Abstract
Interleukin-12 (IL-12) production by human monocytes is stringently regulated through the inducibility of both subunits, p35 and p40, and expression of p35 mRNA is the limiting factor for the secretion of the bioactive IL-12 p70 heterodimer. Optimal induction of p35 mRNA requires priming of the monocytes by interferon-γ (IFN-γ), followed by brief exposure to lipopolysaccharide or other bacterial products. To investigate control of p35 gene expression, we isolated genomic clones containing the human p35 gene and determined the 5′ end of the mRNA expressed in monocytes. We discovered that a unique p35 transcript is induced in monocytes that begins downstream of a consensus TATA box that lies within the 5′ end of the cDNA originally cloned from Epstein-Barr virus (EBV)-transformed B cells. Analysis of p35 mRNA by Northern blotting showed that the message from monocytes is approximately 200 bases shorter than message derived from the EBV-transformed B-cell line VDS. The initiation sites downstream from the TATA box were confirmed by RNase protection and 5′ RACE. The data indicate that p35 transcription can initiate from different sites depending on the cell type and that the shorter inducible transcript in monocytes is the one that accumulates after stimulation. Protein translation of these two forms may result in proteins of different sizes with potential implications for the regulation of IL-12 secretion and function.
INTERLEUKIN-12 (IL-12) is a key mediator of the immune response.1-3 IL-12 is a heterodimeric cytokine (p70) composed of two independently regulated subunits, p35 and p40,4-7 which was originally identified in the supernatants of Epstein-Barr virus (EBV)-transformed B-cell lines by its capacity to stimulate natural killer (NK) cells8 and cytolytic T lymphocytes.9 The primary physiologically relevant source of IL-12 expression is cells of the monocyte/macrophage lineage,5,6,10-13 including dendritic cells.14-16 Recent attention has been focused on the central role of IL-12 in controlling the cytokine profiles of T cells in an immune response that shifts the balance toward a TH1 phenotype.1,2 One of the well-documented biological functions of IL-12 is the induction of interferon-γ (IFN-γ) expression by T and NK cells.17,18 IFN-γ , in turn, has been shown to enhance4,10 or be absolutely required5 6 for expression of IL-12 p70 by cells in which IL-12 is inducible by bacterial products such as lipopolysaccharide (LPS) or fixed Staphylococcus aureus Cowan strain (SAC). IFN-γ and IL-12 therefore form an important autostimulatory loop which is crucial for the development of cell-mediated immunity.
It was originally believed that the regulation of IL-12 expression in lymphoblastoid cell lines and peripheral blood mononuclear cells (PBMCs) occurred primarily at the level of the p40 subunit, whereas the p35 subunit was constitutively expressed and minimally regulated.19 However, we5 and others6 have shown that p35 expression is highly regulated and is, in fact, the limiting factor in controlling production of bioactive IL-12 p70 in monocytes. We showed that p35 mRNA by Northern analysis was undetectable in human monocytes even upon stimulation with LPS, unless the monocytes were primed by prior incubation with IFN-γ5; these results were confirmed by Snidjers et al.6 This evidence provided an explanation for earlier results in which p40 expression often occurred in vast excess to p70 in PBMCs that were treated only with SAC or LPS alone, whereas this high p40/p70 ratio was reduced in cells that were also treated with IFN-γ.10,11 We further showed that not only was a priming signal required for IL-12 p70 expression, but IFN-γ was much more potent as a priming stimulus for p35 than granulocyte-macrophage colony-stimulating factor (GM-CSF), which primed monocytes for expression of p40 and tumor necrosis factor with equivalent potency.5
To investigate the regulation of human IL-12 p35 expression, we isolated genomic clones of the p35 gene from a human placental genomic library and sequenced the 5′ flanking region containing potential promoter elements for this gene. We show that, in human monocytes, there is a strong inducible transcription initiation start site that is located 3′ to the initiation of the cDNAs described for EBV-transformed B-cell lines.20 21 The data suggest that inducible transcription in monocytes occurs from a TATA-containing element located within the reported cDNA sequences, whereas transcription in the EBV cell lines initiates from an upstream promoter characterized by the presence of a CpG island. Our results demonstrate that induction of p35 mRNA expression occurs by a different pathway in stimulated monocytes, the principal physiologic source of IL-12, than previously reported based on analysis of EBV-transformed B cells.
MATERIALS AND METHODS
Monocytes.
Human peripheral blood monocytes were purified from single donor leukapheresis preparations by centrifugal counterflow elutriation as previously described.22 Monocytes were greater than 95% pure by Giemsa and nonspecific esterase staining. VDS cells, an EBV-transformed lymphoblastoid line, were kindly provided by Dr Giovanna Tosato (Division of Hematologic Products, CBER/FDA, Bethesda, MD). Cells were cultured in 35-mm tissue culture plates (Costar, Cambridge, MA) in RPMI1640 supplemented with L-glutamine, gentamicin sulfate, HEPES, and 10% fetal bovine serum (Life Technologies, Gaithersburg, MD). All culture reagents were tested and free of detectable endotoxin.23
Cytokines and reagents.
Recombinant human IFN-γ was kindly supplied by Genentech, Inc (South San Francisco, CA). LPS was derived from Escherichia coli0128:B12 phenol extract (Sigma Chemical Co, St Louis, MO).
Isolation of p35 genomic clones.
Approximately 1 × 106 plaques from a human placental genomic library cloned into the λ FIX II vector (Stratagene, La Jolla, CA) were screened using a 776-bp EcoRI cDNA fragment containing most of the coding region and some 5′ untranslated sequence from a human IL-12 p35 cDNA kindly provided by U. Gubler (Hoffman-LaRoche, Nutley, NJ).20 Five positive clones were rescreened and yielded two overlapping clones (111 and 112) that were plaque-purified and further characterized by restriction mapping and sequencing. Sequencing was performed using Sequenase (Quick-Denature plasmid sequencing kit; Amersham Life Science Inc, Arlington Heights, IL).
RNA analysis.
Reference is made to two reported cDNA sequences for human IL-12 p35 subunit: the cytotoxic lymphocyte maturation factor (CLMF) cDNA (accession no. M65271) and the natural killer cell stimulatory factor (NKSF) cDNA (accession no. M65291). RNA was isolated using the acid phenol-guanidine isothiocyanate method or by a direct poly-A selection procedure (Ambion, Inc, Austin TX). RNA was fractionated with 1% formaldehyde agarose gels and blotted to nylon filters (Life Technologies), followed by UV cross-linking for immobilization. Filters were hybridized with a human IL-12 p35 cDNA probe (see above) using a formamide-based hybridization solution (Fast-Pair; Digene, Silver Spring, MD) and washed under high stringency (0.1× SSC, 0.1% sodium dodecyl sulfate [SDS], 63°C) before autoradiography. RNase protection analysis was performed as follows: an antisense RNA probe was transcribed in the presence of 32P-UTP from the T3 promoter of a Bluescript SK(−) template containing the first 388 bases of the CLMF cDNA (accession no. M65271) after linearization at the 5′ end with HindIII. Labeled transcripts were hybridized with RNA samples overnight at 55°C in 80% formamide/100 mmol/L sodium citrate/300 mmol/L sodium acetate/1 mmol/L EDTA, pH 6.4 (Ambion, Inc). Samples were then diluted in 0.2 mol/L NaCl/20 mmol/L Tris-HCl, pH 7.5, and digested with RNase T1 (1,000 U) for 1 hour at 37°C. After extraction with phenol:chloroform and precipitation with yeast RNA, samples were analyzed on 6% polyacrylamide sequencing gels and subjected to autoradiography.
Rapid amplification of cDNA ends (5′ RACE) was performed using both a TdT-based dCTP-tailing procedure (Life Technologies) and a second-strand blunt end ligation procedure (Marathon cDNA Amplification Kit; Clontech Laboratories, Palo Alto, CA). Poly-A–selected RNA was isolated from monocytes stimulated with IFN-γ (100 ng/mL) for 16 hours followed by LPS (1 μg/mL) for 2 hours or from VDS cells stimulated with PMA (10 ng/mL) and calcium ionophore (25 ng/mL) for 24 hours. This RNA was used as a template for reverse transcriptase extension to the 5′ end from either a gene-specific primer, 5′-CTTGGTTAATTCCAATGGTA-3′ (CLMF cDNA bases 445-426 or NKSF cDNA bases 479-460) or from a lock-docking oligo-dT primer (Clontech). RACE products were derived by polymerase chain reaction (PCR) with the appropriate primers after either dC extension of the first-strand (Life Technologies) or second-strand synthesis by the Gubler-Hoffman method24 and adaptor ligation (Clontech). Antisense gene-specific primers GSP34, 5′-TGGAGTGGCCACGGGGAGGTTTCT-3′ (CLMF cDNA bases 259-236 or NKSF cDNA bases 293-270), and GSP50, 5′-GGTAAACAGGCCTCCACTGTGCTGG-3′ (CLMF cDNA bases 429-405 or NKSF cDNA bases 463-439), were used for amplification of RACE products. Touchdown PCR25 was performed as follows: the first five cycles were performed with an annealing and extension temperature of 72°C for 3 minutes, followed by 5 cycles at 70°C, and then 25 cycles at 68°C; the melting temperature was 94°C, with 30 seconds for all cycles. RACE products were analyzed on a 1.5% agarose gel and cloned using the TA cloning system (Invitrogen Corp, Carlsbad, CA) or the CloneAmp UDG system (Life Technologies). Resulting clones were isolated and sequenced at both ends with Sequenase (Amersham).
RESULTS AND DISCUSSION
To isolate genomic clones containing potential regulatory sequences involved in the control of IL-12 p35 expression, a human placental genomic library was screened using a 32P-labeled human p35 cDNA probe. Of 1 × 106 plaques examined, two overlapping genomic clones were isolated and characterized. Restriction analysis established that one of these clones contained more 5′ flanking sequence than the other, and this clone was selected for further study. Intron-exon boundaries were established by DNA sequencing with primers derived from the human IL-12 p35 cDNA. The restriction map and seven exons are shown in Fig 1A and B. Intron splice sites all conform to the GT-AG rule26 and are located at sites analogous to those in the murine IL-12 p35 gene.27 28
Structure of the human IL-12 p35 gene (A and B). The λ Fix II clone contained three BamHI fragments of approximately 3, 6, and 9 kb, respectively, from the 5′ end. Restriction sites for BamHI (B), EcoRI (E), andPst I (P) are noted. Exons are labeled numerically (I through VII). The transcription start sites (see Fig 5) are labeled as S1 (monocyte start site) and S2 (lymphoblastoid cell start site). Exon boundaries were sequenced from within the exons, and intron distances were estimated by PCR using appropriate exon primers or were determined directly by sequencing through to the next exon. Numbers surrounding the exon boxes (B) refer to amino acids, beginning with the second methionine codon (Met35 in Wolf et al21). Sequence data from this clone are available from GenBank under accession no.AF050083. (C) Comparison of human and mouse (accession no.S82412) p35 gene 5′ flanking sequences. Nucleotide sequences were aligned using GCG BestFit, as described in the text. Sequences are aligned from the 3′ end of exon I (human), which represents the second exon in the mouse. Notations are made for the two transcription start sites in the human gene (S1, the monocyte start site; S2, the lymphoblastoid start site). The TATA box (TATAAA) is overlined, as are the two methionine (ATG) codons Met1 and Met35. The proposed 5′ end of the second exon of the mouse p35 gene is noted; the mouse sequence presented in this figure therefore represents the 3′ end of the first intron and the entire second exon. There is no evidence for an upstream first exon in the human gene.
Structure of the human IL-12 p35 gene (A and B). The λ Fix II clone contained three BamHI fragments of approximately 3, 6, and 9 kb, respectively, from the 5′ end. Restriction sites for BamHI (B), EcoRI (E), andPst I (P) are noted. Exons are labeled numerically (I through VII). The transcription start sites (see Fig 5) are labeled as S1 (monocyte start site) and S2 (lymphoblastoid cell start site). Exon boundaries were sequenced from within the exons, and intron distances were estimated by PCR using appropriate exon primers or were determined directly by sequencing through to the next exon. Numbers surrounding the exon boxes (B) refer to amino acids, beginning with the second methionine codon (Met35 in Wolf et al21). Sequence data from this clone are available from GenBank under accession no.AF050083. (C) Comparison of human and mouse (accession no.S82412) p35 gene 5′ flanking sequences. Nucleotide sequences were aligned using GCG BestFit, as described in the text. Sequences are aligned from the 3′ end of exon I (human), which represents the second exon in the mouse. Notations are made for the two transcription start sites in the human gene (S1, the monocyte start site; S2, the lymphoblastoid start site). The TATA box (TATAAA) is overlined, as are the two methionine (ATG) codons Met1 and Met35. The proposed 5′ end of the second exon of the mouse p35 gene is noted; the mouse sequence presented in this figure therefore represents the 3′ end of the first intron and the entire second exon. There is no evidence for an upstream first exon in the human gene.
Two independently isolated cDNA clones (identical except for their 5′ ends) have been reported for the IL-12 p35 subunit (see the Materials and Methods): the CLMF cDNA (accession no. M65271), cloned by Gubler et al,20 begins 41 bases downstream of the NKSF cDNA (accession no. M65291), cloned by Wolf et al.21 Comparison of the sequence of the genomic clone surrounding exon I with the reported p35 cDNA sequences20 21 within exon I shows the following: (1) exon I contains the cDNA open reading frame with two potential initiator methionine codons separated by 33 amino acid codons; (2) the region immediately upstream of the first initiator methionine codon contains a TATA-like promoter motif (TATAAA) that, if functional, would preclude transcription of this codon; and (3) no other TATA-like motif is discernible within the adjacent 1.7-kb upstream region.
Located within the 5′ flanking sequence (upstream of the reported p35 cDNA sequence) of the human p35 genomic clone is a CpG island, as defined by a GC content greater than 50% and the approximate equivalence of CpG with GpC.29 When this sequence was compared against nucleotide databases, a sequence derived from a generic isolation of CpG islands in the human genome (H sapiens CpG DNA, clone 38d5, accession no. Z65420) was identified that corresponded (94% identity) to a region of the human p35 5′ flanking region (bases 932-1162).30 CpG islands are associated with the promoters of many, if not all, housekeeping genes and about 40% of tissue-restricted genes in the human genome.29 However, this GC-rich region had limited homology to mouse p35 genomic sequences, consistent with the proposal by Tone et al28that the mouse p35 gene was not controlled by a GC-rich promoter.
Tone et al28 provided evidence for the existence of an additional 5′ untranslated exon in the mouse p35 gene; however, there is no evidence for the existence of a similar upstream noncoding exon in the human gene (see below). The sequence of the human 5′ flanking region was compared with the mouse p35 gene using the GCG BESTFIT program (Fig 1C). Using a liberal gap weight of 1.0 with the default gap length of 0.3, a comparison of 1,703 bases of the human sequence was aligned with 1,446 bases from the mouse sequence using the same 3′ end, which represented the end of the first exon in both cases. The overall sequence identity was 85%, although the most homologous region generated by more stringent gapping parameters (gap weight of 5.0) was most striking within the 300 bases of the 3′ end of exon I. The sequence of this otherwise highly homologous region also showed that there is no consensus splice site (AG) located at the position of the splice site at nucleotide 1370 of the mouse (corresponding to base 1599 of the human gene; Fig 1C) for the proposed upstream first exon in the mouse p35 gene.28 In addition, there are regions of high homology within the sequence that would represent this proposed intron between the mouse and human genes. These data, along with 5′ RACE results (see below), provide no compelling evidence for an upstream noncoding exon in the human p35 gene. The high conservation of the region immediately surrounding the consensus TATA box sequence (bases 1579-84, human, and bases 1350-55, mouse) and the first ATG codon immediately following the TATA region (Fig 1C) suggests that this region is functionally critical. In both reports on analyses of murine p35 genes, multiple transcription start sites were proposed.27,28 In one of these reports,28 3 of 14 5′-RACE clones initiated downstream of this TATA-like region.
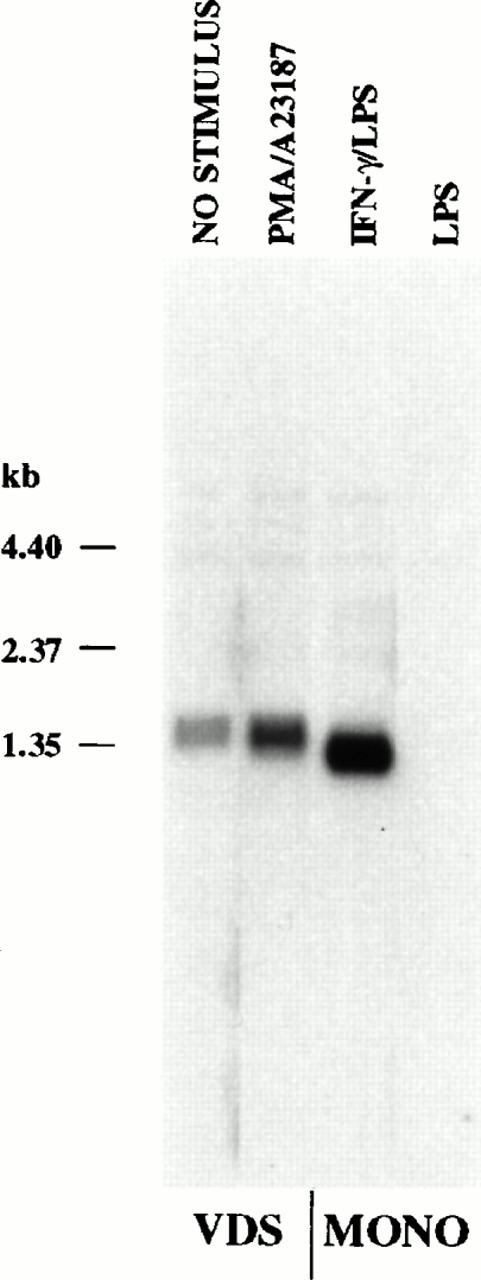
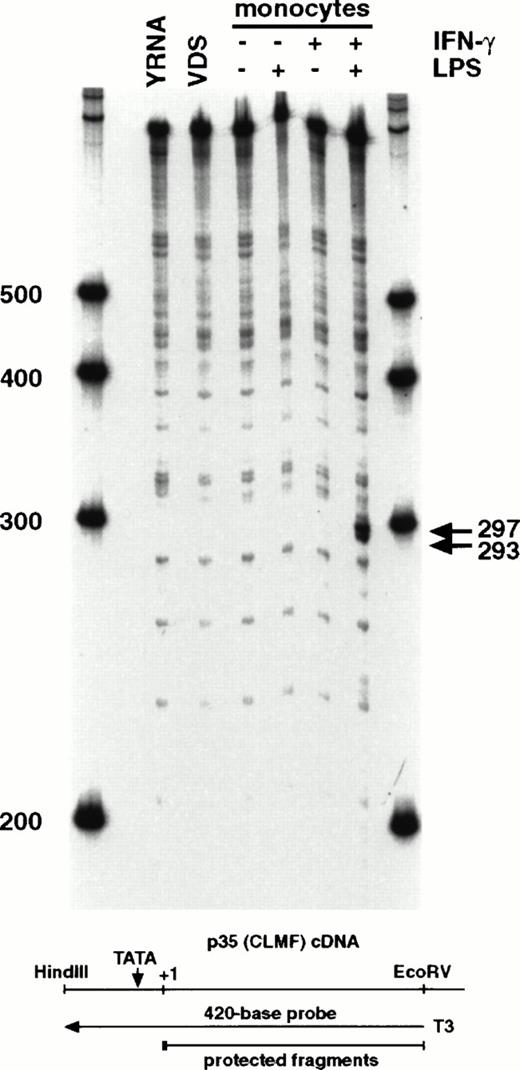
Northern analysis of polyadenylated RNA derived from monocytes and lymphoblastoid cells indicated a difference in size of approximately 200 bases from these two cell types (Fig2). To determine the region of the mRNA that contributed to the size difference seen by Northern analysis, RNA samples were subjected to RNase protection analysis. Using an antisense probe that contained the first 388 bases of the CLMF (IL-12) p35 cDNA, two protected fragments of 293 and 297 bases were detected with mRNA from induced monocytes (IFN-γ followed by LPS) that were not present in any other sample, including RNA from constitutively expressing VDS cells or from monocytes stimulated with LPS alone (Fig3). These fragments corresponded to start sites that begin 25 and 29 bases downstream, respectively, of the TATA motif (TATAAA) located within the 5′ end of the cDNA.20 21 Initiation of transcription at these sites is consistent with TATA-mediated transcription and would produce transcripts containing the second, but not the first, initiator methionine codon reported for lymphoblastoid cell-derived p35 cDNA (see Fig 5).
Steady-state p35 mRNA induced in monocytes is approximately 0.2 kb shorter than constitutive or enhanced mRNA in EBV-transformed lymphoblastoid cells (Northern analysis). Polyadenylated RNA (2 × 106 cell equivalents/lane) was isolated from the EBV-transformed cell line VDS that was either uninduced (lane 1) or stimulated with PMA (10 ng/mL) and A23187 (25 ng/mL) for 24 hours (lane 2), from monocytes primed with IFN-γ (100 ng/mL for 16 hours), followed by LPS (1 μg/mL) for 2 hours (lane 3), or from monocytes stimulated with LPS alone (lane 4). RNA was resolved by electrophoresis on a 1.5% agarose gel and blotted to a nylon filter. The blot was probed with a p35 cDNA probe as described in the Materials and Methods. RNA size was estimated using an RNA ladder (Life Technologies).
Steady-state p35 mRNA induced in monocytes is approximately 0.2 kb shorter than constitutive or enhanced mRNA in EBV-transformed lymphoblastoid cells (Northern analysis). Polyadenylated RNA (2 × 106 cell equivalents/lane) was isolated from the EBV-transformed cell line VDS that was either uninduced (lane 1) or stimulated with PMA (10 ng/mL) and A23187 (25 ng/mL) for 24 hours (lane 2), from monocytes primed with IFN-γ (100 ng/mL for 16 hours), followed by LPS (1 μg/mL) for 2 hours (lane 3), or from monocytes stimulated with LPS alone (lane 4). RNA was resolved by electrophoresis on a 1.5% agarose gel and blotted to a nylon filter. The blot was probed with a p35 cDNA probe as described in the Materials and Methods. RNA size was estimated using an RNA ladder (Life Technologies).
RNase protection analysis using an antisense riboprobe (420 bases) from the 5′ end of the CLMF cDNA results in smaller protected fragments of 293-297 bases from induced monocyte mRNA. RNA was isolated from VDS cells (VDS) or from monocytes stimulated as indicated with nothing, IFN-γ, or LPS, or both (sequentially). YRNA, yeast RNA control. RNA was hybridized overnight with a32P-labeled antisense RNA probe transcribed from theEcoRV site at base 388 of the CLMF p35 cDNA. Products were treated with RNase T1, extracted, precipitated, and loaded onto a 6% sequencing gel with in vitro transcribed Century markers (Ambion). Marker size is indicated in bases. The arrows indicate the two specific bands of 293 and 297 bases protected at the site labeled in Fig 1 as S1.
RNase protection analysis using an antisense riboprobe (420 bases) from the 5′ end of the CLMF cDNA results in smaller protected fragments of 293-297 bases from induced monocyte mRNA. RNA was isolated from VDS cells (VDS) or from monocytes stimulated as indicated with nothing, IFN-γ, or LPS, or both (sequentially). YRNA, yeast RNA control. RNA was hybridized overnight with a32P-labeled antisense RNA probe transcribed from theEcoRV site at base 388 of the CLMF p35 cDNA. Products were treated with RNase T1, extracted, precipitated, and loaded onto a 6% sequencing gel with in vitro transcribed Century markers (Ambion). Marker size is indicated in bases. The arrows indicate the two specific bands of 293 and 297 bases protected at the site labeled in Fig 1 as S1.
Map of transcription start sites in the human IL-12 p35 promoter. The first exon and 5′ flanking sequence (bases 1224-1703) are shown. Notations are made for the start site in lymphoblastoid cells (S2, −165), the cDNA start sites for NKSF (−126) and CLMF (−85), the TATA box (−32), the monocyte start site (S1, +1), and the two initiator methionine (Met) codons.
Map of transcription start sites in the human IL-12 p35 promoter. The first exon and 5′ flanking sequence (bases 1224-1703) are shown. Notations are made for the start site in lymphoblastoid cells (S2, −165), the cDNA start sites for NKSF (−126) and CLMF (−85), the TATA box (−32), the monocyte start site (S1, +1), and the two initiator methionine (Met) codons.
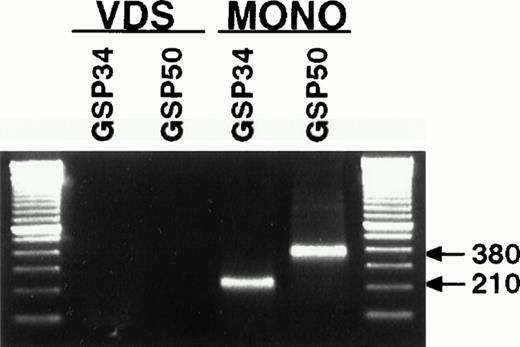
To confirm the sequence of transcription initiation sites, we performed 5′ rapid amplification of cDNA ends (RACE) by two different methods to verify the 5′ ends of the p35 mRNA from monocytes induced with IFN-γ and LPS and from VDS lymphoblastoid cells. We synthesized cDNA using the Marathon system (Clontech), in which second-strand synthesis is performed using the Gubler-Hoffman method and the cDNA is blunt-ended with T4 polymerase before ligation of an adaptor. Adaptor and two different gene-specific primers were then used to amplify the cDNA ends using touchdown PCR (the Materials and Methods). The resulting amplified products were analyzed by agarose gel electrophoresis (Fig 4). A product was observed only in the samples from stimulated monocytes of 210 bases, using antisense primer GSP34 (see the Materials and Methods) or 380 bases, using antisense primer GSP50 (see the Materials and Methods). Adjusting for the adaptor primer extension of 49 bases, a start site at approximately base 132 of the NKSF cDNA, which is 32 bases downstream of the TATA box at bases 95 to 100 of the cDNA, is predicted. A minor band at around 700 bases appeared in the GSP50 amplification reaction from monocytes that, upon sequence analysis, appeared to be an unrelated product (phosphoethanolamine cytidylyltransferase, accession no. D84307) that possessed a sequence homologous to the 3′ end of that primer. Under the same conditions, no distinct RACE product developed from the amplification of cDNA from the lymphoblastoid cells, suggesting that there are heterogeneous transcription start sites in these cells.
5′ RACE analysis of monocyte RNA yields a single amplification product, not detectable from VDS RNA, that maps to a unique transcription start site. Polyadenylated RNA was prepared from PMA-treated VDS cells or from IFN-γ–primed and LPS-stimulated monocytes and used to prepare a cDNA library according to manufacturer's recommendations (Marathon cDNA Amplification Kit; Clontech). Each library (VDS, lanes 1 and 2; monocyte, lanes 3 and 4) was amplified using the Marathon adaptor primer and two different gene-specific primers, GSP34 (lanes 1 and 3) and GSP50 (lanes 2 and 4). RACE products were resolved on a 1.5% agarose gel with ethidium bromide. Markers are multiples of 100 bp (Life Technologies).
5′ RACE analysis of monocyte RNA yields a single amplification product, not detectable from VDS RNA, that maps to a unique transcription start site. Polyadenylated RNA was prepared from PMA-treated VDS cells or from IFN-γ–primed and LPS-stimulated monocytes and used to prepare a cDNA library according to manufacturer's recommendations (Marathon cDNA Amplification Kit; Clontech). Each library (VDS, lanes 1 and 2; monocyte, lanes 3 and 4) was amplified using the Marathon adaptor primer and two different gene-specific primers, GSP34 (lanes 1 and 3) and GSP50 (lanes 2 and 4). RACE products were resolved on a 1.5% agarose gel with ethidium bromide. Markers are multiples of 100 bp (Life Technologies).
Because of the requirement for snapback priming in the Gubler-Hoffman method, absolute determination of the 5′ end of transcripts is often questionable, since the 5′ end of the original message functions to prime second-strand synthesis.24 We therefore sequenced RACE products derived by the dC extension procedure, which is more likely to identify the 5′ end of the mRNA, because it does not rely on snapback priming for second-strand synthesis, but may be subject to specificity problems in regions of high GC content. We generated a number of clones from the shorter transcript downstream of the TATA box using this system, but were able to generate only a limited number of specific products upstream in the GC-rich region 5′ of the TATA box. From the monocyte cDNA, 70% of clones examined began with the A at base 131 in the NKSF (IL-12) p35 cDNA sequence; other clones initiated downstream of this site in the second exon and were likely derived from prematurely terminated reverse transcription products. We designate this defined monocyte transcriptional initiation site as S1 (Fig5). No sequences 5′ to this site in monocyte-derived transcripts were observed. Clones derived from VDS mRNA initiated at various sites (bases 1474, 1591, 1611, and 1644 in Fig 5). This was likely due to premature first-strand termination products in the GC-rich 5′-flanking region, although multiple start sites for p35 transcription have been reported for the mouse gene.27,28The longest VDS transcript began 39 bases upstream of the 5′ end of the NKSF cDNA (S2, 165 bases upstream of the monocyte start site; Fig 5). The sequence at this site (TTAATCC) meets the criteria for a consensus Inr transcriptional initiator site (Py Py A N T/A Py Py).31 Heterogeneity of transcription start sites for p35 was suggested in both reports from the mouse gene, and there was no agreement even between these studies on a single dominant initiation site in the mouse.27,28 It is noteworthy that both of these investigations used cell lines constitutively expressing p35 as the source of mRNA. Our data also support the probability that there is heterogeneity in transcriptional initiation in lymphoblastoid cells. Consistent with the mapping of the cDNA ends derived from human lymphoblastoid cells,20 21 our 5′ RACE data confirms that transcriptional initiation in lymphoid cell lines that constitutively express low levels of p35 occurs upstream of the TATA box whose function in monocytes we have described herein.
The expression of the IL-12 p35 gene is a critical control point for production of the IL-12 p70 heterodimer. The regulation of this gene is therefore an important determinant in the nature (TH subset and cytokine phenotype) and extent of the immune response. We have previously demonstrated that expression of p35 mRNA in human monocytes requires a priming signal, provided specifically by IFN-γ, followed by a bacterial stimulus such as LPS. This is in contrast to the constitutive expression of this gene commonly observed in EBV-transformed lymphoblastoid cell lines that can be enhanced with phorbol ester stimulation.32 The results presented here demonstrate that expression of p35 mRNA can occur from distinct promoters. These data have important implications for the complex regulation of p35 and IL-12 heterodimer expression specifically and for the cytokine phenotype of immune responses in general. Thus, transcription of p35 mRNA in human monocytes differs from that observed in lymphoblastoid cell lines by initiating from a TATA-containing promoter to generate a transcript that lacks the first initiator methionine found in the lymphoblastoid mRNAs.
Alternative promoter usage has been recognized as a common mechanism in the control of gene expression at multiple levels.33 The potential functional relevance of the alternative promoters in the human p35 gene is intriguing. The reported cDNA clones for p35 were derived from RNA isolated from EBV-transformed lymphoblastoid cell lines20 21 that constitutively produce low levels of IL-12 heterodimer and are independent of regulation by physiologic stimuli. In contrast, monocytes produce up to 100-fold higher levels of IL-12, and expression in monocytes requires a priming stimulus provided by IFN-γ (optimal exposure 8 to 24 hours), followed by brief exposure to a secondary signal such as bacterial lipopolysaccharide. IFN-γ priming is therefore a differentiative response that allows subsequent recognition of the TATA-dependent promoter for transcriptional initiation in monocytes. The use of this promoter is restricted both by stimulus, cell lineage, and perhaps differentiation state of the cell.
The inducible monocyte transcript described here begins downstream of the first initiator methionine codon and therefore encodes a protein that lacks the first 34 amino acids encoded by the longer form of p35 mRNA expressed in lymphoblastoid cells. The use of these alternative promoters could provide an explanation for differences in the relative levels of IL-12 production by lymphoblastoid cells and monocytes. The upstream promoter may be poorly transcribed, or the processing, transport, or translation of the upstream transcript may be less efficient than the shorter monocyte transcript. The alternative translation products could differ in their relative secretion or release of a soluble form of the protein. What role the first 34 amino acids may play in the secretion or compartmentalization of the long form of IL-12 is currently unknown. Wolf et al21 speculated that this amino-terminal extension could yield a membrane-bound form of p35/IL-12, which would provide an effective mechanism for localizing proinflammatory responses. In preliminary studies, we have determined that both in vitro translation and transfectants initiate readily from the first methionine when both initiation codons are available.
The regulation of p35 expression is emerging as an important control point for IL-12 production. Recent evidence has indicated that p40 homodimers, which act as potent receptor antagonists for IL-12, are detected in vivo after endotoxin challenge.34 In addition, p40 homodimers appear to have a unique function in stimulating IFN-γ production by CD8+ T cells.35 These data underscore the importance of considering the relative levels of p40 and p35 expression in estimating the presence of active IL-12 heterodimers in different biological systems. Because p40 is capable of dimerizing with either p35 to form active IL-12 or with itself to form a receptor antagonist, the expression of p35 is critical in determining the relative level of active IL-12 produced by a given cell. Further investigation of the regulation of these distinct p35 transcripts and their translation products should provide new insights into the control of this critical host defense cytokine.
ACKNOWLEDGMENT
The authors thank Drs Giovanna Tosato, David Finbloom, and Edward Max for critical review of the manuscript. We also thank Dr Tosato for providing the VDS cell line and Valerie Calvert for purification of human monocytes.
Supported in part by an appointment (F.J.M.) to the Postgraduate Research Participation Program at the Center for Biologics Evaluation and Research administered by the Oak Ridge Institute for Science and Education.
Address reprint requests to Mark P. Hayes, PhD, Division of Cytokine Biology, Food and Drug Administration, 1401 Rockville Pike, HFM-508, Rockville, MD 20852-1448; e-mail: hayesm@a1.cber.fda.gov.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal