Abstract
The translocation t(10;11)(p13;q14) has been observed in acute lymphoblastic leukemia (ALL) as well as acute myeloid leukemia (AML). A recent study showed a MLL/AF10 fusion in all cases of AML with t(10;11) and various breakpoints on chromosome 11 ranging from q13 to q23. We recently cloned CALM (Clathrin Assembly Lymphoid Myeloid leukemia gene), the fusion partner of AF10 at 11q14 in the monocytic cell line U937. To further define the role of these genes in acute leukemias, 10 cases (9 AML and 1 ALL) with cytogenetically proven t(10;11)(p12-14;q13-21) and well-characterized morphology, immunophenotype, and clinical course were analyzed. Interphase fluorescence in situ hybridization (FISH) was performed with 2 YACs flanking the CALM region, a YAC contig of the MLLregion, and a YAC spanning the AF10 breakpoint. Rearrangement of at least one of these genes was detected in all cases with balanced t(10;11). In 4 cases, including 3 AML with immature morphology (1 AML-M0 and 2 AML-M1) and 1 ALL, the signals of the CALM YACS were separated in interphase cells, indicating a translocation breakpoint within the CALM region. MLL was rearranged in 3 AML with myelomonocytic differentiation (2 AML-M2 and 1 AML-M5), including 1 secondary AML. In all 3 cases, a characteristic immunophenotype was identified (CD4+, CD13−, CD33+, CD65s+).AF-10 was involved in 5 of 6 evaluable cases, including 1 case without detectable CALM or MLL rearrangement. In 2 complex translocations, none of the three genes was rearranged. All cases had a remarkably poor prognosis, with a mean survival of 9.6 ± 6.6 months. For the 7 AML cases that were uniformly treated according to the AMLCG86/92 protocols, disease-free and overall survival was significantly worse than for the overall study group (P = .03 and P = .01, respectively). We conclude that the t(10;11)(p13;q14) indicates CALM and MLL rearrangements in morphologically distinct subsets of acute leukemia and may be associated with a poor prognosis.
MALIGNANT TRANSFORMATION is a multistep process that involves the sequential alteration of genes regulating cell growth and/or differentiation. Many of the genes involved in leukemogenesis have been identified by the molecular analysis of nonrandom chromosome abnormalities.1
The rare, but recurring translocation t(10;11)(p13;q14) has been observed in acute lymphoblastic leukemia (ALL) as well as acute myeloid leukemia (AML).2-5 A recent study showed a MLL/AF10fusion in all cases of AML with t(10;11) and various breakpoints on chromosome 11q, including 6 cases with rearrangement of chromosomal band 11q13.6MLL (also calledALL1, Htrx1, and HRX) is involved in translocations with up to 40 different translocation partner genes in AML as well as acute lymphoid leukemia, including the t(10;11)(p13;q23) in AML.7-12 In this translocation, the fusion partner genesAF-10 and MLL are often found to be involved in complex rearrangements (inversions/insertions), because they are transcribed in opposite direction relative to the telomere/centromere orientation.6 These complex rearrangements may be interpreted as t(10;11) (p13;q14-21).
The molecular characterization of the t(10;11)(p13;q14) in the monocytic cell line U937 identified a different breakpoint on chromosome 11, involving the previously unknown geneCALM.13,14CALM has a very high homology to the murine clathrin assembly protein ap-3.15Interestingly, both translocations t(10;11) have the same fusion partner gene on chromosome 10, the putative transcription factorAF10.11,14 16
To further define the involvement of the three genes AF10,CALM, and MLL in acute leukemias, we analyzed 10 cases (9 AML and 1 ALL) with cytogenetically proven t(10;11)(p12-14;q13-21) and well-characterized morphology, immunophenotype, and clinical course. Only cytogenetic material stored in fixative for several years was available, which was not suitable for an analysis at the RNA level. An interphase fluorescence in situ hybridization (FISH) approach was chosen.17 Nonchimeric YACs from the genomic region of the three genes were used. A translocation was assumed when one of the YAC signals was split resulting in three hybridization signals in a diploid interphase nucleus.
MATERIALS AND METHODS
Patient samples.
This study was designed and performed according to the updated declaration of Helsinki. Patient material was collected from 725 cases that entered the studies of the German AML Cooperative Group 86 and 92 and that were subjected to cytogenetic analysis.18 Based on the cytogenetic data and the availability of material, 8 cases with t(10;11)(p12-14;q13-21) were identified (Table 1). In addition, 1 adult ALL (no. 10) and 1 infant AML (no. 6) with t(10;11) were included in this series. All cases were morphologically classified according to the French-American-British (FAB) criteria and were reviewed by an independent reference hematologist (H.L.).19Immunophenotyping was performed according to the consensus protocol for flow cytometric immunophenotyping of hematopoietic malignancies.20,21 Clinical data were obtained from the AMLCG study center or directly from the patients' physicians (patients no. 6 and 10). Patients no. 3 and 7 had been previously treated for malignant T-cell lymphoma and were in complete remission. All AML patients received a uniform chemotherapy according to the AMLCG86/92 protocols comprising double induction, followed by intensive consolidation and 3 years of monthly maintenance therapy.18Patient no. 6 (pediatric patient) and patient no. 8 (initially misclassified as ALL) were treated differently (Table 1).
Characteristics of Acute Leukemias With t(10;11)
| No. . | Sex/ Age (yr) . | Leuko . | Diagnosis . | Induction Therapy . | Resp . | Survival (mos) . | Immunophenotype . | Karyotype . | FISH . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EventFree . | Overall . | B-Cell . | T-Cell . | Myeloid . | Other . | AF10 . | CALM . | MLL . | |||||||
| 1 | M/51 | 60,800 | AMLM5A | TAD-HAM | CR | 14 | 15 | CD19− | CD2− CD4+ CD7+ | CD13− CD14− CD15− CD33+ CD65s+ | CD10(+) CD34− HLA-DR+ TDT− | 46,XY, t(10;11)(p13;q14) | Split | N | Split |
| 2 | F/46 | 130,300 | AMLM2 | TAD-TAD | CR | 15 | 19 | CD19− | CD2− CD4+ CD7− | CD13− CD14− CD15− CD33+ CD65s+ | CD10− CD34+ HLA-DR+ TDT− | 48,XX, +6,+8, t(10;11)(p12-13;q14-21) | ND | N | Split |
| 3 | F/39 | 13,800 | sec. AMLM2 | TAD-HAM | ED | <1 | <1 | CD19− | CD2− CD4+ CD7− | CD13− CD14− CD15− CD33+ CD65s+ | CD10− CD34− HLA-DR+ TDT− | 45, X, −X, der(10)t(10;11)(p12-13;q13.1) inv(11)(q13.1q23.1), der(11)t(10;11)(p12-13;q13.1) | ND | N | Split |
| 4 | F/74 | 200 | AMLM5 | TAD-HAM* | CR | 2 | 14 | CD19− | CD2− CD4+ CD7− | CD13− CD14− CD15− CD33+ CD65s+ | CD10− CD34+ HLA-DR+ TDT(+) | 46, XX, t(10;11;17)(p13;q13;q23) | ND | N | ND |
| 5 | F/37 | 4,000 | AMLM1 | TAD-HAM | CR | 8 | 16 | CD19− | CD2+ CD4− CD7− | CD13+ CD14− CD15+ CD33− CD65s+ | CD10− CD34+ HLA-DR+ TDT− | 46, XX, der(10)t(10;11)(p12-13;q21) der(11)t(10;11;11;16) (p12-13;q21;p14-15;q13-22) del(11)(p13), del(12)(p11.2) | N | N | N |
| 6 | M/2 | NA | AMLM5 | AML-BFM87 | NA | NA | 5 | CD19− | CD2− CD4− CD7− | CD13− CD14− CD15+ CD33+ CD65s+ | CD10− CD34− HLA-DR+ TDT− | 46, XY, t(10;11)(p12;q21) | Split | N | N |
| 7 | M/19 | 1,300 | sec. AMLM1 | TAD-HAM | ED | 1 | 1 | CD19− | CD2− CD4− CD7(+) | CD13(+) CD14− CD15− CD33− CD65s− | CD10− CD34+ HLA-DR+ TDT− | 46, XY, t(10;11)(p13;q14) | Split | Split | N |
| 8 | M/47 | 20,000 | AMLM0 | T-ALL | ED | 3 | 3 | CD19− | CD3− | CD13+ CD33+ | ND | 48, XY, +3,add(7)(q33-35), der(9), der(9), +der(9), t(10;11)(p13;q21), der(12)t(12;18)(p11;q11), −18, +mar | Split | Split | N |
| 9 | F/21 | 60,800 | AMLM1 | TAD-HAM | CR | 7 | 11 | ND | ND | ND | ND | 47, XX, +19, t(10;11)(p13-14;q14-21) | Split | Split | N |
| 10 | M/31 | NA | T-ALL | CHOEP | NA | 7 | 11 | CD22− | CD1+ CD3+ CD4+ CD5+ | CD11c− | CD10+ HLA-DR− | 46, XY, del(7)(q34-q35), i(9)(q10), t(10;11)(p1?4;q14), der(17)t(1;17)(p13;p11) | ND | Split | N |
| No. . | Sex/ Age (yr) . | Leuko . | Diagnosis . | Induction Therapy . | Resp . | Survival (mos) . | Immunophenotype . | Karyotype . | FISH . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EventFree . | Overall . | B-Cell . | T-Cell . | Myeloid . | Other . | AF10 . | CALM . | MLL . | |||||||
| 1 | M/51 | 60,800 | AMLM5A | TAD-HAM | CR | 14 | 15 | CD19− | CD2− CD4+ CD7+ | CD13− CD14− CD15− CD33+ CD65s+ | CD10(+) CD34− HLA-DR+ TDT− | 46,XY, t(10;11)(p13;q14) | Split | N | Split |
| 2 | F/46 | 130,300 | AMLM2 | TAD-TAD | CR | 15 | 19 | CD19− | CD2− CD4+ CD7− | CD13− CD14− CD15− CD33+ CD65s+ | CD10− CD34+ HLA-DR+ TDT− | 48,XX, +6,+8, t(10;11)(p12-13;q14-21) | ND | N | Split |
| 3 | F/39 | 13,800 | sec. AMLM2 | TAD-HAM | ED | <1 | <1 | CD19− | CD2− CD4+ CD7− | CD13− CD14− CD15− CD33+ CD65s+ | CD10− CD34− HLA-DR+ TDT− | 45, X, −X, der(10)t(10;11)(p12-13;q13.1) inv(11)(q13.1q23.1), der(11)t(10;11)(p12-13;q13.1) | ND | N | Split |
| 4 | F/74 | 200 | AMLM5 | TAD-HAM* | CR | 2 | 14 | CD19− | CD2− CD4+ CD7− | CD13− CD14− CD15− CD33+ CD65s+ | CD10− CD34+ HLA-DR+ TDT(+) | 46, XX, t(10;11;17)(p13;q13;q23) | ND | N | ND |
| 5 | F/37 | 4,000 | AMLM1 | TAD-HAM | CR | 8 | 16 | CD19− | CD2+ CD4− CD7− | CD13+ CD14− CD15+ CD33− CD65s+ | CD10− CD34+ HLA-DR+ TDT− | 46, XX, der(10)t(10;11)(p12-13;q21) der(11)t(10;11;11;16) (p12-13;q21;p14-15;q13-22) del(11)(p13), del(12)(p11.2) | N | N | N |
| 6 | M/2 | NA | AMLM5 | AML-BFM87 | NA | NA | 5 | CD19− | CD2− CD4− CD7− | CD13− CD14− CD15+ CD33+ CD65s+ | CD10− CD34− HLA-DR+ TDT− | 46, XY, t(10;11)(p12;q21) | Split | N | N |
| 7 | M/19 | 1,300 | sec. AMLM1 | TAD-HAM | ED | 1 | 1 | CD19− | CD2− CD4− CD7(+) | CD13(+) CD14− CD15− CD33− CD65s− | CD10− CD34+ HLA-DR+ TDT− | 46, XY, t(10;11)(p13;q14) | Split | Split | N |
| 8 | M/47 | 20,000 | AMLM0 | T-ALL | ED | 3 | 3 | CD19− | CD3− | CD13+ CD33+ | ND | 48, XY, +3,add(7)(q33-35), der(9), der(9), +der(9), t(10;11)(p13;q21), der(12)t(12;18)(p11;q11), −18, +mar | Split | Split | N |
| 9 | F/21 | 60,800 | AMLM1 | TAD-HAM | CR | 7 | 11 | ND | ND | ND | ND | 47, XX, +19, t(10;11)(p13-14;q14-21) | Split | Split | N |
| 10 | M/31 | NA | T-ALL | CHOEP | NA | 7 | 11 | CD22− | CD1+ CD3+ CD4+ CD5+ | CD11c− | CD10+ HLA-DR− | 46, XY, del(7)(q34-q35), i(9)(q10), t(10;11)(p1?4;q14), der(17)t(1;17)(p13;p11) | ND | Split | N |
Clinical, immunophenotypic, and cytogenetic characteristics and FISH results of acute leukemias (9 AML and 1 ALL) with t(10;11).
Abbreviations: Leuko, leukocytes per microliter at presentation; Resp, response; CR, complete response; ED, early death; NA, not available; (+), weakly positive; N, not split; ND, not done/evaluable (no material left for patients no. 4 and 10; poor sample quality for patients no. 2 and 3); ALL-BFM87, combined chemotherapy following the BFM87 protocol20; T-ALL, intensified combined chemotherapy for adult T-cell lymphoblastic leukemia; CHOEP, combined chemotherapy (Cyclophosphamide/Adriamycin/Vincristin/Etoposide/Prednisolon).
*Reduced dose.
Mononuclear cells of patient samples were grown in short-term culture or processed directly, harvested using standard cytogenetic techniques, and stored in Carnoy's solution until final analysis, as previously published.22
FISH probes.
YAC-derived FISH probes were generated either by a sequence-independent amplification technique or nick translation (MLLYACs).23 24
For AF10, a YAC (807b3, 1,050 kb) from the Centre d'Etude Polymorphisme Humain (CEPH) library had been previously identified that spans the breakpoint region on 10p and reliably detects the breakpoint in U937.14
For MLL, we used a pool of two YACs (785c6 and 856b9) that span the MLL gene and approximately 1.0 mega-bp of distally flanking sequences, because 11q23 translocations are often associated with interstitial deletions.17,25 26
For CALM, the Mega-YAC Library from CEPH was screened with the previously reported CALM primers NG.T45 and NG.B362.14 In addition, 5 flanking YACs of the region were identified using the Whitehead web pages (www.genome.wi.mit.edu).27 28 A total of 9 YACs were evaluated by FISH for chimerism (hybridization with normal peripheral blood cells) and were mapped relative to theCALM breakpoint (hybridization with U937). Based on the intensity of hybridization signal, 2 YACs (proximal 785c1, 1,390 kb, and distal 914D9, 1,190 kb) were chosen for interphase analysis.
Hybridization.
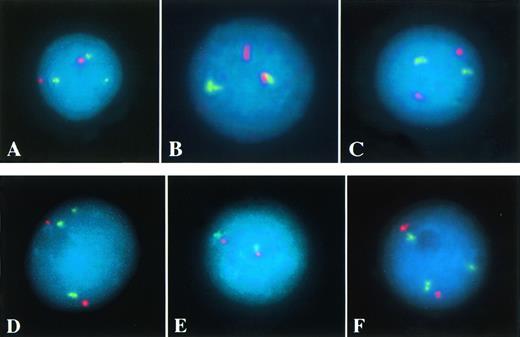
Dual-color FISH was performed with the two YAC-derived probes of theCALM region as well as the AF10 YAC or the MLLprobe and the corresponding centromeric probe (CEP10 and CEP11 Spectrum Orange; Vysis, Downers Grove, IL) to exclude numerical chromosomal aberrations (Fig 1).
(A through C) Interphase FISH of patient no. 1 (AML withAF10 and CALM rearrangement, Table 1). Three AF10 signals (green), indicating a split of one YAC signal, and two centromere 10 signals (red) are detectable in (A). In (B), the proximal (green) and the distal CALM YAC (red) are separated, indicating a breakpoint within this genomic region. Two MLL signals (green) and two centromere 11 signals (red) suggest no rearrangement of the MLL region in (C). (D through F) Interphase FISH of patient no. 8 (AML with AF10 and MLL rearrangement). Three AF-10 signals (green), indicating a split of one YAC signal, and two centromere 10 signals (red) are detectable in (D). In (E), the proximal (green) and the distal CALM YAC (red) are colocalized, excluding a rearrangement of this genomic region. In (F), the detection of three MLL signals (green), but only two centromere 11 signals (red) indicate a split of one MLL probe (C).
(A through C) Interphase FISH of patient no. 1 (AML withAF10 and CALM rearrangement, Table 1). Three AF10 signals (green), indicating a split of one YAC signal, and two centromere 10 signals (red) are detectable in (A). In (B), the proximal (green) and the distal CALM YAC (red) are separated, indicating a breakpoint within this genomic region. Two MLL signals (green) and two centromere 11 signals (red) suggest no rearrangement of the MLL region in (C). (D through F) Interphase FISH of patient no. 8 (AML with AF10 and MLL rearrangement). Three AF-10 signals (green), indicating a split of one YAC signal, and two centromere 10 signals (red) are detectable in (D). In (E), the proximal (green) and the distal CALM YAC (red) are colocalized, excluding a rearrangement of this genomic region. In (F), the detection of three MLL signals (green), but only two centromere 11 signals (red) indicate a split of one MLL probe (C).
FISH was performed as previously described.29 Briefly, the hybridization solution contained approximately 0.1 μg of each probe, 1 μg human Cot1-DNA, 0.6 μg human placental DNA, and 1 μg salmon sperm DNA/slide in a 10 μL volume. The probes were either directly labeled with Cy-3-dUTP (Vysis) or biotinylated and detected with fluorescein isothiocyanate (FITC)-conjugated avidin and amplified with antiavidin-FITC (16-Bio-dUTP [Boehringer Mannheim, Germany]; FITC-avidin and FITC anti-avidin [Vector, Burlingame, CA]). The slides were counterstained with 4′,6′-diamidino-2-phenylindole dihydrochloride (DAPI) and were analyzed using a fluorescence microscope (Zeiss, Jena, Germany). For each case, at least 100 single, intact interphase cells were analyzed.
For Fig 1, separate gray scale images of DAPI-stained cells and fluorescence signals were captured using a charge-coupled device camera (Xillix, Richmond, British Columbia, Canada) and were pseudocolored and merged using Adobe Photoshop software (Adobe Systems, Mountain View, CA).
Statistical analysis.
Survival data were analyzed using the χ2 test using PC Statistik software (Topsoft, Hannover, Germany).
RESULTS
Patients' characteristics.
Our study group included 1 adult T-ALL (no. 10, Table 1), 1 infant AML (no. 6), and 8 adult AML. The morphology of the myeloid leukemias were classified as immature (FAB M0) in 1 case (no. 8) and as leukemia with minimal maturation (FAB M1) in 3 cases (no. 5, 7, and 9). Significant maturation at or beyond the promyelocyte stage (FAB M2) was diagnosed in 2 cases (no. 2 and 3) and monocytic differentiation (FAB M5) in 3 cases (no. 1, 4, and 6).
Leukocyte counts at presentation were high, but varied significantly within the study group (mean, 23,900/μL; range, 200 to 60,800/μL). All patients with t(10;11) were rather young (36.7 ± 19.9 years old). There was no gender bias (5 male and 5 female).
FISH/cytogenetics.
Rearrangements of AF10 and/or either CALM orMLL were detected in all cases with balanced t(10;11).MLL rearrangements were detected in 3 of 8 cases (no. 1, 2, and 3; Table 1), including the only cytogenetically identified inversion 11.
CALM was rearranged in another 4 patients (no. 7 through 10) with balanced translocation t(10;11).
All 5 cases of 7 with MLL or CALM rearrangements, which were evaluable, had a rearrangement of AF10. In addition, in 1 case (no. 6) with a balanced translocation t(10;11), the AF10probe was split, indicating a breakpoint within this region, but noMLL or CALM rearrangements were detected.
No rearrangements were detected in the 2 patients with complex rearrangements (no. 4 and 5). However, for case no. 4, cell material was only sufficient for hybridization with the CALMprobes.
Morphology and immunophenotype.
All cases with MLL rearrangement showed a myeloid maturation at or beyond the promyelocyte stage or a monocytic morphology according to the FAB classification (M2 or M5), including 1 secondary AML after treatment for high malignant T-cell lymphoma.19
In contrast, cases with CALM rearrangement showed a rather immature phenotype (M0 or M1). In fact, case no. 8 was initially misdiagnosed as ALL and treated accordingly.
A CALM rearrangement was detected in the only ALL in this series. The morphological diagnosis was confirmed by the typical immunophenotype with expression of various T-cell antigens and coexpression of CD10, but absence of a myeloid marker (CD11c) or B-cell antigens.
A complete immunophenotype was available in only 1 of 3 AML withCALM rearrangement (no. 7). In agreement with the undifferentiated morphology of this case, leukemic blasts expressed antigens often associated with immature morphologic subtypes (ie, CD7 and CD34). Of the panmyeloid antigens tested, only CD13 was weakly expressed. Additionally, however, strong positivity of myeloperoxidase was found.
All 3 cases with MLL rearrangement were positive for the panmyeloid antigens CD33 and CD65s but lacked CD13 and CD15 and coexpressed CD4. Expression of markers associated with immaturity varied. In addition, 1 case (no. 1) showed coexpression of CD7 and CD10.
Clinical course.
In agreement with previous reports, hyperleukocytosis was detected in all cases with MLL rearrangements (mean, 35,000/μL; range, 13,800 to 60,800/μL).30-33 Leukocyte counts were also high in cases with CALM rearrangements (mean, 27,400/μL; range, 1,300 to 60,800/μL). However, the number of cases is too small for statistical evaluation.
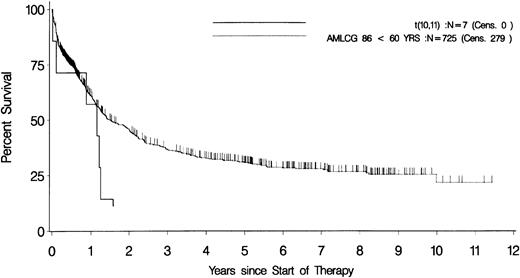
The overall survival of the total group was remarkably poor (9.6 ± 6.6 months), although, initially, 5 of 8 evaluable patients reached a complete remission. There was no significant difference between patients with MLL or CALM rearrangements. For the 7 patients who were treated according to the AMLCG86/92 protocol, disease-free survival and overall survival were 5 ± 4.9 months and 11 ± 7.2 months, respectively. The survival data of these patients were significantly worse than for the overall study group. After 14 months, none of the t(10;11) patients but 40% of the overall group were still disease-free (P = .03, χ2), and after 19 months, none of the t(10;11) patients but 49% of the overall group were still alive (P = .01, χ2; Fig 2).18
Overall survival in AML with t(10;11). Kaplan-Meier plots of the overall survival of patients with AML with t(10;11)(p13;q14) (n = 7; solid line) and the total study group with identical treatment (n = 725; dotted line).18
Overall survival in AML with t(10;11). Kaplan-Meier plots of the overall survival of patients with AML with t(10;11)(p13;q14) (n = 7; solid line) and the total study group with identical treatment (n = 725; dotted line).18
DISCUSSION
The translocation t(10;11)(p13;q14) is a recurring chromosomal abnormality that has been observed in acute lymphoblastic as well as AML.2-5 On the molecular level, two types of t(10;11)(p12-p14;q13-q21) have been characterized so far. One type results in a MLL/AF10 fusion, the other causes aCALM/AF10 fusion.11,14 We performed this study to define the relevance of MLL and CALM rearrangements in acute leukemia with t(10;11)(p13;q13-21). Both types of rearrangements were detected with similar frequency in cases with balanced translocation t(10;11). CALM was rearranged in AML as well as in ALL. Interestingly, the AML cases with CALM rearrangement had a rather immature morphology, and 1 case was initially misdiagnosed as ALL. This may have been also the case for some of the initially reported ALL cases with t(10;11) when immunophenotyping was not yet routinely performed.3 However, our series included a well-characterized lymphoblastic leukemia as well as myeloid leukemias, suggesting that CALM may play a critical role in the early progenitor/stem cell before either myeloid or lymphoid differentiation takes place. CALM has a very high homology to ap-3, a murine clathrin assembly protein.15 So far, CALM is the only protein interacting with clathrin that has been found to be involved in malignant transformation. A transactivation domain that was recently identified in CALM could be critical for the transformation potential of CALM/AF10.34 However, further studies are necessary to define the precise mechanism by which the CALM/AF10 fusion contributes to malignant transformation.
In contrast to the rather undifferentiated morphology of myeloid leukemias with CALM rearrangement, cases with MLLrearrangement showed a monocytic morphology or a myeloid maturation beyond the promyelocyte stage. Accordingly, all previously reported leukemias with t(10;11) and rearrangement of MLL either detected by FISH or by other molecular techniques, were AML-M4 or M5/M5a.6,35,36 Similarly, MLL rearrangements in general are strongly correlated with monocytic or myelomonocytic phenotypes.30,36 In contrast, Poirel et al37detected MLL rearrangements in 20% of AML-M1. However, these translocations did not involve chromosome 10. In fact, althoughMLL rearrangements are detected in AML as well as ALL, different translocation partner genes are usually involved.36 For example, t(4;11) and t(11;19)(q23;p13.3) are detected in ALL, whereas t(6;11), t(9;11), and t(11;19)(q23;p13.1) are common translocations in various subtypes of AML.36Therefore, the results of Poirel et al37 that included t(6;11),t(9;11), and an uncharacterized t(11;19) might not be applicable to leukemias with t(10;11).
In accordance with a previously published series of AML withMLL rearrangements, in our series, all 3 cases with MLLrearrangement were CD4+, CD13−, CD33+, CD65s+.20 The latter three markers are panmyeloid markers that are rather broadly expressed.21 Therefore, this immunophenotype does not represent an aberrant expression of different lineage- or maturity-specific markers as detected in the majority of leukemias.38 Hence, we do not consider this immunophenotype as leukemia-specific for MLL translocation, although the only completely characterized AML with CALM rearrangement had a clearly distinct immunophenotype (CD4−, CD13+, CD33−, CD65s−).
AF10 is the first fusion partner of MLL, which is fused to another gene in a different translocation, emphasizing its important role in malignant transformation. It is thought that AF10functions as a transcription factor and that the zinc finger might constitute a DNA binding domain.12,16 Interestingly, 1 case with a balanced translocation t(10;11) (no. 6) showed a breakpoint within the AF10 region, but did not have a CALM orMLL rearrangement. Translocations of 11q23 are often associated with distal interstitial deletions that would make it difficult to detect the translocation by FISH. However, our MLL probe, which encompasses approximately 1 mega-bp of 3′ flanking sequences, was able to detect these kinds of translocations in previous studies.17 35 Alternatively, another partner gene on chromosome 11 may be involved.
So far, MLL rearrangements have been shown to be a negative prognostic factor, especially in childhood ALL.31-33 For AML, data are sparse and contradictory. In a series of pediatric AML, patients with t(9;11)(p22;q23) had a better outcome, whereas Lo Coco et al39 identified MLL rearrangements as a negative prognostic factor in ALL and AML.39,40 In infant AML, 11q23/MLL abnormalities had no effect on survival.31
In our study, all cases with t(10;11) had a remarkably poor prognosis. This holds up for the subpopulation of 7 AMLCG86/92 patients whose event-free and overall survival was significantly worse than the survival data of the overall study group.18 Therefore, our results suggest that cytogenetic/molecular analysis may be used to identify this subgroup with poor prognosis.
In summary, we were able to identify a MLL or a CALMrearrangement, each in approximately half of the leukemias with a t(10;11)(p13,q13-21). Moreover, MLL or CALMrearrangements were found in leukemias with different morphology.MLL translocations were identified in leukemias with monocytic morphology or maturation beyond the promyeloid stage, whereasCALM rearrangements were detectable in immature leukemias as well as ALL. Further studies are necessary to characterize the biological characteristics of these molecular alterations.
Recently, Kobayashi et al41 reported 4 cases of acute leukemia with CALM rearrangements that were studied by FISH. However, the cases were not classified according to the FAB criteria.
ACKNOWLEDGMENT
The authors thank H. Döhner for generously providing the MLL YACs and C. Sauerland for assistance in statistical analysis.
Address reprint requests to M.H. Dreyling, MD, Department of Hematology/Oncology, University of Göttingen, Robert Koch-Street 40, D-37075 Göttingen, Germany.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal