-HCl, pH 8.3, 75 mmol/L KCl, 3 mmol/L MgCl2, 500 μmol/L of each deoxynucleotide-5′-triphosphate, and 300 U of Superscript II RT (GIBCO-BRL) in a final volume of 25 μL. Reverse transcription was performed at 37°C for 60 minutes. After heat-inactivation of the reverse transcriptase at 95°C for 2 minutes, half of the cDNA was amplified by PCR. The final reaction mix contained 10 mmol/L Tris-HCl, pH 9, 50 mmol/L KCl, 1.5 mmol/L MgCl2, 200 μmol/L of each deoxynucleotide-5′-triphosphate, 0.1% Triton X-100, 25 pmol of each primer (5′NPM and 3′ALK for NPM-ALK amplification and 5′ALK2 and 3′NPM2 for ALK-NPM amplification), and 1.5 U of Taq polymerase (Promega, Madison, WI) in a final volume of 100 μL overlaid with 75 μL of mineral oil. Tubes were heated at 94°C for 5 minutes and then were subjected to 40 cycles of PCR by denaturing at 94°C for 1 minute, annealing at 60°C for 1 minute, and extending at 72°C for 2 minutes with a touch-down protocol that decreased the annealing temperature by 1°C every 6 cycles from 60°C to 55°C. One microliter of the standard PCR product was used as template for the nested PCR with the following nested primers: 5′NPMint, 3′ALKint for NPM-ALK amplification and 5′ALK2int, 3′ALK2int for ALK-NPM amplification. Each of the 36 amplification cycles was composed of a 1-minute denaturation step at 94°C, a 1-minute annealing step at 60°C, and a 1-minute elongation step at 72°C. The standard and nested RT-PCR products (10 μL) were electrophoresed on a 2% NuSieve Agarose gel (FMC, Rockland, MA), stained with ethidium bromide, and photographed under UV light.
PCR amplification of t(2;5) breakpoints on derivative chromosomes 2 and 5.
DNA was extracted from frozen skin biopsies using a standard phenol:chloroform protocol.37 The reaction mix contained 250 ng of genomic DNA, 10 mmol/L Tris-HCl, pH 8.3, 50 mmol/L KCl, 1.5 mmol/L MgCl2, 200 μmol/L of each deoxynucleotide-5′-triphosphate, 25 pmol of each primer (5′NPM and 3′ALK for NPM-ALK or 5′ALK2 and 3′ALK2 for ALK-NPM), and 1.25 U of AmpliTaq polymerase (Perkin-Elmer, Norwalk, CT) in a final volume of 50 μL overlaid with 75 μL of mineral oil. The tubes were heated at 94°C for 5 minutes and then subjected to 36 cycles of PCR by denaturing at 94°C for 1 minute, annealing at 60°C for 1 minute, and extending at 72°C for 3 minutes with a touch-down protocol that decreased the annealing temperature by 1°C every 6 cycles from 60°C to 55°C. Three microliters of a 1:8,000 dilution of the standard PCR products was used as template for 36 cycles of nested PCR with the internal primers (5′NPMint and 3′ALKint for NPM-ALK or 5′ALK2int, and 3′ALK2int for ALK-NPM). Standard or nested PCR products (10 μL) were electrophoresed on a 1% NuSieve Agarose gel, stained with ethidium bromide, visualized, and photographed under UV light.
Controls.
For the RT-PCR and DNA-PCR assays, the cDNA and genomic DNA of SU-DHL1 cell line (gift of Dr M. Cleary, Stanford University, Stanford, CA) were used as positive controls. Titration studies were performed for the DNA-PCR on derivative chromosomes 2 and 5, using SU-DHL1 DNA diluted in reactive lymph node DNA. The amplification of a 3,016-bp β-globin gene fragment was performed with primers 5′Globin (5′-GAAGAGCCAAGGACAGGTAC-3′) and 3′Globin (5′-GTTTGATGTAGCCTCACTTC-3′) for all DNA samples to check the feasibility of long-range PCR. To avoid cross-contamination, extraction of nucleic acids was performed in an independent laboratory. Amplification, electrophoresis, and nested procedure were performed in separate rooms. Nested RT-PCR without the reverse transcription step and amplification of the reaction mix without template were performed as negative controls. All results were reproduced by two independent experiments for each sample.
Southern blot analysis.
PCR products (10 μL) electrophoresed on agarose gels were blotted onto nylon membranes (Hybond-N+; Amersham International, Buckinghamshire, UK). Membranes were prehybridized and then hybridized at 42°C overnight in a solution of 5× standard saline citrate (SSC), 5× Denhardt's solution, 0.5% sodium dodecyl sulfate (SDS), 0.2 g/L of salmon testes sonicated denatured DNA (Sigma, St Louis, MO), and the appropriate α32P-dATP oligonucleotide probe labeled at the 3′end using terminal transferase (GIBCO-BRL). Blots were washed twice in 2× SSC, 0.1% SDS for 10 minutes at room temperature and then twice in 1× SSC, 0.1% SDS for 20 minutes at 5°C below the theoretical Tm. Blots were exposed to x-ray film (Kodak X-Omat, Rochester, NY) with intensifying screens at −80°C. Probes were removed from membranes by the dehybridization procedure described by the manufacturer to be analyzed with another probe after a radioautographic control.
Sequencing.
PCR products (35 μL) were purified through MicroSpin S-300 Columns (Pharmacia Biotech, Uppsala, Sweden) and sequenced on both DNA strands using the ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction Kit (Perkin Elmer Applied Biosystems, Foster City, CA) on an automated Applied ABI 377A DNA sequencer (Perkin Elmer Applied Biosystems). Nucleotide sequence data were analyzed using the Sequence Navigator software (Perkin Elmer Applied Biosystems). Sequence comparisons were made with the Genbank database by using the Wisconsin Package (Genetics Computer Group, Inc, Madison, WI), FASTA38and BLAST39 programs.
RESULTS
The age of the patients with CD30+ cutaneous lymphoproliferations (20 men and 17 women) ranged from 5 to 92 years (median, 49 years; Table 1). Thirty-four cases had a T-cell phenotype and 1 case of secondary cutaneous lymphoma had a B-cell phenotype. Two cases of CD30+ primary cutaneous lymphoma had a null phenotype, but the genomic study showed a monoclonal rearrangement of the TCRγ chain gene in both cases (data not shown).
Correlation Between DNA-PCR for the Derivative Chromosome 5 and Chromosome 2, RT-PCR for the Reciprocal Translocation, and Immunohistochemistry Using ALK1 and Anti-p80 Antibodies in the Cases With Amplifiable NPM-ALK Transcripts by RT-PCR
| . | Case No. . | Chromosome 5 . | IHC ALK1 . | IHC p80 . | Chromosome 2 . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| RT-PCR . | Nested RT-PCR . | DNA-PCR . | Nested DNA-PCR . | RT-PCR . | Nested RT-PCR . | DNA-PCR . | Nested DNA-PCR . | ||||
| Secondary CLCL | 1 | + | + | + | + | + | + | − | − | +* | − |
| 2 | + | + | + | + | + | + | − | + | + | + | |
| 3 | + | + | + | + | + | + | − | + | + | + | |
| Lymphomatoid papulosis | 4 | − | + | − | −-151 | − | + | − | − | − | − |
| Primary CLCL | 5 | − | + | − | − | − | + | − | − | − | − |
| 6 | − | + | − | − | − | + | − | − | − | − | |
| 7 | − | + | − | +-151 | − | − | − | − | − | − | |
| 8 | + | + | + | + | + | + | − | + | + | + | |
| 9 | − | + | − | +-151 | − | + | − | − | − | − | |
| 10 | + | + | − | − | − | − | − | − | − | − | |
| 11 | − | + | − | − | − | − | − | − | − | − | |
| . | Case No. . | Chromosome 5 . | IHC ALK1 . | IHC p80 . | Chromosome 2 . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| RT-PCR . | Nested RT-PCR . | DNA-PCR . | Nested DNA-PCR . | RT-PCR . | Nested RT-PCR . | DNA-PCR . | Nested DNA-PCR . | ||||
| Secondary CLCL | 1 | + | + | + | + | + | + | − | − | +* | − |
| 2 | + | + | + | + | + | + | − | + | + | + | |
| 3 | + | + | + | + | + | + | − | + | + | + | |
| Lymphomatoid papulosis | 4 | − | + | − | −-151 | − | + | − | − | − | − |
| Primary CLCL | 5 | − | + | − | − | − | + | − | − | − | − |
| 6 | − | + | − | − | − | + | − | − | − | − | |
| 7 | − | + | − | +-151 | − | − | − | − | − | − | |
| 8 | + | + | + | + | + | + | − | + | + | + | |
| 9 | − | + | − | +-151 | − | + | − | − | − | − | |
| 10 | + | + | − | − | − | − | − | − | − | − | |
| 11 | − | + | − | − | − | − | − | − | − | − | |
*Positive Southern hybridization with NPM-2P but not with ALK-2P.
Positive Southern hybridization with ALK-5P but not with NPM-5P.
RT-PCR for NPM-ALK transcripts.
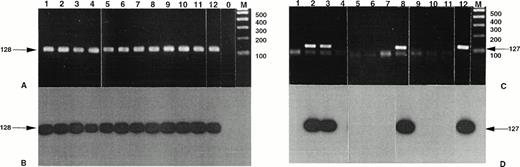
Standard PCR of the RT-PCR assay allowed the detection of NPM-ALK transcripts in 2 of the 15 CD30+ primary CTCL (cases no. 8 and 10) and in 3 of the 11 CD30+ secondary cutaneous lymphoma (cases no. 1, 2, and 3; Fig 2 and data not shown). Furthermore, the nested PCR allowed the detection of the NPM-ALK transcript in 1 of the 11 LyP (case no. 4); in 7 (cases no. 5 through 11) of the 15 CD30+ primary CTCL, including the above-mentioned 2 cases; and in the 3 previously detected cases of the 11 CD30+ secondary cutaneous lymphomas. After ethidium bromide staining showing the same sized amplicons, these results were confirmed both by direct sequencing and by Southern blot hybridization with the junction-specific oligoprobe NPM-ALK-J. The study of MF and BID samples did not show any NPM-ALK transcript after the standard PCR of the RT-PCR assay. However, nested amplification showed NPM-ALK specific amplicons in 6 of the 27 MF and in 1 eczema of the 16 BID samples (data not shown).
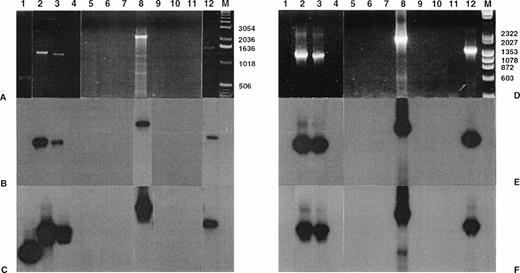
Detection of NPM-ALK and reciprocal ALK-NPM transcripts by nested RT-PCR. Total RNA was extracted from frozen skin biopsies or cultured cells and submitted to nested RT-PCR analysis, followed by electrophoresis on 2% agarose gel. Lanes 1, 2 and 3, cases no. 1, 2, and 3, respectively, CD30+ secondary CLCL; lane 4, case no. 4, LyP; lanes 5 through 11, cases no. 5 through 11, CD30+ primary CLCL; lane 12, t(2;5)+SU-DHL-1 cell line; lane 0, no template; lane M, molecular weight marker 100-bp DNA ladder (GIBCO-BRL). Detection of NPM-ALK transcripts: (A) ethidium bromide staining and (B) radioautography. Detection of ALK-NPM transcripts: (C) ethidium bromide staining and (D) radioautography. The gels were transferred to a nylon membrane, hybridized either with the NPM-ALK-J probe (B) or the ALK-NPM-J probe (D), and radioautographed. Sizes are indicated in bases.
Detection of NPM-ALK and reciprocal ALK-NPM transcripts by nested RT-PCR. Total RNA was extracted from frozen skin biopsies or cultured cells and submitted to nested RT-PCR analysis, followed by electrophoresis on 2% agarose gel. Lanes 1, 2 and 3, cases no. 1, 2, and 3, respectively, CD30+ secondary CLCL; lane 4, case no. 4, LyP; lanes 5 through 11, cases no. 5 through 11, CD30+ primary CLCL; lane 12, t(2;5)+SU-DHL-1 cell line; lane 0, no template; lane M, molecular weight marker 100-bp DNA ladder (GIBCO-BRL). Detection of NPM-ALK transcripts: (A) ethidium bromide staining and (B) radioautography. Detection of ALK-NPM transcripts: (C) ethidium bromide staining and (D) radioautography. The gels were transferred to a nylon membrane, hybridized either with the NPM-ALK-J probe (B) or the ALK-NPM-J probe (D), and radioautographed. Sizes are indicated in bases.
Immunohistochemistry.
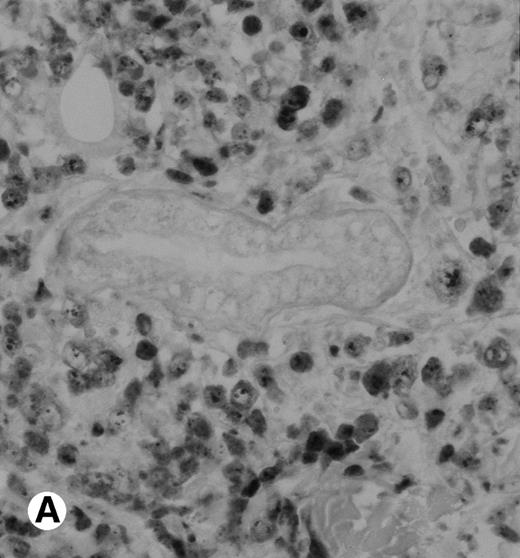
The polyclonal anti-p80 provided a staining of some large cells of 1 of the 11 LyP (case no. 4). A cytoplasmic staining of lymphomatous cells was also seen in 4 of the 15 CD30+ primary CTCL (cases no. 5, 6, 8, and 9) and in 3 of the 11 CD30+ secondary cutaneous lymphomas (cases no. 1, 2, and 3). Keratinocytes or dendritic cells of the dermis were sometimes stained by anti-p80. All p80+ cases were previously shown to contain NPM-ALK transcripts by nested RT-PCR. However, not all cases with NPM-ALK chimeric transcripts were stained by p80+. No p80+ cells were detected in MF and BID sections. The staining with the monoclonal ALK1 antibody was cytoplasmic and nucleolar and restricted to tumoral cells of CD30+lymphomas. Only 1 case of CD30+ primary CTCL was ALK1+ (case no. 8; Fig 3). This case was 1 of the 2 cases with a positive standard RT-PCR amplification, whereas the other 1 was found to be negative for both p80 and ALK1 immunostaining (case no. 10). The 3 cases of CD30+ secondary CTCL with NPM-ALK transcripts were stained for ALK1 (cases no. 1, 2, and 3; Fig 3). No ALK1-immunoreactive cell was found in LyP sections and no labeling of the epidermis was observed. None of the cases with a negative NPM-ALK detection by RT-PCR and none of the MF and BID was found to be stained for ALK1.
Immunohistochemical detection of chimeric NPM-ALK protein (×400). A case of CD30+ primary CLCL (case no. 8; A) and a case of CD30+ secondary CLCL (case no. 1; B) both with a positive NPM-ALK amplification by both RT-PCR and DNA-PCR were stained with the monoclonal ALK1 antibody. A granular cytoplasmic and strong nucleolar staining was observed on the large lymphomatous cells in these 2 cases.
Immunohistochemical detection of chimeric NPM-ALK protein (×400). A case of CD30+ primary CLCL (case no. 8; A) and a case of CD30+ secondary CLCL (case no. 1; B) both with a positive NPM-ALK amplification by both RT-PCR and DNA-PCR were stained with the monoclonal ALK1 antibody. A granular cytoplasmic and strong nucleolar staining was observed on the large lymphomatous cells in these 2 cases.
Amplification of t(2;5) breakpoint on derivative chromosome 5.
After standard DNA-PCR, 4 of the 11 cases with a chimeric transcript detected by nested RT-PCR showed a specific amplicon (Fig 4). The size of the chimeric amplicons varied from case to case, ranging between 0.8 and 2.9 kb, according to variable intronic breakpoints.23,40 These cases, which were previously found to contain NPM-ALK transcripts by standard RT-PCR, corresponded to 3 CD30+ secondary CTCL (cases no. 1, 2, and 3) and 1 CD30+ primary CTCL (case no. 8). The nested amplification allowed the detection of size-specific amplicons in the same cases (no. 1, 2, 3, and 8) and in 2 additional CD30+primary CTCL (cases no. 7 and 9). Titration experiments showed that the sensitivity of DNA-PCR was 10−3 for the standard PCR and 10−4 for the nested PCR (data not shown). Southern blot hybridization of PCR products confirmed the specificity of these results, giving a positive signal with both NPM-5P and ALK-5P in the 3 CD30+ secondary CTCL and in one CD30+primary CTCL (case no. 8). However, the 2 other CD30+primary CTCL with a visible amplicon after nested DNA-PCR (cases no. 7 and 9) hybridized only with ALK-5P but not with NPM-5P. Moreover, 1 case of LyP (case no. 4) was shown to give a positive hybridization only with ALK-5P, although no signal was detectable on ethidium bromide-stained gel. The nested DNA-PCR amplicon of case no. 7 was sequenced and found to contain the 3′ end of the ALK exon targeted by the 3′ALKint oligonucleotide and the flanking intron, but lacked the 5′ end of the NPM exon targeted by the 5′NPMint oligonucleotide. In cases no. 4 and 9, the amount of amplicon was too low to perform sequencing. Size-specific amplicons of cases no. 1, 2, 3, and 8 were sequenced and shown to contain chimeric intronic sequences between the flanking 5′NPM and 3′ALK exons. Multiple alignment analysis of these sequences with the t(2;5) genomic nucleotide sequence of SU-DHL-122 showed a perfect homology of the ranging from both exonic extremities over a variable intronic area from case to case depending on the location of the breakpoint (data not shown). In contrast, all MF and BID cases were negative even after nested DNA-PCR.
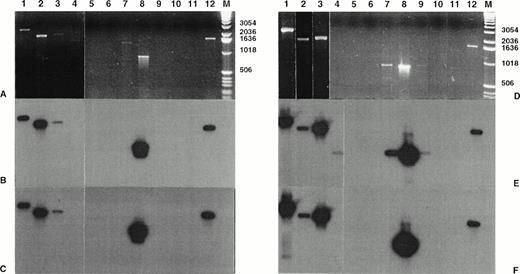
Amplification of genomic breakpoint on derivative chromosome 5. Genomic DNA was subjected to standard and nested amplification, followed by product separation on a 1% agarose gel. Ethidium bromide staining of standard DNA-PCR products (A) and of nested DNA-PCR products (D). Lanes 1 through 11, cases no. 1 through 11; lane 12, t(2;5)+ SU-DHL-1 cell line; lane M, molecular weight marker 1-kb DNA ladder (GIBCO-BRL). The gels were transferred to a nylon membrane and hybridized either with the ALK-5P (radioautographies B and E) or the NPM-5P (radioautographies C and F). The sizes are indicated in bases.
Amplification of genomic breakpoint on derivative chromosome 5. Genomic DNA was subjected to standard and nested amplification, followed by product separation on a 1% agarose gel. Ethidium bromide staining of standard DNA-PCR products (A) and of nested DNA-PCR products (D). Lanes 1 through 11, cases no. 1 through 11; lane 12, t(2;5)+ SU-DHL-1 cell line; lane M, molecular weight marker 1-kb DNA ladder (GIBCO-BRL). The gels were transferred to a nylon membrane and hybridized either with the ALK-5P (radioautographies B and E) or the NPM-5P (radioautographies C and F). The sizes are indicated in bases.
Amplification of t(2;5) reciprocal breakpoint on derivative chromosome 2.
Standard DNA-PCR allowed the detection of amplicons (0.6 to 2.2 kb) by gel staining in 3 CD30+ secondary CTCL (cases no. 1, 2, and 3) and in 1 CD30+ primary CTCL (case no. 8; Fig 5). These cases (no. 1, 2, 3, and 8) were also positive at the DNA-PCR level on derivative chromosome 5 and by both standard and nested RT-PCR analysis. Size-specific amplicons were also obtained by nested DNA-PCR for cases no. 2, 3, and 8 but not for case no. 1. Titration study showed that the sensitivity of this DNA-PCR was 10−2 for the standard PCR and 10−3 for the nested PCR (results not shown). Southern blot hybridization with both NPM-2P and ALK-2P confirmed these results in 3 of the 4 cases. In case no. 1 (CD30+ secondary CTCL), a positive hybridization of standard PCR products, was obtained only with NPM-2P, but not with ALK-2P. Sequence analysis showed that the standard DNA-PCR amplicon of case no. 1 contained the 3′ end of the NPM exon targeted by the 3′NPM2int oligonucleotide and the flanking intron, but lacked the 5′ end of the ALK exon targeted by the 5′ALK2int primer. Amplicons of the nested DNA-PCR in cases no. 2, 3, and 8 and the SU-DHL-1 cell line were purified and sequenced (Fig 6). Multiple alignment analysis of these sequences showed a perfect homology of chimeric intronic sequences ranging from both exonic extremities over a variable area from case to case depending on the position of the breakpoint (data not shown). The complete sequence of the reciprocal breakpoint DNA fragment of the SU-DHL1 was characterized (Genbank accession no. AF032882). No genomic breakpoint was amplified in the MF and BID cases.
Amplification of genomic breakpoint on derivative chromosome 2. Genomic DNA was subjected to standard and nested amplification, followed by product separation on a 1% agarose gel. Ethidium bromide staining of standard DNA-PCR products (A) and of the nested DNA-PCR products (D). Lanes 1 through 11, cases 1 through 11; lane 12, t(2;5)+ SU-DHL-1 cell line; lane M, molecular weight marker 1-kb DNA ladder (GIBCO-BRL). The gels were transferred to a nylon membrane and hybridized either with the ALK-2P (radioautographies B and E) or the NPM-2P (radioautographies C and F). The sizes are indicated in bases.
Amplification of genomic breakpoint on derivative chromosome 2. Genomic DNA was subjected to standard and nested amplification, followed by product separation on a 1% agarose gel. Ethidium bromide staining of standard DNA-PCR products (A) and of the nested DNA-PCR products (D). Lanes 1 through 11, cases 1 through 11; lane 12, t(2;5)+ SU-DHL-1 cell line; lane M, molecular weight marker 1-kb DNA ladder (GIBCO-BRL). The gels were transferred to a nylon membrane and hybridized either with the ALK-2P (radioautographies B and E) or the NPM-2P (radioautographies C and F). The sizes are indicated in bases.
Genomic nucleotide sequence of the reciprocal translocation on the derivative chromosome 2 in SU-DHL1. The shaded boxes at the 5′ end and at the 3′ end of the sequence contain, respectively, the ALK exon sequence and the NPM exon sequence flanking the breakpoint.
Genomic nucleotide sequence of the reciprocal translocation on the derivative chromosome 2 in SU-DHL1. The shaded boxes at the 5′ end and at the 3′ end of the sequence contain, respectively, the ALK exon sequence and the NPM exon sequence flanking the breakpoint.
Detection of ALK-NPM reciprocal transcript.
The standard RT-PCR assay showed no amplicon after electrophoresis staining (Fig 2 and data not shown). Nested PCR allowed the amplification of the same-sized products (127 bp) visible on gel staining for the same 3 cases positive with the DNA-PCR assay on derivative chromosome 2 (cases no. 2, 3, and 8; 2 CD30+secondary CTCL and 1 CD30+ primary CTCL). The specificity of the results was confirmed by Southern blotting with ALKNPM-J probe and by DNA sequencing of the nested RT-PCR products. No reciprocal transcript was detected in the other cases including MF and BID cases even after nested RT-PCR and Southern blotting.
DISCUSSION
Our study demonstrates that lymphoproliferative or inflammatory cutaneous diseases may contain NPM-ALK transcripts in some instances in which no genomic breakpoints or expression of the chimeric NPM-ALK protein can be detected. PCR contamination or artifacts were ruled out by extensive controls such as negative amplification without the reverse transcription step, hybridization, and sequencing of NPM-ALK amplicons. Differences in the detection threshold of the different PCR techniques may give a rationale for these discrepancies and the differences between our previous results21,41 and those of other groups that did not detect NPM-ALK transcripts or breakpoints in CD30+ primary cutaneous lymphoproliferations.17,20,22,24 These groups did not perform a nested RT-PCR amplification that appeared in our hand as the most sensitive assay, reaching a sensitivity threshold of 1:106. In addition, cells carrying t(2;5) theoretically contain only one copy of the NPM-ALK chimeric gene on the derivative chromosome 5, which is transcribed in several copies of mRNA. Therefore, nested RT-PCR probably allows the detection of chimeric transcripts in a higher number of cases than DNA-PCR. Such a nested RT-PCR proved to be a reliable technique for the detection of NPM-ALK transcripts in nodal or systemic CD30+lymphomas.15,31 42 In cutaneous samples, a small number of CD30+ cells may be intermingled within normal or inflammatory cells, especially in LyP cases. However, ALK immunoreactivity and t(2;5) breakpoints were detected in only 1 of 7 CD30+ cutaneous lymphoproliferations and in no BID or MF cases containing NPM-ALK transcripts.
The existence of scarce cells expressing chimeric transcripts could also explain our results. Whether these bystander cells are normal or tumoral cells cannot be addressed but similar results have been obtained by nested PCR assay for the detection of t(14;18) and t(9;22) in normal tissues or blood samples.43-46 Therefore, the detection of chimeric transcripts may overevaluate the implication of t(2;5) in the genesis of CD30+ cutaneous lymphoproliferations, because ALK immunoreactivity was not found in most cases. Moreover, NPM-ALK transcripts were detected in some MF or BID cases that did not contain CD30+ cells. Similarly, NPM-ALK transcripts have frequently been detected in peripheral blood cells of healthy donors by using nested RT-PCR assay.47Such data are to be likened to the discordant results obtained by several groups in Hodgkin's disease.26,27,33 48
Immunohistochemistry with monoclonal ALK1 and polyclonal anti-p80 antibodies provided a concordant staining of tumoral CD30+cells in 1 CTCL and 3 secondary CTCL. These cases were further characterized by t(2;5) breakpoints. In addition, the p80 antibody also stained large cells in 1 LyP case and in 3 CTCL. In the latter cases, p80 staining was not correlated with the detection of the (2;5) breakpoint. We previously observed a p80 staining of large dendritic cells of the dermis.21 Moreover, several groups found p80 staining difficult to interpret in some instances.31,34This may depend on the fixative or the fixation time, especially for small specimens such as skin biopsies. Nonetheless, the ALK1 antibody was restricted to CD30+ lymphoid cells.3,31However, as for bcl2 immunoreactivity,49 ALK expression is not specific for the 2;5 chromosomal translocation. ALK1 recognizes a part of the tyrosine kinase domain of ALK, and other molecular events such as t(1;2) or inversion on chromosome 2 may lead to an aberrant ALK immunoreactivity of T-cell lymphomas.31,34,50 In addition, a new subtype of large B-cell lymphomas was found to express the entire ALK protein in the absence of t(2;5) translocation.51 Both clinical and biological differences may exist between NPM-ALK+ and ALK+ lymphoma. Moreover, NPM sequences proved to be necessary for the oncogenicity of the chimeric protein.12,13 Therefore, the identification of t(2;5)+ lymphomas requires a combination of molecular and immunohistochemical evidence in the lack of conventional cytogenetics. In a few recent studies,19,22 DNA-PCR has been proposed as an alternative procedure to conventional cytogenetics beside RT-PCR. Indeed, genomic breakpoints are specific for each tumor and the detection on both derivative chromosomes 2 and 5 could be a specific PCR assay for the diagnosis of t(2;5). The size of the NPM and ALK introns was confirmed to be short, approximately 1 and 2 kb, respectively, suggesting that the largest possible size for the chimeric NPM-ALK intron is about 3 kb.36 Indeed, the sum of the sizes of the DNA-PCR products in our cases was about 3 kb. However, our study points to the need to check size-specific amplicons obtained by DNA-PCR, either by Southern hybridization with probes complementary to both breakpoint sides or by sequencing. Size-specific amplicons may be generated by PCR mispriming, as observed on derivative chromosome 5 for case 7 that contained only exonic and intronic ALK sequences, thus hybridizing only with ALK-5P.
Amplification of the reciprocal ALK-NPM breakpoint on derivative chromosome 2 was correlated with the ALK1+ immunophenotype and with the amplification of the NPM-ALK breakpoint in all cases but 1. In this case (case no. 1), characterized by the presence of a derivative chromosome 5 breakpoint, a small reciprocal chromosome fragment, here 5q35, could have been lost on derivative chromosome 2, as described for reciprocal translocations involving chromosome.15 52 A mispriming of 5′ALK2 primer within intronic sequences of either NPM genes or NPM pseudogenes, both on a nontranslocated chromosome 5, could explain this result. Therefore, DNA-PCR for the amplification of derivative chromosome 2 breakpoint provided an amplicon containing NPM exonic and intronic sequences but no ALK sequences. Furthermore, the amplification of the derivative chromosome 2 breakpoint allowed us to further characterize the genomic nucleotide sequence of the SU-DHL1 cell line.
The reciprocal breakpoint was also shown to encode for chimeric reciprocal ALK-NPM transcripts in all cases with positive DNA-PCR on derivative chromosome 2. However, these transcripts were detected only by nested RT-PCR, which may indicate a low level of expression. This would be expected, because ALK promoter is normally silent in normal lymphoid cells.10 Whether reciprocal ALK-NPM transcripts have a biological function needs to be elucidated as for other reciprocal translocations, such as t(9;22) or t(12;21).53 54
Finally, our findings may be reconciled with those obtained by several groups in cutaneous CD30+ disorders. Firstly, t(2;5)-positive CD30+ primary CTCL may exist but appear to be very rare as opposed to t(2;5)-positive CD30+ secondary cutaneous lymphomas (1/26 v 3/11). The t(2;5) appeared not to be implicated in LyP (no amplification of genomic breakpoints, no ALK1 immunoreactivity), as previously reported.22,23 34Secondly, the presence of NPM-ALK transcripts, detected both in cutaneous lymphoproliferative and inflammatory disorders, does not allow the distinction between CD30+ primary and secondary cutaneous lymphomas by nested RT-PCR alone. Thirdly, ALK1 immunostaining was found to be specific for cutaneous lymphomas that harbor t(2;5) by genomic PCR. In the absence of conventional cytogenetics, the amplification of chromosomal breakpoints on either derivative chromosomes 2 or 5 provided tumor-specific molecular evidence for the presence of t(2;5) among lymphomas expressing NPM-ALK transcripts. Our study clearly demonstrates the specificity of such DNA-breakpoint amplification versus the detection of NPM-ALK transcripts for the diagnosis and monitoring of patients with t(2;5)-positive lymphomas.
ACKNOWLEDGMENT
The following members of the French Study Group of Cutaneous Lymphoma have contributed to this study: M.F. Avril and J. Bosq (Villejuif), M. Bagot and J. Wechsler (Creteil), L. Vaillant and A. de Muret (Tours), C. Beylot (Pessac-Bordeaux), M. Delaunay (Bordeaux), S. Dalac and T. Petrella (Dijon), P. Joly and E. Thomine (Rouen), and C. Bodemer and S. Fraitag (Necker-Paris). We also thank J. Ferrer, C. Bartoli, J.C. Garroste, and M. Turmo for their expert technical assistance and J.P. Javerzat (Genetic Laboratory CNRS 9026) for his contribution. We thank Dr M.L. Cleary for the gift of the SU-DHL1 cell line.
Supported by grants from the Région Aquitaine and Comité de Gironde et de Charente de la Ligue Contre le Cancer. K.P. was supported by the Leukaemia Research Fund.
Address reprint requests to M. Beylot-Barry, MD, Equipe Histologie et Pathologie du Système Immunitaire, Laboratoire d'Histologie-Embryologie, UFR 3, Université de Bordeaux 2, 33076 Bordeaux Cedex, France; e-mail:Marie.Beylot-Barry@histo.u-bordeaux2.fr.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal