Abstract
Previous studies have indicated that cytidine deaminase (CDD) is a potent growth inhibitor of granulocyte-macrophage colony-forming cells (GM-CFC). In this study, we have undertaken molecular cloning and purification of recombinant human CDD to elucidate the growth regulatory potential and mechanism behind the growth suppressive effect. The purified protein had a specific activity of 1.35 × 105 U/mg and a Km value of 30 μmol/L. In the GM-CFC assay, the recombinant protein was shown to reduce colony formation to 50% at 16 pmol/L concentration. Similarly, as was observed with CDD derived from granulocyte extract, the effect depended on the presence of thymidine (≥ 4 × 10-5 mol/L). These results imply that CDD is an extremely potent inhibitor of GM-CFC and that no additional factor from the granulocyte extract is required for the growth inhibitory effect. Modification of CDD by truncation from the C-terminal end, or by amino acid substitution of an active site glutamate residue, eliminated both the enzyme activity and the growth regulatory potential of CDD. Furthermore, CDD fromEscherichia coli was found to be even more effective than human CDD in growth suppression of GM-CFC, with 10-fold higher inhibitory activity corresponding to a 10-fold higher enzymatic activity. Taken together, these results show that the catalytic nucleoside deaminating function of the protein is essential for the growth suppressive effect of CDD. Most probably, CDD exerts growth inhibition by depleting the cytidine and deoxycytidine pool required for DNA synthesis, as addition of deoxycytidine monophosphate, which is not a substrate for CDD, neutralizes the inhibiting effect.
THE DEVELOPMENT of blood cells from hematopoietic stem cells in the bone marrow is a complex process dependent on the microenvironment constituted by stromal cells, cytokines, and the extracellular matrix.1-4 The cytokines mediate communication between the cells and exert their biological functions mostly through specific receptors expressed on the surface of the target cells. These signal substances have the capacity to stimulate, enhance, or suppress the proliferation of the stem and progenitor cells. Some cytokines even have pleiotropic effects, with different activities depending on the target cell or assay system used.5 The inhibitors constitute a rather heterogeneous group of molecules, ranging from higher molecular weight protein factors like the transforming growth factor-β, tumor necrosis factor-α and -β, macrophage inflammatory protein-1α, and the interferons6 to shorter oligopeptides like pEEDCK7,8 and AcSDKP.9 10
We have previously shown that mature human blood granulocytes produce an inhibitor of granulopoiesis in diffusion chambers11 and granulocyte-macrophage colony-forming cells (GM-CFC) in agar assays.12 The inhibitor was identified as cytidine deaminase (CDD), as judged from different experimental criteria.13 The inhibitor copurified with CDD activity, the molecular weight of the inhibitor was found to be in the same size range of that reported for CDD (≈50 kD), and the growth suppressive effect of granulocyte extract was abolished by 3,4,5,6-Tetrahydrouridine (THU), a well-known inhibitor of CDD activity. CDD was found to be a species nonspecific, but seemingly lineage-specific suppressor11 that requires thymidine (≥3 × 10-5 mol/L for human mononuclear cells [MNC] in agar assays) to exert a strong inhibitory effect in vitro.14 CDD is considered to be an intracellular cytosolic enzyme, which deaminates cytidine and deoxycytidine (dC) to their respective uridine derivatives.15 It has been suggested that CDD mRNA expression may serve as a marker for myeloid differentiation because the expression level increases with maturation.16-18 Mature granulocytes also have a markedly higher CDD mRNA expression than chondrocytes, fibroblasts, T-cell lines, and B-cell lines.16 Several studies have indicated that CDD is actively released from viable granulocytes. Leukemic granulocytes with prolonged survival were found to inhibit G-CFC in human bone marrow more than rapidly dying leukemic cells.11 Granulocyte-conditioned medium causes a significant inhibition of GM-CFC.12 Other investigators have reported that polymorphonuclear cells show augmented release of CDD on activation,19,20 correlated to the granulocyte specific lactoferrin release from secondary granules.20 These findings support a negative feedback inhibition of granulopoiesis by released CDD. However, the mechanism of action has not been resolved. One interpretation is that CDD acts as a classical signal transducer with growth regulatory effects. An alternative model is that CDD exerts its action through its enzymatic properties.
To distinguish between these possibilities and further characterize the molecular effects of CDD on granulopoiesis, we have undertaken molecular cloning, purification, and characterization of human CDD. It is shown that human recombinant CDD (rhCDD) is as effective as authentic CDD from mature granulocytes in suppressing GM-CFC. Furthermore, site-directed mutagenesis of active site residues of CDD indicates that the growth inhibitory function of CDD in granulopoiesis is correlated to the enzymatic activity of the protein and is the result of depleting the pool of cytidine and dC required for genome replication.
MATERIALS AND METHODS
Materials.
Random priming kit and T4 DNA ligase were obtained from Boehringer Mannheim (Mannheim, Germany). α-32P-deoxycytidine triphosphate (dCTP), α-35S-deoxyadenosine triphosphate (dATP) (Easytide), Deoxy[5-3H]cytidine, and Rainbow colored molecular weight markers were obtained from Amersham Pharmacia (Uppsala, Sweden). Sequencing kit was obtained from United States Biochemical (Cleveland, OH). RPMI 1640, L-glutamine, and penicillin-streptomycine mixture were obtained from Bio-Whittaker (Walkersville, MD). Fetal calf serum (FCS) was from GIBCO (Grand Island, NY). Lymphoprep was provided by NYCOMED (Oslo, Norway). Chemicals and other reagents were obtained from Sigma (St Louis, MO), Fluka (Buchs, Switzerland), Bio-Rad Laboratories (Hercules, CA), and New England Biolabs (Beverly, MA).
Synthesis of DNA probe and screening of cDNA library.
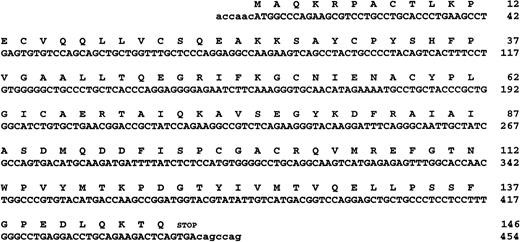
The incomplete CDD nucleotide sequence of Kühn et al16 was used to synthesize the 5′- and 3′-primers (P1 and P2) for amplification of a specific DNA probe from a human polymorphonuclear leukocyte cDNA library (λgt11; Clontech, Palo Alto, CA). The sequences of these primers were: P1, 5′-TGCTGGTTTGCTCCCAGG (nucleotides 59-76, Fig 1A) and P2, 3′-CAGTACTGCCAGGTCCTC (nucleotides 382-399, Fig 1A). The DNA probe was radiolabeled using a random oligonucleotide priming kit, and the probe purified through a Nick column (Pharmacia, Uppsala, Sweden). A human blood cDNA library in the ZAP ExpressEcoRI Vector (Stratagene, La Jolla, CA) was screened with the radiolabeled DNA probe. Plaques were blotted on Nylon filters (BA 85; Schleicher & Schuell, Keene, NH), the λ-DNA denatured in 1.5 mol/L NaCl/0.5 mol/L NaOH and neutralized in 1.5 mol/L NaCl/0.5 mol/L Tris-HCl, pH 8. The filters were prehybridized in 6 × standard sodiumcitrate (SSC), 5 × Denhardt's solution, 0.5% sodium dodecyl sulfate (SDS), and 100 μg/mL denatured salmon sperm for 1.5 hours at 68°C. The denatured32P-labeled DNA probe was added to the hybridization solution to give about 1 × 106 cpm/mL and the hybridization was performed overnight at 68°C. The filters were washed four times for 30 minutes at 42°C in 1 × SSC and 0.1% SDS. The filters were dried and wrapped in plastic before exposure to an AGFA CURIX x-ray film in a Kodak X-Omatic cassette (Eastman Kodak Co, Rochester, NY) with intensifying screen at −70°C. Isolated positive clones were converted to the pBK-CMV phagemid by in vivo excision as described (Stratagene).
(A) The nucleotide and predicted amino acid sequences of human CDD (GenEMBL Accession no. AJ000474). (B) Multiple alignment of CDD from different species as indicated: BACSU; Bacillus subtilis (P19079), ECOLI; Escherichia coli (P13652), HAEIN; Haemophilus influenzae, (P44325); HUMAN; Homo sapiens (P32320), MYCGE; Mycoplasma genitalium(P47298), MYCPI; Mycoplasma pirum (P47718), MYCPN;Mycoplasma pneumoniae (P75051), BRUMA; Brugia malayi, (U80980). Exposed regions indicate the active site domains and arrows denote the conserved cysteines/histidines involved in the Zn binding. The conserved glutamate referred to in the text is in bold within the first exposed region. Asterisks represent conserved residues found in all members of the protein family.
(A) The nucleotide and predicted amino acid sequences of human CDD (GenEMBL Accession no. AJ000474). (B) Multiple alignment of CDD from different species as indicated: BACSU; Bacillus subtilis (P19079), ECOLI; Escherichia coli (P13652), HAEIN; Haemophilus influenzae, (P44325); HUMAN; Homo sapiens (P32320), MYCGE; Mycoplasma genitalium(P47298), MYCPI; Mycoplasma pirum (P47718), MYCPN;Mycoplasma pneumoniae (P75051), BRUMA; Brugia malayi, (U80980). Exposed regions indicate the active site domains and arrows denote the conserved cysteines/histidines involved in the Zn binding. The conserved glutamate referred to in the text is in bold within the first exposed region. Asterisks represent conserved residues found in all members of the protein family.
Enzyme activity analysis by complementation.
Escherichia coli (E coli) strain JF611 (pyrE60 cdd thi-1 argE3 his-4 proA2 thr-1 leu-6 mtl-1 xyl-5 ara-14 galK2 lacY1 rpsL supE44) lacks CDD activity and pyrimidine de novo synthesis and is phenotypically unable to use cytidine or dC as sole pyrimidine source.21 Thus, the double mutant requires either uracil or uridine for growth. The strain was used to check plasmid constructs for functional expression of human CDD by growth tests in the absence of uracil or uridine. The pBK-CMV transformed bacteria was induced with 0.2 mmol/L isopropyl-β–D-thiogalactoside (IPTG) and cultured in minimal medium supplemented with 0.2% amino acids, 40 μg/mL dC, and 50 μg/mL kanamycin.
Isolation of bacterial cell extracts by plasmolysis.
The procedure was performed on ice.22 A total of 4 mL of cell culture was centrifuged at 3,500 rpm and the cell pellet washed in 0.04 mol/L Tris-HCl, pH 8. The cells were pelleted, resuspended in 84% sucrose solution (50 μL 0.04 mol/L Tris-HCl, pH 8), and incubated 10 minutes before the addition of 250 μL 50 mmol/L 4-morpholinepropanesulfonate (pH 7.5)/1 mmol/L EDTA/100 mmol/L KCl/1 mmol/L dithiothreitol (DTT)/125 μg/mL lysozyme. After a 45-minute incubation, cell debris and DNA was pelleted at 13,000 rpm for 15 minutes and the supernatant frozen on dry ice/ethanol.
CDD assays.
CDD activity in cell extract isolated by plasmolysis was determined by radiochemical and spectrophotometric assays. The radiochemical assay is described previously,13 with the modification that the enzyme was diluted in buffers containing 200 μg/mL bovine serum albumin (BSA) and 1 mmol/L DTT to stabilize the enzyme. The spectrophotometric assay is based on the decrease of absorbance when dC is converted to deoxyuridine (dU) at 235 nm where Δε = 2,250 mol/L-1 for a 1-cm light path and the initial rate is vi = (ΔA/Δt)/Δε. The assay with diluted enzyme was performed in 100 μmol/L dC, 100 mmol/L Tris-HCl (pH 7.5), 200 μg/mL BSA, and 1 mmol/L DTT. One unit of enzyme activity is defined as the amount of enzyme that catalyzes the deamination of 1 nmol dC/min at room temperature under the above conditions.
DNA sequencing.
The insert in pBK-CMV was sequenced on both strands using Sequenase version 2.0 (United States Biochemical, Cleveland, OH). The primers used were: P1, P2, P3 (complementary to P1), P4 (5′-TCTCCATGTGGGGCCTGC, nucleotides 295-312 in Fig 1A), P5 (complementary to P4), and the T3 and T7 primers in the respective promoter regions of the pBK-CMV vector.
Expression of rhCDD in E coli from pT7-SC.
The CDD cDNA was cloned into the vector pT7-SC (USB) and used for transformation of E coli BL21 (E coli B F-dcm ompT hsdS(rB-mB-) gal). The T7 RNA polymerase (encoded on a lysogenic lambda bacteriophage), and hence, CDD, were induced with 1 mmol/L IPTG for 2 hours. Expressed proteins were analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) as described by Schägger and von Jagow23 using a 16.5% T separating gel made from the 49.5% T/3% C acrylamide-bisacrylamide mixture and a 4% T stacking gel.
Purification of rhCDD.
All purification steps were performed at 4°C. The cell extract, from 4 L of bacterial cells prepared as described,22 was applied to an Affi-Gel Blue column (Bio-Rad) preequilibrated with buffer A (20 mmol/L Tris-HCl, pH 7.5; 50 mmol/L KCl; 1 mmol/L EDTA; 1 mmol/L DTT; and 20% glycerol). The column was washed with buffer A and proteins were eluted with 1 mol/L KCl in buffer A. The eluate was desalted by PD-10 chromatography (Pharmacia), and applied to a Mono Q anion exchange column (0.7 × 5.5 cm; Pharmacia Fast Protein Liquid Chromatography, FPLC). After washing, the column was developed with a linear gradient to 0.2 mol/L KCl in buffer A at a flow rate 0.2 mL/min. CDD active fractions were desalted by PD-10 and applied to a Mono S cation exchange column (0.7 × 5.5 cm; Pharmacia). The column was eluted with 0.2 mol/L KCl in buffer A. The final step yielded a single ultraviolet (UV)-absorbing (280 nm) peak of rhCDD. Protein concentration was determined by the Bio-Rad protein assay (Bio-Rad), with BSA as a standard.
Purification of E coli CDD.
The E coli CDD gene was amplified by polymerase chain reaction (PCR) from strain AB1157 (wild-type). The primers were selected from the CDD sequence entered in the EMBL Data Library (Accession no. X63144). The 5′-primer sequence, P6, was 5′-GGAATTCCATATGCATCCACGTTTTCA (theNdeI-site with flanking sequence in italic letters, the translational start codon underlined), the 3′-primer sequence, P7, 3′-CATCGGACTACCTTTAAACTTAAGG (nucleotides from the 3′-nontranslated region and the EcoRI-site with flanking sequence in italic letters). The PCR product was cloned into the pT7-SC vector after restriction cleavage with NdeI and EcoRI and used for transformations of BL21. Expression of E coli CDD after IPTG induction was examined by enzyme activity analysis and by SDS-PAGE. The possible incorporation of incorrect nucleotides during amplification was checked by sequence analysis; however, no mutations were found. Purification of the enzyme was performed in a three-step procedure at 4°C. The extract isolated by plasmolysis was applied to a diethylaminoethyl (DEAE)-Sephacel (Pharmacia) anion exchange column preequilibrated with buffer A. The column was step-eluted with 1 mol/L KCl in the same buffer. Salts were removed by PD-10 chromatography and the protein fraction was subjected to FPLC MonoQ chromatography. The column was eluted by a linear gradient of 0.2 mol/L KCl in buffer A at a flow rate of 0.2 mL/min. After desalting, active fractions were applied to a Mini Q anion exchange column of the SMART system (Pharmacia), eluted with 0.25 mol/L KCl at a flow rate of 0.1 mL/min. The protein concentration was measured by the Bio-Rad protein assay.
Enzyme kinetics.
For the determination of Km in a Lineweaver-Burk plot, initial rates (vi) were measured as a function of substrate concentrations covering the range at least from 0.5 to 5 × Km. Quartz cuvettes of 1-cm light path were used.
Cells.
Bone marrow cells (BMC) were obtained from the femurs of female (C57BL/6JxDBA/2) F1 hybrid mice (Bomholdt Gaard, Ejby, Denmark). MNC from human peripheral blood and buffy coat samples were separated as previously described.13 24 A human bladder carcinoma cell line (5637) was kindly provided by Dr Andrew King (SmithKline Beecham, Philadelphia, PA). The cell line was cultured in RPMI 1640 medium with 10% FCS and subcultured once or twice weekly. The medium conditioned by the carcinoma cell line is a rich source of human granulocyte colony-stimulating factor (G-CSF) (CM 5637).
GM-CFC assay.
Mouse BMC (5 × 104/mL) were cultured in 0.3% agar (Bacto-Agar; DIFCO, Detroit, MI) or 1% methylcellulose (Methocel MC 4000; Fluka) in McCoy's 5A medium with 16% FCS, 1 mL per culture dish. A total of 10% CM 5637 was used as stimulator in the routine agar assay, 5 ng/mL interleukin (IL)-1β, IL-3, and stem cell factor in the methylcellulose assay. Colonies (>50 cells) were counted after 7 days of incubation at 37°C with 7.5% CO2 in humidified air. Human peripheral blood MNC (5 × 105/mL) were cultured for 14 days without stimulator added. Aggregates of more than 40 cells were counted as colonies. The cultures were run in triplicates.
Site-directed in vitro mutagenesis of CDD.
cDNAs expressing truncated forms of CDD were constructed by PCR using primers with translation stop codons introduced at defined positions. The sequence of the PCR primers were: P8, 5′-GGAATTCGGCACGAGACCAACATG (EcoRI-site with flanking sequence in italic letters, linker sequence underlined and nucleotides 1-9 in Fig 1A); P9, 3′-CCGTGGTTGACCGGGCACATTCGAACCC (nucleotides 334-353, the stop codon underlined and HindIII-site with flanking sequence in italic letters); P10, 3′-TGGTTCGGCCTACCATGCATTCGAACCC (nucleotides 358-377, otherwise as for P9) and P11, 3′-CAGGTCCTCGACGACGGGATTCGAACCC (nucleotides 391-409, otherwise as for P9). CDD cDNA in pT7-SC was used as template for the primers in the amplification reaction. The PCR product was cleaved with EcoRI and HindIII and recloned in the pT7-SC vector.
Site-directed mutations of CDD were produced by using Altered Sites in vitro Mutagenesis System (Promega, Madison, WI). The system uses a unique mutagenesis vector (pAlter phagemid) and a simple procedure for selection of oligonucleotide-directed mutants. The sequence of the mutated oligonucleotides was: P12, 5′-TGTGCTGACCGGACCGCT (nucleotides 199-216 in Fig 1A, substitution underlined), P13, 5′-ATCTGTGCTCAACGGACC (nucleotides 196-213, substitution underlined), P14, 5′-ATCTGTGCTGCACGGACCGCT (nucleotides 196-216, substitution underlined). All mutants were checked by sequencing. Truncated and mutagenic forms of CDD were expressed in E coliBL21 and purified through the Affi Gel step as described above.
RESULTS
Isolation of human CDD cDNA clones.
Approximately 5 × 105 plaques from a human blood cDNA library constructed in the λ ZAP Express vector were screened for CDD sequences by hybridization with a CDDprobe (see Materials and Methods) and 44 positive clones were isolated. Several clones were converted to the pBK-CMV phagemid by in vivo excision to allow insert characterization in a plasmid system. Restriction cleavage identified CDD cDNA inserts of varying lengths, which were subsequently subcloned for sequence and enzyme activity analysis. Three of four clones transforming E coliJF611 (cdd-, pyr-) complemented the double mutant for growth on minimal medium without added uracil. Radiochemical assays of dC to dU conversion by crude extract from the three clones confirmed expression of CDD activity from the plasmid clones. Sequence analysis disclosed 5′- and 3′-nontranslated sequences of different lengths, and the clone with the shortest 5′-nontranslated sequence (6 bp) was selected for expression and purification of rhCDD (Fig 1A). The coding region of CDD include 146 codons and is only 1 codon larger (start codon) than the original incomplete CDD cDNA reported by Kühn et al.16 In addition, at amino acid 27, we identified a substitution from Q → K, and a silent point mutation at nucleotide 441 from C → T.
Expression of human CDD in E coli.
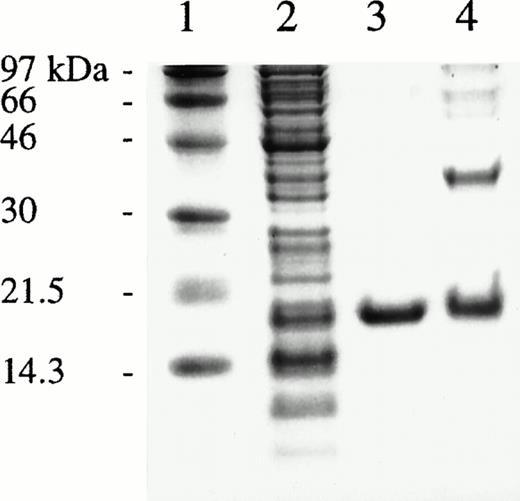
CDD cDNA excised from the pBK-CMV phagemid was ligated into pT7-SC, a T7 RNA polymerase-based expression system useful for high production of toxic proteins in E coli. After 2 hours of IPTG induction, a strong band of about 17.5 kD was observed by SDS-PAGE analysis (Fig 2, lane 2), which is in reasonable agreement with the predicted monomeric molecular mass of CDD of 16.2 kD. Enzyme activity analysis of crude extract showed 80-fold higher activity than in bacteria transformed with the CDD cDNA in the original pBK-CMV vector.
SDS-PAGE of fractions obtained during purification of rhCDD and of cross-linked enzyme. Lanes from left: 1, molecular weight markers; 2, crude extract; 3, purified rhCDD; 4, cross-linked rhCDD. Proteins were visualized by Coomassie Blue staining.
SDS-PAGE of fractions obtained during purification of rhCDD and of cross-linked enzyme. Lanes from left: 1, molecular weight markers; 2, crude extract; 3, purified rhCDD; 4, cross-linked rhCDD. Proteins were visualized by Coomassie Blue staining.
Purification of rhCDD.
The recombinant CDD was purified by a three-step procedure (Table 1). The Affi-Gel Blue column was quite effective, removing 85% of the protein with only 8% loss of the enzyme activity, and yielding a fraction producing only one distinct protein band on the gels. However, minor contaminants were removed by additional Mono Q and Mono S chromatography steps (Fig 2, lane 3). At the final step, the enzyme was purified 18-fold with a 31% recovery implying that the rhCDD constitute nearly 6% of the soluble protein in the extract. Cross-linking of rhCDD in 0.01% glutaraldehyde overnight at room temperature followed by SDS-PAGE suggested a homotetramer composition of the enzyme (Fig 2, lane 4). The final specific activity of the pure enzyme was about 1.35 × 105 U/mg. In a Lineweaver-Burk plot the characteristic Km-value for CDD was measured to 30 μmol/L at pH 7.5 and room temperature.
Purification of rhCDD Expressed in E coli
| Step . | Total Protein (mg) . | Total Activity (U × 10−3) . | Specific Activity (U × 10−3/mg) . | Purification (fold) . | Activity Recovery (%) . |
|---|---|---|---|---|---|
| E coli extract | 225 | 1,720 | 7.6 | 1 | 100 |
| Affi-Gel Blue | 35 | 1,590 | 45.4 | 6 | 92 |
| Mono Q | 7 | 820 | 117.0 | 15 | 48 |
| Mono S | 4 | 540 | 135.0 | 18 | 31 |
| Step . | Total Protein (mg) . | Total Activity (U × 10−3) . | Specific Activity (U × 10−3/mg) . | Purification (fold) . | Activity Recovery (%) . |
|---|---|---|---|---|---|
| E coli extract | 225 | 1,720 | 7.6 | 1 | 100 |
| Affi-Gel Blue | 35 | 1,590 | 45.4 | 6 | 92 |
| Mono Q | 7 | 820 | 117.0 | 15 | 48 |
| Mono S | 4 | 540 | 135.0 | 18 | 31 |
Recombinant CDD is an inhibitor of GM colony formation.
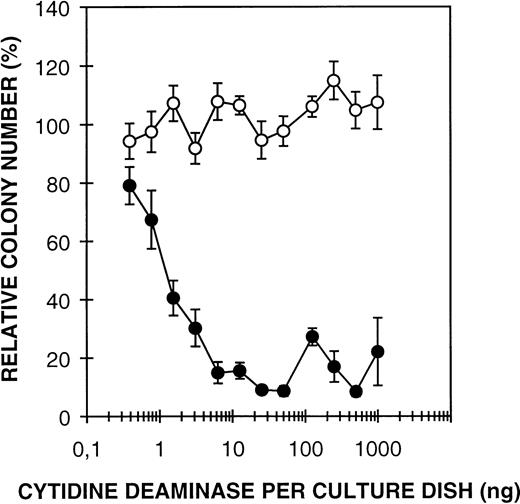
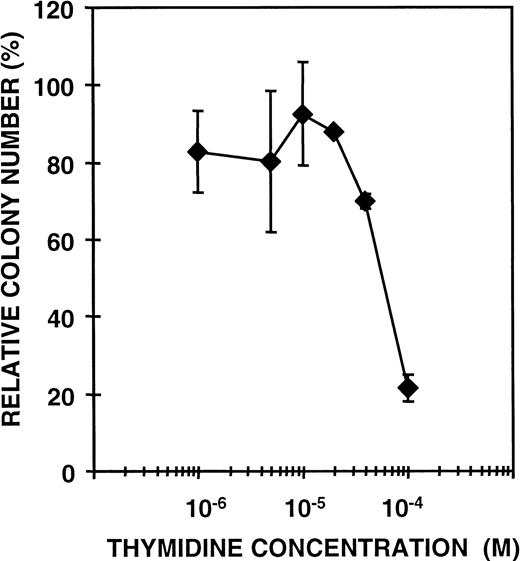
Previous studies have shown that CDD in granulocyte extract acts as an inhibitor of GM-CFC13; an effect dependent on the presence of thymidine in the growth medium.13 14 In those experiments, partially purified CDD from granulocytes was investigated, and it could not be excluded that additional factors in the extract would be required for the CDD effect. However, rhCDD mediates efficient inhibition of GM colony formation of both mouse BMC and human MNC, thus excluding that any additional protein factor is needed (Fig 3). These results imply that CDD produced in bacteria is fully active and can be used in further studies of the growth regulatory effect. RhCDD appears to be a very potent inhibitor of GM-CFC; 1 ng/mL (16 pmol/L) enzyme was found to reduce the GM colony formation of mouse BMC to approximately 50%, whereas 6 ng/mL (96 pmol/L) produced almost a complete inhibition. Thymidine at greater than or equal to 4 × 10-5 mol/L concentration is required for significant effects of recombinant CDD on colony formation (Fig 4) in agreement with results obtained previously from similar experiments with granulocyte extract preparations. Thymidine alone at these concentrations has no effect on GM-CFC (data not shown).
Effect of rhCDD on mouse bone marrow GM-CFC in the presence (•) or absence (○) of 10-4 mol/L thymidine. The cells were cultured for 7 days in a GM-CFC agar assay with 10% CM 5637 as stimulator. Colonies of more than 50 cells were counted. Mean values ± standard deviation (SD) are given as percent of controls.
Effect of rhCDD on mouse bone marrow GM-CFC in the presence (•) or absence (○) of 10-4 mol/L thymidine. The cells were cultured for 7 days in a GM-CFC agar assay with 10% CM 5637 as stimulator. Colonies of more than 50 cells were counted. Mean values ± standard deviation (SD) are given as percent of controls.
Effect of rhCDD (50 ng/plate) on colony formation of mouse BMC as a function of thymidine concentration added. The cells were cultured in a GM-CFC methylcellulose assay with 5 ng/mL of recombinant mouse IL-1β, IL-3, and stem cell factor as stimulators. Colonies of more than 50 cells were counted after 7 days. Mean values ± SD are given as percent of controls.
Effect of rhCDD (50 ng/plate) on colony formation of mouse BMC as a function of thymidine concentration added. The cells were cultured in a GM-CFC methylcellulose assay with 5 ng/mL of recombinant mouse IL-1β, IL-3, and stem cell factor as stimulators. Colonies of more than 50 cells were counted after 7 days. Mean values ± SD are given as percent of controls.
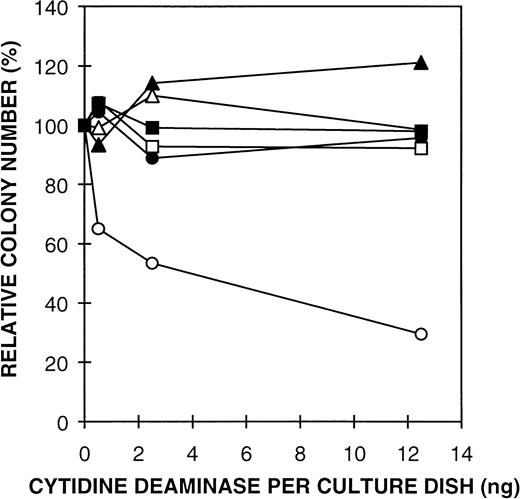
The effect of mutant CDD proteins on GM colony formation.
The mechanism of action for the growth suppressive effect of CDD has not been studied in any detail. We have investigated the possible enzyme activity requirement for the growth regulatory function of CDD by producing different modified forms of the enzyme. Three different truncated forms of the enzyme were made, deleted from the C-terminal end, including 115, 123, and 134 amino acids of the 146 amino acid CDD sequence, respectively. Possible enzymatic activity of the three truncated forms of CDD was measured in cell extracts from IPTG-induced BL21, transformed by the expression plasmids, using both the spectrophotometric and radiochemical assay methods. The truncated enzymes were all inactive, the measured activity being equivalent to the background value caused by E coli CDD in the crude extract (data not shown). The truncated forms were purified through the Affi Gel step, quantified by immunologic staining (Gran and Seeberg, unpublished observations, January 1997), and analyzed for possible growth suppressive effect on GM-CFC. The mutant proteins were all unable to inhibit colony formation (Fig5, and data not shown). These results strongly suggest a correlation between the enzyme activity and the growth regulatory effect of CDD. However, the possibility remained that the truncated CDD fragments did not fold properly and therefore were inactive due to an altered structural conformation. Therefore, we also constructed three point mutant forms of CDD; E67D (primer P12), E67Q (P13), and E67A (P14). E67 is positioned in a conserved sequence cluster common to all the CDD sequences known (Fig 1B) and is localized to the active site of the enzyme interacting with the substrate.25 Replacing the corresponding residue in the E coli enzyme by alanine has previously been shown to reduce kcat by 8 orders of magnitude.26 Spectrophotometric CDD assays showed no enzyme activity above background in extract from mutant forms expressed inE coli by IPTG induction. GM-CFC assays with the site-specific CDD mutant proteins confirmed that the catalytic function of CDD is essential for the growth suppressive function of CDD (Fig5).
Effect of mutant rhCDD on mouse bone marrow GM-CFC. Thymidine (10-4 mol/L) was added to all culture plates in the GM-CFC assay. The colonies (≥50 cells) were counted after 7 days in culture with 10% CM 5637 as stimulator. Mean values ± SD are given as percent of controls. Symbols: CDD truncated 12 amino acids from the C-terminus (□), Mutant E67D (•), Mutant E67Q (▵), Mutant E67A (▴), rhCDD (○), and extract from E coli BL21 transformed by the vector (pT7-SC) (▪).
Effect of mutant rhCDD on mouse bone marrow GM-CFC. Thymidine (10-4 mol/L) was added to all culture plates in the GM-CFC assay. The colonies (≥50 cells) were counted after 7 days in culture with 10% CM 5637 as stimulator. Mean values ± SD are given as percent of controls. Symbols: CDD truncated 12 amino acids from the C-terminus (□), Mutant E67D (•), Mutant E67Q (▵), Mutant E67A (▴), rhCDD (○), and extract from E coli BL21 transformed by the vector (pT7-SC) (▪).
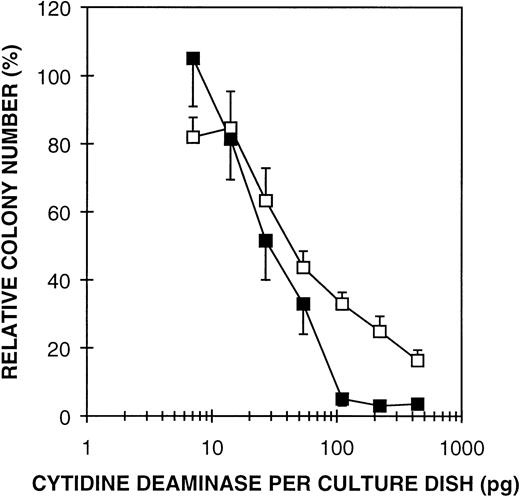
E coli CDD is a potent inhibitor of GM-CFC.
Another approach to investigate the significance of enzyme activity in regulating GM-CFC is to compare growth suppressive effects of CDD from different organisms relative to the specific activities of the enzymes. The coding region of E coli CDD was PCR-amplified and expressed with the pT7-SC system. High expression of a 31-kD protein was observed by SDS-PAGE and the enzyme was purified to homogeneity by a three-step purification procedure (Table 2). The specific activity of pure E coli CDD was measured to 1.25 × 106 U/mg, an approximately 10-fold higher activity than for the human enzyme. The Km value was estimated to 130 μmol/L compared with 30 μmol/L for rhCDD. The potential inhibiting effect of E coli CDD was tested in a GM-CFC assay using both human MNC and mouse BMC. The bacterial form of the enzyme appeared to be a more potent inhibitor of GM-CFC than the human enzyme (Fig 6). In combination with thymidine (10-4 mol/L), an enzyme concentration of 50 pg/mL (0.8 pmol/L) yielded a 50% reduction in GM colony formation of human MNC from peripheral blood, 400 pg/mL (6.5 pmol/L) producing nearly complete inhibition. This is about 1/15 of the amount of the rhCDD required to produce the same growth inhibitory effect. When considering the 10-fold higher specific activity of E coli CDD, these results strongly argue that the catalytic function of CDD is essential for the growth regulatory effect.
Purification of E coli CDD
| Step . | Total Protein (mg) . | Total Activity (U × 10−3) . | Specific Activity (U × 10−3/mg) . | Purification (fold) . | Activity Recovery (%) . |
|---|---|---|---|---|---|
| E coli extract | 32.0 | 11,000 | 340 | 1.0 | 100.0 |
| DEAE-Sephacel | 25.0 | 10,000 | 400 | 1.2 | 91.0 |
| Mono Q | 2.7 | 2,800 | 1,040 | 3.1 | 25.5 |
| Mini Q | 0.6 | 750 | 1,250 | 3.7 | 6.8 |
| Step . | Total Protein (mg) . | Total Activity (U × 10−3) . | Specific Activity (U × 10−3/mg) . | Purification (fold) . | Activity Recovery (%) . |
|---|---|---|---|---|---|
| E coli extract | 32.0 | 11,000 | 340 | 1.0 | 100.0 |
| DEAE-Sephacel | 25.0 | 10,000 | 400 | 1.2 | 91.0 |
| Mono Q | 2.7 | 2,800 | 1,040 | 3.1 | 25.5 |
| Mini Q | 0.6 | 750 | 1,250 | 3.7 | 6.8 |
Effect of purified E coli CDD on GM-CFC from human MNC (□) and mouse BMC (▪) in the presence of thymidine (10-4 mol/L). Cells were cultured in a GM-CFC assay for 14 and 7 days, respectively. The mean values ± SD are given as percent of controls.
Effect of purified E coli CDD on GM-CFC from human MNC (□) and mouse BMC (▪) in the presence of thymidine (10-4 mol/L). Cells were cultured in a GM-CFC assay for 14 and 7 days, respectively. The mean values ± SD are given as percent of controls.
The effect of nucleotide precursors on the inhibitory activity of CDD.
In view of the results showing that the catalytic activity of CDD is essential for the growth suppressive function, we tested the effect of different DNA precursors on GM-CFC growth inhibition. Addition of deoxyadenosine (dA) and deoxyguanosine (dG) up to 10-4mol/L concentration had no effect or even enhanced growth inhibition (Table 3). Addition of dC produced variable results depending on the concentration of dC and rhCDD (data not shown). Because dC is the natural substrate for CDD, any results obtained with dC will be difficult to interpret, as addition of dC in any case will compete for the catalytic function of CDD. Therefore, to evaluate if growth inhibition is due to cytidine nucleotide deficiency, experiments with deoxycytidine monophosphate (dCMP) additions were performed (Fig 7). DeoxyCMP is not a substrate for CDD,27 but will be taken up by the cells to an extent sufficient for restoring DNA synthesis if the availability of cytidine nucleotides is the limiting factor for DNA synthesis and cell growth. Addition of 10-3 mol/L dCMP together with thymidine reversed the inhibitory activity of CDD, indicating that the growth inhibitory effect of CDD can be explained by depletion of the cytidine and dC pool.
The Effect of Nucleotide Precursors on the Inhibitory Activity of CDD
| dT + CDD . | dT + CDD + dA (10−5 mol/L) . | dT + CDD + dA (10−4 mol/L) . | dT + CDD + dG (10−5 mol/L) . | dT + CDD + dG (10−4 mol/L) . |
|---|---|---|---|---|
| 42% | 43% | 33% | 30% | 15% |
| dT + CDD . | dT + CDD + dA (10−5 mol/L) . | dT + CDD + dA (10−4 mol/L) . | dT + CDD + dG (10−5 mol/L) . | dT + CDD + dG (10−4 mol/L) . |
|---|---|---|---|---|
| 42% | 43% | 33% | 30% | 15% |
Mouse BMC were cultured for 7 days in a GM-CFC agar assay with 10% CM 5637 as stimulator. Thymidine (dT) was added at 10−4 mol/L concentration and rh CDD at 25 ng/plate. Colonies of more than 50 cells were counted, and the values are given as percent of controls. No effect was obtained with dT or CDD alone.
The effect of dCMP on the growth inhibition mediated by CDD and thymidine. Thymidine was added at 10-4 mol/L concentration and rhCDD at 50 ng/plate. Mouse BMC were cultured in a GM-CFC agar assay and colonies (≥ 50 cells) were counted on day 7. Mean values ± SD are given as percent of controls. Symbols: dCMP (⧫), dCMP + rhCDD (▪), dCMP + rhCDD + thymidine (▴).
The effect of dCMP on the growth inhibition mediated by CDD and thymidine. Thymidine was added at 10-4 mol/L concentration and rhCDD at 50 ng/plate. Mouse BMC were cultured in a GM-CFC agar assay and colonies (≥ 50 cells) were counted on day 7. Mean values ± SD are given as percent of controls. Symbols: dCMP (⧫), dCMP + rhCDD (▪), dCMP + rhCDD + thymidine (▴).
DISCUSSION
In the present work, we have isolated human CDD cDNA from a blood cDNA library and expressed and purified rhCDD to homogeneity. The nucleotide sequence of the open reading frame of the CDD cDNA clone is in agreement with the recently published sequence of Laliberté and Momparler.28 Enzyme activity measurements showed a functional product with a specific activity of 1.35 × 105 U/mg and a Km value of 30 μmol/L, which agrees with values reported previously.18,28-30 In a GM-CFC assay, we have confirmed the growth suppressive action of CDD on progenitor cells as previously described by Bøyum et al13 and excluded the need for other cofactors, apart from thymidine.14 Approximately 16 pmol/L of rhCDD in combination with an optimal concentration of thymidine (10-4 mol/L, Fig 4) reduced GM colony formation of mouse BMC to approximately 50% (Fig 3). This implies that CDD exerts its effect at very low concentrations, within the same range or even less than required for inhibition by transforming growth factor-β (80 pmol/L).31 Thymidine alone had no significant effect on GM-CFC (data not shown). When granulocyte extract was used as the source of inhibitor in GM-CFC assays, a bell shaped dose-response curve was observed.14 However, high doses of purified rhCDD did not alleviate inhibition of colony formation, suggesting that crude granulocyte extract contains factors with growth stimulatory or antagonistic activity when added at higher concentrations.
Until now, the mechanism responsible for the growth regulatory potential of CDD has not been investigated in any detail. However, experiments with sorted primitive cells have indicated a direct suppressive effect on high potential proliferative colony-forming cells (Løvhaug, unpublished observations, February 1995). One hypothesis is that growth inhibition requires the conversion of dC or cytidine to dU or uridine, respectively, which in combination with added thymidine, results in an imbalance of the nucleotide pool sufficient to prevent DNA synthesis. A fine balance of all four deoxyribonucleotides is required for the optimum rate of DNA synthesis and normal growth.32 Reduction of dCTP levels by deamination of dC/cytidine through the salvage pathway may not by itself be sufficient for growth suppression. Thymidine might contribute by altering the DNA progenitor balance as well as by preventing the de novo synthesis of dCTP by changing the substrate specificity of ribonucleotide reductase.32-34 Ribonucleotide reductase catalyzes the formation of deoxyribonucleotides from ribonucleotides, and the complex feedback network regulation of ribonucleotide reductase has been thoroughly studied.35,36 Accumulation of deoxythymidine triphosphate (dTTP) will shut off the production of deoxypyrimidine nucleotides and further activate deoxyguanosine triphosphate (dGTP) and subsequently dATP production.37 Alternatively, we cannot completely exclude that the growth inhibition is a receptor-mediated process, which is the most common way for cells to communicate.5 CDD binding to a receptor on the cell surface could induce signal transduction that leads to growth suppression of GM-CFC. Thymidine could be an allosteric factor promoting the binding to the receptor.
Our studies show a correlation between the enzyme activity and the growth regulatory effect of CDD. Truncation of the CDD sequence 12 amino acids from the C-terminus abolishes both the enzyme activity and the growth suppressive effect, as does the substitution of E67 with D, Q, and A (Fig 5). E67 is within the active site of the enzyme and important for stabilizing the hydrated substrate in the transition state for deamination (Fig 1B).25,26 Results obtained withE coli CDD showing more growth inhibition by an enzyme with higher specific activity also support that the catalytic function of the CDD protein is essential. E coli CDD is a dimer where each subunit is composed of two core domains and has a molecular mass approximately equal to 31 kD.25 The human enzyme is most probably a homotetramer of subunits with Mr 16,200 (Fig2).30 Each subunit of human CDD is suggested to have an underlying similarity to the core domain of the bacterial enzyme, and sequence homologies are found at the topological switch point and the Zn-binding site (Fig 1B).25 The fact that CDD activity is essential for the growth suppressive function supports that the nucleotide balance in the cells is impaired to prevent normal cell growth. This conclusion is further substantiated by the results showing that addition of dCMP reverses the growth inhibitory effect (Fig 7). This would be the case if dCTP levels were considerably reduced as a consequence of cytidine and dC deamination and further inhibition of deoxypyrimidine nucleotide synthesis by dTTP. In contrast, addition of dG or dA, which are converted to dATP or dGTP, respectively, do not reverse inhibition, consistent with the expectation that the deoxypurine nucleotide pool will be increased rather than decreased as a consequence of thymidine addition.37 The indication of a synergistic effect of dG in combination with CDD and thymidine (Table3) can be nicely explained by the known regulation of ribonucleotide reductase where dGTP will inhibit the production of deoxypyrimidine nucleotides, as well as itself.35 According to the regulatory mechanisms of ribonucleotide reductase, high concentrations of dG (or dA) could even to some extent be expected to replace the requirement of thymidine for growth inhibition.
There is recent evidence to suggest that the plasma and serum levels of CDD in healthy persons are relatively high, about 100 ng/mL (Bøyum, Brandtzag, Tennfjord, Gran, Løvhaug, in preparation). The extracellular CDD descends most probably from mature granulocytes, which express high amounts of the enzyme.11,16-18 The release of CDD must either occur from lysis of damaged cells or by an active secretion mechanism.11,12,19,20 One might suggest that released CDD could function enzymatically as a feedback inhibitor of the granulopoietic pathway, both in blood and bone marrow. As shown in Fig 4, the enzyme requires thymidine (≥ 4 × 10-5mol/L) for the regulatory process in vitro (GM-CFC assays) and the concentration in human plasma is relatively low (≈ 0.2 μmol/L).38 However, this theory is supported by previous work with double diffusion chambers containing granulocytes and mouse BMC implanted intraperitoneal in mice.11 These experiments showed significant depression of granulopoiesis without any addition of thymidine. Such a system resembles the in vivo conditions more closely than the colony assays. Furthermore, in bone marrow where huge numbers of erythroid nuclei are continuosly being phagocytosed and degraded by macrophages,39 the nucleoside concentration may also be much higher than in blood.40 This might then provide conditions for a physiological role of CDD in homeostatic control of granulopoiesis. The enzyme may alter the extracellular pool of nucleosides or act intracellularly of the progenitor cells. One could speculate that the enzyme would be transiently active in the progenitor's lysosomes before degradation, after compartmentalization by endocytosis.
Mature granulocytes do not affect blastoid transformation and proliferation of lymphocytes, nor do they inhibit growth of cells already committed to granulopoiesis.11,12 Thus, it appears to be a growth regulatory effect directed specifically towards the granulopoiesis in diffusion chambers11 and GM-CFC in agar assays.12 To achieve cell specificity, one would assume that a receptor-mediated effect of CDD was a more likely mechanism. However, there could well be a combined mechanism where both a receptor is involved and the enzyme activity is required, similarly as has been reported for the observed effects of adenosine deaminase (ADA).41 ADA has generally been considered to be a cytosolic enzyme deaminating adenosine/dA to the respective inosine metabolites by a mechanism similar to that of CDD.25Recently, ADA was found to be associated with the extracellular domain of a T-cell activation molecule, CD26 or dipeptidyl peptidase IV, producing a costimulatory response in the T-cell activation events.41-43 It has been suggested that the role of ecto-ADA may be the regulation of the local level of extracellular adenosine, which can modulate the signal transduction in T cells.41,44,45 On the other hand, the presence of ADA itself might alter the intracellular concentration of the enzyme and in turn modify the intracellular adenosine metabolism. Actually, compartmentalization of ADA within lysosomes by endocytosis has been shown in fibroblasts.46 Similarly, there may be a specific receptor for CDD on GM-CFC. Alternatively, the specificity of the growth suppressive effect of CDD may be due to a highly variable nucleotide profile of human blood cells.47 Lymphocytes have a lower purine/pyrimidine nucleotide ratio than granulocytes. The nucleotide profile could also change through the differentiation stages. The promyelocytic leukemia cell line HL-60 has reciprocal alterations of guanosine monophosphate (GMP) reductase and inosine monophosphate dehydrogenase activities during differentiation48 and activation of a Na+-dependent nucleoside transport system.49-51On the other hand, the unresponsiveness of cells already committed to granulopoiesis could be explained simply by the increased expression of CDD mRNA during maturation of granulocytes.16 Another possible explanation for the cell specificity observed may be that some cells are more dependent on the salvage pathway than the de novo biosynthesis of pyrimidines for cell growth. CDD, which is committed to the salvage pathway, will not affect the de novo synthesis of dCTP produced by the CTP synthase-mediated amination of uridine triphosphate.
Finally, we would like to point out the interesting similarity between cytidine deaminases and RNA editing proteins, eg, apolipoprotein B.52 both at the sequence level and with respect to enzymatic activity. Perhaps some hitherto unknown deaminating reaction may also contribute to the growth inhibitory effect.
ACKNOWLEDGMENT
We are grateful to Dr Jan Neuhard for the gift of E coli strain JF611, to Dr Anders Høgset for stimulating discussions and support, and to Vivi-Ann Tennfjord for technical assistance with some of the growth experiments.
Supported by NYCOMED Imaging (Oslo, Norway) and the Norwegian Cancer Society (Oslo, Norway).
Address reprint requests to Erling C. Seeberg, PhD, Institute of Medical Microbiology, Department of Molecular Biology, University of Oslo, The National Hospital N-0027 Oslo, Norway.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal