Abstract
We have analyzed by immunocytochemistry (ICC) the frequency of p53 protein expression in 181 cases of B-cell chronic lymphocytic leukemia (CLL) followed at a single institution to assess the relationship between p53 and the clinical and morphological features of the disease, as well as the possible involvement of this protein in the pathogenesis of the more aggressive forms of CLL. The overall frequency of p53 protein positivity in CLL was 15% (27 of 181 cases). There were no significant differences in age, sex, absolute lymphocyte count, or lymphocyte doubling time between p53-positive and -negative patients. By contrast, p53-positive patients had a significantly higher percentage of prolymphocytes (P = .002) and a significantly lower percentage of residual CD3-positive T lymphocytes (P = .0001). No correlation was found between the percentage of p53-positive cells and the percentage of cells in cycle assessed by the monoclonal antibody Ki-67. When the percentage of p53 positivity was correlated with the clinical stage of the disease, the proportion of p53-positive cases increased significantly from Binet's stage A (8 of 108; 7.4%), to stage B (12 of 49; 24.4%) and C (7 of 24; 29.2%) (P = .002). p53 positivity correlated also with the phase of the disease, showing a low expression at diagnosis (8 of 112; 7.1%) and a significantly higher expression in patients studied during the course of the disease (7 of 35; 20%) and, to a further extent, with disease progression (12 of 34; 35.3%) (P = .0001). The association of p53 protein expression with mutations in the gene was confirmed by direct sequence of the entire cDNA in 15 of the 17 ICC positive cases tested (88%). A significantly shorter treatment-free interval from diagnosis (P = .003) and a poorer response to therapy (P = .007) was observed in p53-positive compared with p53-negative patients. Overall survival from the time of diagnosis, as well as from the time of p53 protein analysis, was significantly shorter in patients with p53 protein expression (P = .03 and .0001, respectively). Moreover, in multivariate analysis, p53 expression and stage C were independently associated with a short survival. The results of this study indicate that in CLL the expression of the p53 protein, analyzed by a simple and reliable immunocytochemical method, is strongly associated with p53 gene mutations, a morphological variant (CLL with >10% prolymphocytes), advanced clinical stage, progressive disease, poor response to therapy, and short survival.
B-CELL CHRONIC LYMPHOCYTIC leukemia (CLL) is the most common leukemia in the Western world. It is characterized by a highly variable clinical course, with some patients living several years untreated without changes in their clinical status and others showing a more rapid disease progression and a significantly shorter survival.1 The biological mechanisms underlying such variability in clinical behavior remain largely unclear. The issue of identifying in CLL parameters, which bear predictive implications, is becoming of greater relevance in view of the progressive change in the management of a disease for which, until recently, observation and conservative treatment was the strategy of choice for the majority of patients. Several considerations have contributed to this modified attitude, including the knowledge that about 20% of patients are diagnosed with CLL at 55 years or younger, that 60% to 70% of patients at the time of diagnosis have an early stage disease, that the biological age of patients in their 60s has dramatically improved, that the overall life expectancy is progressively increasing, and that we now have a broader therapeutic armamentarium for patients with CLL.2
The p53 tumor suppressor gene, located on chromosome 17 band p13.1, is a transcription factor that is involved in the cell cycle arrest and induction of apoptosis in genetically damaged cells. Mutations or deletions of the p53 gene may facilitate the transmission of a genetic damage and the emergence of neoplastic clones with a survival advantage.3,4 p53 is the most frequently altered gene in human cancer, being mutated in approximately 50% of all human tumors.5 This gene is known to be altered in a number of hematologic malignancies, but the frequency of p53 gene mutations tends to be low in most lymphoid malignancies and is found mainly in aggressive non-Hodgkin's lymphoma (NHL),6-11 progressive CLL,12-18 and B-cell chronic prolymphocytic leukemia (PLL).19 Cases of indolent lymphoma and most cases of CLL have so far been reported to be negative for p53 mutations.
The p53 gene encodes a p53-kD phosphoprotein that is normally present in the nucleus of the cells. The wild-type p53 protein has a short half-life and cannot be detected in the cell nucleus of most normal human tissues. In contrast, mutated p53 has a prolonged half-life and becomes detectable by immunologic techniques using anti-p53 monoclonal antibodies (MoAb).20,21 For several years the immunologic identification of the p53 protein in human tumors has been considered a marker of p53 gene mutation.3,21 However, more recent studies in high grade NHL, CLL, and PLL have shown that p53 expression may not be associated with detectable gene mutations, whereas a mutant p53 gene can have an undetectable protein,8,19 22-29indicating that gene mutation and protein detection may not be associated.
In the present study, we have analyzed by immunocytochemistry (ICC) the frequency of p53 protein expression in 181 CLL patients followed at a single institution to assess the relationship between p53 and the clinical and morphological features of the disease, the possible involvement of this protein in the pathogenesis of the more aggressive forms of CLL, and its impact on the response to treatment and overall survival. To evaluate whether p53 positivity to ICC was due to mutations in the p53 gene or to other mechanisms of p53 stabilization, a direct sequence of the entire protein coding region was performed in the majority of ICC-positive cases. p53 expression increases in association with cell proliferation.30,31 A relationship between proliferation and p53 expression has been reported in NHL11,32 in which the degree of p53 expression correlated with prognosis, histological grade, and resistance to treatment. Because a correlation between the percentage of leukemic cells in cycle and the stage and clinical behavior of CLL has been documented,33 we have also investigated a possible relationship between p53 expression and positivity with Ki-67, a MoAb that recognizes a nuclear antigen expressed during most phases of the cell cycle.
MATERIALS AND METHODS
Patients.
Peripheral blood samples from 181 cases of CLL referred to our institution were studied. Informed consent was obtained from all patients. Diagnosis, clinical staging, and response were based on the criteria recommended by the International Workshop on CLL.34 Cases were classified as CLL (n = 147) or CLL with greater than 10% prolymphocytes (CLL/PL) (n = 34) on the basis of May-Grunwald Giemsa–stained peripheral blood films.35 All samples that entered the study were CD19+, CD20+, CD5+, and CD23+. B-cell clonality was established using anti-κ and anti-λ immunoglobulin (Ig) light chain reagents. Due to the weak surface Ig expression, staining of fixed cells by immunoperoxidase was necessary in most cases to show the presence of monoclonal Ig light chains. One hundred eight patients were men and 73 were women. The mean age was 66 ± 10 years. According to Binet's staging system, 108 patients were in stage A, 49 in stage B, and 24 in stage C. Within CLL stage A, two groups were considered A' (Hb>12 g/dL and lymphocytes < 30 × 109/L) (n = 74) and A” (Hb <12 g/dL and/or lymphocytes > 30 × 109/L) (n = 34).36According to the phase of the disease, three groups were identified: (1) patients at diagnosis (n = 112), (2) patients with stable disease (n = 35),37 and (3) patients with progressive disease (n = 34).38 Groups 2 and 3 included patients studied 2 to 228 months (median, 41) from diagnosis. The treatment-free interval (TFI), defined as the mean time (months) from diagnosis to the start of treatment, was calculated for the 112 patients studied at diagnosis. Resistance to treatment was defined as the failure to achieve a partial remission (PR)34 after therapy with intermittent chlorambucil plus prednisone, fludarabine plus prednisone or CHOP (cyclophosphamide, doxorubicin, vincristine, and prednisone). Overall, 22 patients were on treatment at the time of this analysis, but all of them had greater than 80% peripheral blood leukemic B lymphocytes.
ICC.
Mononuclear cells were isolated from heparinized peripheral blood by Lymphoprep density gradient centrifugation (Nycomed Pharma AS, Oslo, Norway). Cytospins were prepared with a concentration of 5 × 104 cells per slide, air dried overnight, wrapped in aluminum foil, and stored at −20°C until immunostaining. Because we have observed a progressive decrease in p53 protein expression within a few months from the cytospin preparations, all cases were studied in the 2 months subsequent to storage at −20°C.
The MoAb used for the immunocytochemical detection of p53 were DO-7 (Dako, Glostrup, Denmark) and DO-1 (Oncogene Science, Uniondale, NY), which recognize two different N-terminal epitopes of the human p53 protein. These two antibodies react with both wild-type and mutant p53 protein. DO-7 was used at a final concentration of 5 μg/mL and DO-1 at a final concentration of 2 μg/mL. The immunocytochemical reaction was performed with the immunoperoxidase technique using Dako reagents, as previously described.33The Raji cell line and normal peripheral blood lymphocytes were used as positive and negative controls, respectively. The Ki-67 MoAb (Dako) was used to detect proliferating cells and the UCHT1 (CD3) MoAb (Dako) to recognize T lymphocytes. The proportion of p53, Ki-67, and CD3-positive cells was evaluated by light microscopy with oil immersion (magnification × 1,000) examining 500 lymphoid cells per sample.
DNA sequencing.
Total RNA was isolated from frozen pellets of mononuclear blood cells using the RNeasy mini kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. Reverse transcription was performed on 1 to 2 μg of total RNA using 200 U/sample of Moloney murine leukemia virus reverse transcriptase (Pharmacia Biotech AB, Milan, Italy) and random primers. Each cDNA preparation was amplified with 4 U of Taq polymerase (Ampli Taq; Perkin-Elmer AB, Milan, Italy) according to the manufacturer's instructions, using a Perkin Elmer 9600 PCR equipment programmed to perform 38 cycles. DNA sequencing primers were synthesized according to the cDNA sequence of p53 messenger RNA. Polymerase chain reaction (PCR) primers were prepared by Pharmacia Biotech AB. Four sets of primers were used to cover the complete protein coding region of the p53 cDNA.39 Sequencing reactions were performed as described39 using streptavidin-coupled Sepharose HP attached to the teeth of plastic combs (solid-phase sequencing combs). The combs were removed from the sequencing reaction mixtures and inserted into the wells of an automatic laser fluorescence (ALF) DNA sequencer (Pharmacia Biotech, Uppsala, Sweden). After 10 minutes, the comb was carefully removed from the gel apparatus and electrophoresis initiated. Evaluation of the p53 sequences was performed with the aid of the DNAstar (DNAStar Inc, London, England) software program.
Statistical analysis.
Two-sided χ2 or Student's t-test were used to compare age, sex, absolute lymphocyte count, CLL/PL morphology, lymphocyte doubling time (LDT), percentage of Ki-67–positive and CD3-positive lymphocytes, stage and phase of the disease, TFI, and response to therapy between patients with and without p53 expression. The Kruskal-Wallis test was used to compare the percentage of p53-positive cells in the three groups of patients subdivided according to the clinical stage and phase of the disease. Survival was defined as the time from diagnosis or from p53 analysis to death or to the last observation. Comparison between curves was performed by the two-sided log-rank test and actuarial curves were constructed according to the Kaplan-Meier method. Survival multivariate analysis was performed according to Cox's proportional hazard model.
RESULTS
p53 expression in CLL.
With the immunocytochemical method used to identify p53-positive cells, intense brown nuclear staining of the positive population was obtained, with good preservation of morphological details (Fig 1A). The reaction was always confined to the nucleus. Cytospins with a mixture of Raji cells (p53 positive) and normal peripheral blood lymphocytes (p53 negative) were used as controls (Fig 1B). Because no p53-positive cells were observed in the 10 normal peripheral blood samples used as negative controls, CLL was considered positive when at least 1% of lymphoid cells showed a strong nuclear staining with the anti-p53 MoAb.
Immunoperoxidase staining for p53 protein: the positive population shows intense brown nuclear staining. (A) Cytospin of CLL lymphocytes: the immunostaining shows the coexistence of p53-positive and p53-negative cells within the same leukemic population. (B) Cytospin containing a mixture of Raji cells (p53 positive) and normal peripheral blood lymphocytes (p53 negative) used as control.
Immunoperoxidase staining for p53 protein: the positive population shows intense brown nuclear staining. (A) Cytospin of CLL lymphocytes: the immunostaining shows the coexistence of p53-positive and p53-negative cells within the same leukemic population. (B) Cytospin containing a mixture of Raji cells (p53 positive) and normal peripheral blood lymphocytes (p53 negative) used as control.
The overall frequency of p53 protein positivity in CLL was 15% (27 of 181 cases). The mean percentage of p53-positive cells is reported in Table 1. There was no significant difference between the percentage of DO-7–positive and DO-1–positive cells. Only one case was DO-7 positive and DO-1 negative. For this reason, the results presented will refer to the DO-7 MoAb immunostaining. Five cases had less than 5% p53-positive cells and in six all of the leukemic cells were p53 positive. In most cases, the proportion of p53-positive cells was lower than the percentage of leukemic B cells evaluated on the basis of the percentage of monoclonal κ or λ light chain positive lymphocytes.
p53-Positive CLL: Percentage and Absolute Number of Positive Cells Assessed by ICC Staining
| MoAb . | (%) . | (×109/L) . |
|---|---|---|
| DO-7 | 44.2 ± 6.9 | 34.6 ± 10.6 |
| DO-1 | 42.7 ± 6.8 | 34.2 ± 10.8 |
| Ki-67 | 4.7 ± 1.2 | 3.6 ± 1.7 |
| CD3-150 | 7.3 ± 1.2 | 2.5 ± 0.5 |
| MoAb . | (%) . | (×109/L) . |
|---|---|---|
| DO-7 | 44.2 ± 6.9 | 34.6 ± 10.6 |
| DO-1 | 42.7 ± 6.8 | 34.2 ± 10.8 |
| Ki-67 | 4.7 ± 1.2 | 3.6 ± 1.7 |
| CD3-150 | 7.3 ± 1.2 | 2.5 ± 0.5 |
Results represent the mean ± SEM.
Residual T lymphocytes.
Patients' characteristics according to p53 immunostaining.
Based on the immunostaining pattern, the 181 patients were subdivided into two groups: (1) p53-negative (n = 154) and (2) p53-positive (n = 27) CLL. There were no significant differences in age, sex, absolute lymphocyte count, or LDT between the two groups. By contrast, p53-positive cases had a significantly higher percentage of prolymphocytes and a significantly lower percentage of residual CD3-positive T lymphocytes (Table 2).
Patients Characteristics According to p53 Immunostaining
| Characteristics . | p53 Negative (n = 154) . | p53 Positive (n = 27) . | P Value . |
|---|---|---|---|
| Age (yrs) | 65.2 ± 0.8 | 67.7 ± 1.9 | NS |
| Sex (M/F) | 92/62 | 16/11 | NS |
| CLL/PL (%) | 15 | 41 | .002 |
| Lymph (×109/L) | 42.8 ± 7.2 | 60.7 ± 11.3 | NS |
| CD3 (%) | 15.6 ± 0.9 | 7.3 ± 1.2 | .0001 |
| LDT <1 year (%) | 31.6 | 50 | NS |
| Characteristics . | p53 Negative (n = 154) . | p53 Positive (n = 27) . | P Value . |
|---|---|---|---|
| Age (yrs) | 65.2 ± 0.8 | 67.7 ± 1.9 | NS |
| Sex (M/F) | 92/62 | 16/11 | NS |
| CLL/PL (%) | 15 | 41 | .002 |
| Lymph (×109/L) | 42.8 ± 7.2 | 60.7 ± 11.3 | NS |
| CD3 (%) | 15.6 ± 0.9 | 7.3 ± 1.2 | .0001 |
| LDT <1 year (%) | 31.6 | 50 | NS |
Results represent the mean ± SEM.
Abbreviation: NS, not significant.
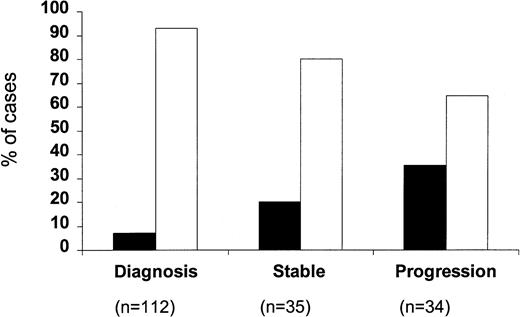
When the percentage of p53-positive cases was correlated with the clinical stage of the disease (Fig 2), only a very small minority of stage A patients were p53 positive (8 of 108; 7.4%). This increased to 24.5% for stage B patients (12 of 49) and to 29.2% for stage C patients (7 of 24) (P = .002). Within stage A CLL, four of 74 stage A' and four of 34 stage A” patients were p53 positive (5.4% v 11.7%). A difference between stages was also observed with regard to the percentage of p53-positive cells, which were 31.2% ± 13.2% in stage A, 39.6% ± 8.1% in stage B, and 66.9% ± 15.6% in stage C, respectively; this difference, however, did not reach significance. When p53 positivity was related to the phase of the disease (Fig 3), it was found that only eight of the 112 cases studied at the time of diagnosis (7.1%) were p53 positive, whereas 7 of 35 (20%) patients studied during the stable course of their disease were p53 positive. The percentage of p53-positive cases increased further when patients with progressive disease were investigated (12 of 34; 35.3%) (P = .0001).
Percentage of p53-positive (▪) and p53-negative (□) patients subdivided according to the clinical stage.
Percentage of p53-positive (▪) and p53-negative (□) patients subdivided according to the clinical stage.
Percentage of p53-positive (▪) and p53-negative (□) patients subdivided according to the phase of the disease.
Percentage of p53-positive (▪) and p53-negative (□) patients subdivided according to the phase of the disease.
p53 sequence.
Among the 27 cases positive for p53 staining by ICC, 17 were studied to verify whether positive ICC was due to gene mutations or to other mechanisms of p53 stabilization. Total RNA was extracted from blood mononuclear cells, reverse transcribed, and the entire protein coding region sequenced. p53 mutations were found in 15 cases (88%) (Table 3). Missense mutations were identified in all 15 samples. One out of frame mutation was found in patient no. 4 together with other two missense mutations. No nonsense mutations or insertions were found. The most frequent mutation, found in 10 cases, was a C → T transition at codon 143 (exon 5), which resulted in a substitution of a valine with an alanine. More than one mutation was found in eight samples. Moreover, four of the 14 different mutations found were located outside the evolutionary conserved regions of the p53 coding for the subdomains I-V. The percentage of p53-positive cells stained by ICC was high in the majority of cases studied by cDNA direct sequence; however, p53 mutations were detected also in four cases (no. 9, 12, 13, and 14), which had a low percentage of p53-positive cells.
p53 Positivity by ICC and Corresponding p53 Gene Mutations Detected by cDNA Sequence Analysis
| Case No. . | p53 Protein (% positive cells) . | Exon . | Codon . | Nucleotide Substitution . | AA Substitution . |
|---|---|---|---|---|---|
| 1 | 72 | 5 | 129 | T → C | Ala → Val |
| 2 | 97 | 4 | 72* | G → T | Pro → Arg |
| 5 | 143 | C → T | Val → Ala | ||
| 3 | 64 | 7 | 201 | A → T | Met → Lys |
| 4 | 59 | 5 | 143 | C → T | Val → Ala |
| 9 | 301 | Del A | Out of frame | ||
| 10 | 314* | T → C | Ser → Phe | ||
| 5 | 97 | 4 | 64* | G → C | Pro → Ala |
| 4 | 72* | G → T | Pro → Arg | ||
| 5 | 143 | C → T | Val → Ala | ||
| 6 | 55 | 5 | 143 | C → T | Val → Ala |
| 7 | 95 | 5 | 110 | T → C | Arg → Pro |
| 8 | 89 | WT | WT | ||
| 9 | 3 | 5 | 143 | C → T | Val → Ala |
| 9 | 286 | G → A | Glu → Gly | ||
| 10 | 85 | WT | WT | ||
| 11 | 97 | 5 | 129 | T → C | Ala → Val |
| 5 | 143 | C → T | Val → Ala | ||
| 12 | 6 | 5 | 143 | C → T | Val → Ala |
| 13 | 1 | 5 | 143 | C → T | Val → Ala |
| 6 | 189 | G → T | Pro → Ala | ||
| 10 | 328* | T → C | Thr → Isol | ||
| 14 | 1.5 | 5 | 143 | C → T | Val → Ala |
| 5 | 140 | G → T | Cys → Gly | ||
| 15 | 14 | 5 | 143 | C → T | Val → Ala |
| 16 | 19 | 5 | 132 | A → T | Met → Aspar |
| 7 | 209 | C → A | Aspar → Hist | ||
| 17 | 29 | 5 | 132 | A → T | Met → Aspar |
| Case No. . | p53 Protein (% positive cells) . | Exon . | Codon . | Nucleotide Substitution . | AA Substitution . |
|---|---|---|---|---|---|
| 1 | 72 | 5 | 129 | T → C | Ala → Val |
| 2 | 97 | 4 | 72* | G → T | Pro → Arg |
| 5 | 143 | C → T | Val → Ala | ||
| 3 | 64 | 7 | 201 | A → T | Met → Lys |
| 4 | 59 | 5 | 143 | C → T | Val → Ala |
| 9 | 301 | Del A | Out of frame | ||
| 10 | 314* | T → C | Ser → Phe | ||
| 5 | 97 | 4 | 64* | G → C | Pro → Ala |
| 4 | 72* | G → T | Pro → Arg | ||
| 5 | 143 | C → T | Val → Ala | ||
| 6 | 55 | 5 | 143 | C → T | Val → Ala |
| 7 | 95 | 5 | 110 | T → C | Arg → Pro |
| 8 | 89 | WT | WT | ||
| 9 | 3 | 5 | 143 | C → T | Val → Ala |
| 9 | 286 | G → A | Glu → Gly | ||
| 10 | 85 | WT | WT | ||
| 11 | 97 | 5 | 129 | T → C | Ala → Val |
| 5 | 143 | C → T | Val → Ala | ||
| 12 | 6 | 5 | 143 | C → T | Val → Ala |
| 13 | 1 | 5 | 143 | C → T | Val → Ala |
| 6 | 189 | G → T | Pro → Ala | ||
| 10 | 328* | T → C | Thr → Isol | ||
| 14 | 1.5 | 5 | 143 | C → T | Val → Ala |
| 5 | 140 | G → T | Cys → Gly | ||
| 15 | 14 | 5 | 143 | C → T | Val → Ala |
| 16 | 19 | 5 | 132 | A → T | Met → Aspar |
| 7 | 209 | C → A | Aspar → Hist | ||
| 17 | 29 | 5 | 132 | A → T | Met → Aspar |
Abbreviation: WT, wild-type.
Codons located outside the evolutionary conserved p53 regions.
Frequency of Ki-67 expression in p53-positive CLL.
With the immunocytochemical method used to identify Ki-67–positive cells, intense brown nuclear staining of the positive population was obtained, with good preservation of morphologic details. The results of immunostaining are summarized in Table 1. The percentage of Ki-67 positivity in the p53-positive cases was 4.7% ± 1.2%. When the percentage of Ki-67–positive cells was correlated with the morphology, a significantly higher percentage of cells in cycle was found in CLL/PL (8.8% ± 2.5%) compared with CLL (2.0% ± 0.4%) cases (P = .004). There was no correlation between the percentage of p53- and Ki-67–positive cells.
Response to therapy and survival.
Of the 154 p53-negative patients by ICC, 82 (53%) have so far never required treatment, 10 (7%) were lost to follow-up, and 62 (40%) were treated with chlorambucil plus prednisone (n = 42), fludarabine plus prednisone (n = 18), or CHOP (n = 2) as first line therapy. Nineteen of the 27 p53-positive patients (70%) were treated with chlorambucil plus prednisone (n = 16), fludarabine and prednisone (n = 2), and CHOP (n = 1). A significantly poorer response to therapy was observed in the p53-positive patients (P = .007) (Table 4). A similar poor response to therapy was observed in the p53-positive group when the analysis was focused on the 112 patients studied at diagnosis (data not shown). The same difference in response rate was observed between the 16 (10%) p53-negative and nine (33%) p53-positive patients who required treatment within 3 months from diagnosis (no response: 31%v 78%, respectively; P = .04).
Relationship Between p53 Expression and Response to Therapy
| . | p53 Negative n = 62 . | p53 Positive n = 19 . | ||
|---|---|---|---|---|
| CR + PR (%) . | NR (%) . | CR + PR (%) . | NR (%) . | |
| Chlorambucil | 27 (64) | 15 (36) | 5 (31) | 11 (69) |
| Fludarabine | 16 (89) | 2 (11) | 2 (100) | 0 |
| CHOP | 1 (50) | 1 (50) | 0 | 1 (100) |
| Total | 44 (71) | 18 (29) | 7 (37) | 12 (63) |
| . | p53 Negative n = 62 . | p53 Positive n = 19 . | ||
|---|---|---|---|---|
| CR + PR (%) . | NR (%) . | CR + PR (%) . | NR (%) . | |
| Chlorambucil | 27 (64) | 15 (36) | 5 (31) | 11 (69) |
| Fludarabine | 16 (89) | 2 (11) | 2 (100) | 0 |
| CHOP | 1 (50) | 1 (50) | 0 | 1 (100) |
| Total | 44 (71) | 18 (29) | 7 (37) | 12 (63) |
Abbreviations: CR, complete response; PR, partial response; NR, no response.
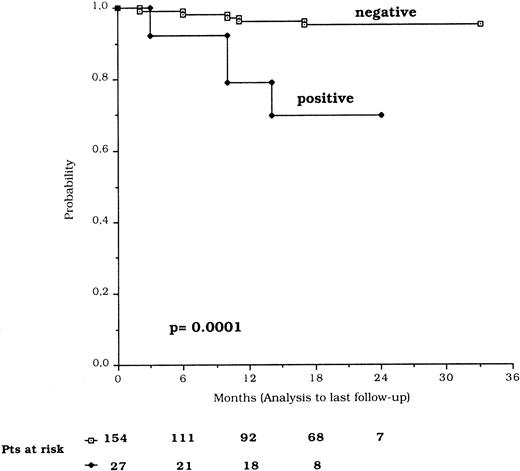
TFI, calculated for the 112 patients studied at diagnosis, showed a mean time of 19.3 ± 1.1 months for the p53-negative and 4.0 ± 2.2 months for the p53-positive patients (P = .003). The overall survival from the time of diagnosis was significantly shorter in p53-positive compared with p53-negative patients (P = .03) (Fig 4). The difference was highly significant (P = .0001) also when survival was considered from the date of p53 protein expression analysis (Fig 5). The Cox proportional hazard model results show that p53 expression and clinical stage C were independently associated with a short survival (Table5).
DISCUSSION
Structural alterations and point mutations of the p53 tumor suppressor gene have been shown in 10% to 15% of CLL; they have been associated with poor survival and nonresponse to therapy and reported in rare cases of high-grade lymphoma evolved from CLL (Richter's transformation), suggesting that p53 may play a role in the clinical course of the disease and in the transformation of some cases of CLL.12-18 Less attention has been paid to the significance of p53 protein expression in CLL, although an association with poor survival and nonresponse to therapy has been observed in small series of CLL.21,40 p53 expression has been shown to be a fairly common feature in high-grade NHL, significantly associated with a short survival and not always secondary to p53 gene mutation. Several studies have, in fact, shown that in some high grade NHL the occurrence of positive immunostaining does not reflect point mutations in the p53 gene and vice versa.8,11,22-24,26-28,41-44 Therefore, it is evident that the relationship between p53 protein detection and the existence of gene mutations is more complex than initially expected and that other mechanisms of p53 stabilization are frequently operating in NHL.27
To determine the frequency of p53 protein expression in CLL, we have examined 181 patients by immunoperoxidase and have found a strong expression of the protein in 15% of cases. Attention was paid to include in the study only patients with a typical CLL phenotype and thus to exclude patients with a leukemic manifestation of NHL. The percentage of cells stained by p53 antibodies was in the majority of cases lower than the percentage of leukemic cells. The coexistence of p53-positive and -negative cells within the same leukemic population supports the hypothesis that p53 disregulation can be a late event in the progression of the disease. Although the p53 alteration may occur early in the course of the disease, even in the so-called smouldering CLL, as shown by the p53 positivity in a proportion of patients studied at diagnosis and in stage A', the highest frequency of p53 expression, as well as the highest percentage of p53-positive cells, has been observed in stages B and C, and in patients with progressive disease. These findings are in agreement with previous studies, which have shown a strong correlation between p53 mutations and progression in hematologic malignancies.7,45,46 The expansion of an initially minor subclone with a mutated p53 during disease progression has indeed been shown in brain tumors47 and in acute myeloid leukemia.48 We have observed this type of clonal evolution in one patient with few p53-positive cells at diagnosis and in whom the percentage of DO-7–positive cells increased progressively during the evolution of the disease (data not shown). A statistical correlation between p53 deletion and presence of lymphadenopathy in CLL has been suggested.15 18 The significantly higher frequency of p53 expression hereby recorded in stages B and C at diagnosis suggests a possible relationship between lymphnode enlargement and p53 expression in CLL.
A strong correlation between p53 expression and atypical CLL morphology, chiefly an increased proportion of prolymphocytes, was found. The immunocytochemical technique used to identify p53-positive cells, which allows a morphological evaluation of the positive population, showed that both small lymphocytes and prolymphocytes were p53 positive, but no clear-cut correlation between the percentage of prolymphocytes and of p53-positive cells could be established. CLL, with an increased proportion of prolymphocytes, seems to be a disease with a high frequency of p53 protein expression. This is in agreement with the high frequency of p53 mutations recently reported in PLL.19 These findings further underline that in CLL a detailed morphological assessment represents a useful prognostic parameter in the context of an otherwise heterogeneous condition.
A significantly lower percentage of CD3-positive lymphocytes was found in the p53-positive group. We have previously shown that the proportion of CD3 lymphocytes in stage A', which included patients with long-standing stable disease, was significantly higher than in the other stages.33 It is possible that in CLL an important immunologic control mechanism may be mediated by residual T cells and that a low percentage of these cells may identify a less controlled disease that can behave more aggressively.
Overexpression of wild-type p53 has been described in highly proliferating cells and reactive tissues.22,26,31 Moreover, p53 mRNA increases in association with cell proliferation.30 The existence of a relationship between proliferation and p53 expression has also been observed in NHL.32 A number of studies have shown that the proliferative activity of neoplastic cells is a good indicator of the biological behavior of the tumor with both prognostic and therapeutic implications. We have reported that the percentage of Ki-67–positive cells in CLL increases with the stage of the disease and correlates with the proportion of prolymphocytes.33 In the present study, we have found among p53-positive cases a percentage of Ki-67–positive cells comparable to the values reported in CLL with advanced clinical stage, atypical morphology,33 and trisomy 12,49 which are higher than those observed in indolent CLL. However, this elevated percentage of cells in cycle is due to the high frequency of CLL/PL within the p53-positive patients. Moreover, no direct correlation was observed between the percentage of Ki-67–positive and p53-positive cells, p53-positive cases having a significantly lower percentage of cells in cycle than the percentage of p53-expressing cells.
Although concordance between ICC and DNA analysis of p53 has been reported in hematologic malignancies,21 it has been suggested that p53 protein expression observed in progressive CLL may be due to posttranscriptional modifications that induce functional and conformational alterations.40 To verify whether the aberrant p53 expression observed in our CLL series was due to mutations in the p53 gene or to other types of modification, a cDNA sequence analysis was performed in 17 of the 27 p53-positive cases. Because it has been reported that sequences restricted to exons 5 through 9 leave undetected a consistent portion of mutations,39 we sequenced the entire p53 protein coding region. A strong correlation was found (88%) between p53 ICC positivity and p53 mutations detected by cDNA direct sequence, showing that p53 expression was mainly due to gene mutations. Four of the 14 mutations found were outside the hot spots. This underlines the importance of analyzing also outside the conserved regions of the p53 gene to compare the sensitivity of the immunologic and molecular techniques in identifying p53 alterations. We found the same mutation at codon 143, the C → T transition, in 10 of the 15 cases with p53 gene alterations. This type of mutation is particularly frequent in lung, head, and neck cancers.50With regard to the correlation between the percentage of p53-positive cells and gene mutations, we detected p53 mutations also in cases with a low percentage of cells stained by p53 antibodies. Moreover, eight cases had more than one mutation. This raises the issue of the mechanism of acquisition of these mutations and whether they occur in one or both of the p53 genes. Further studies are in progress to clarify this question. Although ICC represents a sensitive method of p53 mutation detection in CLL, giving concordant results with direct sequence analysis in a high proportion of cases, ICC may be positive in the absence of detectable p53 mutations and could correspond to overexpression of a nonmutated p53, confirming that mechanisms of p53 stabilization other than p53 gene alteration operate in CLL.
p53 protein expression bears strong implications in the clinical course of the disease. p53-positive patients showed a significantly shorter TFI from diagnosis and poorer response to therapy compared with p53-negative patients. Moreover, when the analysis was centered on patients with active disease at diagnosis who required treatment within 3 months from diagnosis, p53-negative patients showed a significantly higher response rate than p53-positive patients. p53 gene deletions have been associated with nonresponse to therapy with purine analogs in CLL.16 In our hands, 16 of 18 p53-negative patients treated with fludarabine achieved a PR (n = 9) or CR (n = 7), whereas the two p53-positive patients treated with this purine analog obtained only a short-lived PR. Among the presently available prognostic factors, clinical stage is considered the strongest predictor of survival in CLL. We have shown a significant relationship between p53 expression and short survival. Moreover, p53 expression and stage C were independently associated with a short survival. The same relationship has been found in NHL27 and in other types of tumors, including colon,51 breast,52bladder,53 gastric,54 lung,50,55and prostatic56 cancer.
In conclusion, our findings indicate that p53 expression in CLL is strongly associated with p53 gene mutations, a morphological variant (CLL/PL), advanced clinical stage, progressive disease, poor response to therapy, and short survival. In the context of a heterogeneous condition like CLL, this simple, inexpensive, and reliable immunocytochemical method appears to offer a useful prognostic tool capable of identifying patients with aggressive disease and who may be considered upfront for more intensive therapeutic strategies. This is of particular relevance for younger patients for whom more eradicative approaches are becoming more frequently used.
Supported by Istituto Superiore di Sanità, Italy-US Project on “Therapy of Tumors,” Rome and by ROMAIL (Italian Association against Leukemia, Section of Rome), Italy. I.C. is the recipient of a fellowship from Istituto Superiore di Sanità, Rome, Italy.
Address reprint requests to Iole Cordone, MD, Dipartimento di Biotecnologie Cellulari ed Ematologia, Università “La Sapienza,” Via Benevento 6, 00161 Rome, Italy.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal