Abstract
Uptake of vitamin B12 (cyanocobalamin) is facilitated by the cobalamin-binder gastric intrinsic factor (IF), which recognizes a 460-kD receptor, cubilin, present in the epithelium of intestine and kidney. Surface plasmon resonance analysis of ligand-affinity-purified human cubilin demonstrated a high-affinity calcium- and cobalamin-dependent binding of IF-cobalamin. Complete cDNA cloning of the human receptor showed a 3597 amino acid peripheral membrane protein with 69% identity to rat cubilin. Amino-terminal sequencing of the receptor indicates that the cDNA sequence encodes a precursor protein undergoing proteolytic processing due to cleavage at a recognition site (Arg7-Glu8-Lys9-Arg10) for the trans-Golgi proteinase furin. Using fluorescence in situ hybridization, radiation hybrid mapping, and screening of YAC clones, the human cubilin gene was mapped between the markers D10S1661 and WI-5445 on the short arm of chromosome 10. This is within the autosomal recessive megaloblastic anemia (MGA1) 6-cM region harboring the unknown recessive-gene locus of juvenile megaloblastic anemia caused by intestinal malabsorption of cobalamin (Imerslund-Gräsbeck's disease). In conclusion, the present molecular and genetic information on human cubilin now provides circumstantial evidence that an impaired synthesis, processing, or ligand binding of cubilin is the molecular background of this hereditary form of megaloblastic anemia.
MEGALOBLASTIC anemia and neurological disturbances are common symptoms during deficiency of the coenzyme vitamin B12 (cyanocobalamin). The cellular uptake of the vitamin and its modified forms depends on the binding to the carrier proteins, intrinsic factor (IF) produced in the stomach, and transcobalamin, present in the circulation and various tissue fluids. Hereditary forms of cobalamin deficiency are known to relate to qualitatively abnormal IF,1 to decreased synthesis of transcobalamin,2 and to a defect of the intestinal epithelium leading to decreased uptake of IF-cobalamin and failure to absorb cobalamin.3-5 The latter condition is known as Imerslund-Gräsbeck's disease, and a recent gene linkage analysis of 38 patients diagnosed in Norway and Finland localized the unknown autosomal recessive megaloblastic anemia (MGA1) gene to the short arm of chromosome 10 between markers D10S548 and D10S466.6 The molecular defect in this group of cobalamin-deficient patients is hitherto unknown, but it has been proposed that the cobalamin malabsorption might be related to an abnormal epithelial translocation of cobalamin, perhaps due to decreased receptor function/expression.
Studies in rodents have shown that uptake of cobalamin in complex with IF is facilitated by an intestinal 460-kD protein7,8designated cubilin.9 Immunohistochemical studies and Northern blotting of various tissues of rodents have so far only detected cubilin in the epithelia of the intestine, yolk sac, and the renal proximal tubules,7,9 the same tissues shown to have IF-cobalamin binding activity.10 Cubilin is suggested to traffic by means of megalin, a 600-kD endocytic receptor expressed in the same tissues and mediating uptake of a number of ligands, including transcobalamin-cobalamin complexes.11
Cubilin has as megalin a significant higher expression in the renal proximal tubules compared with the intestine, and, because IF is only present in minute amounts in nongastrointestinal tissues, cubilin might also have multiligand properties. It is in this context interesting that cubilin recently has been shown to bind receptor-associated protein (RAP),7 a 40-kD endoplasmic reticulum protein also binding with high affinity to the multiligand giant receptors (eg, megalin) belonging to the low-density lipoprotein receptor family protein. RAP is suggested to function as a chaperone during folding of the receptors.12 13
The complete primary structure of rat cubilin was very recently solved,9 and it shows that almost the entire sequence is accounted for by a cluster of eight epidermal growth factor repeats (EGF) followed by a large cluster of 27 CUB* domains leading to the present name of the receptor.
The present study is a molecular characterization of human cubilin and chromosomal mapping of the encoding gene. The data show that the processed ligand-binding active human receptor is a furin-cleaved protein sharing structural homology with the rat receptor. The cubilin gene was mapped within the 6-cM region pointed out to harbor the recessive-gene locus of Imerslund-Gräsbeck's disease.6
MATERIALS AND METHODS
Purification of human IF and human IF-cobalamin receptor/cubilin.
Human IF was purified from gastric juice and porcine IF from lyophilized porcine gastric mucosa extract from GEA14(Copenhagen, Denmark). The human receptor was purified by IF-cobalamin affinity chromatography of Triton X-100–solubilized renal cortex membranes prepared from legal human autopsy material taken 6 to 24 hours postmortem. The renal cortex was dissected and membranes were prepared according to the previously published procedure for preparation of rabbit renal cortex membranes.15 The solubilized membranes were then pumped onto a 10-mL column of CNBr-activated Sepharose coupled with porcine IF-cobalamin (3 mg/mL Sepharose). After extensive washing with running buffer containing 10 mmol/L NaH2PO4, 150 mmol/L NaCl, 2 mmol/L CaCl2, 0.6% 3-[(3-cholamidopropyl)dimethylammonio]-propane sulfonic acid, bound protein was eluted by adjusting pH to 4.0 and the addition of 5 mmol/L EDTA. For protein sequencing, approximately 5 μg of the 460-kD IF-cobalamin–binding human receptor was electroblotted from a 4% to 16% polyacrylamide reducing sodium dodecyl sulfate (SDS)-gel to a polyvinylidene diflouride membrane (Problot; Applied Biosystems, Foster City, CA). The electroblotted band was cut out and subjected to Edmann degradation using an Applied Biosystems 477 A sequencer equipped with a 120 A online chromatograph. A cross-flow reaction and the Doublot reaction and conversion cycles were used.
SDS-gel electrophoresis of ligand affinity-purified human IF-cobalamin receptor/cubilin and determination of its amino-terminal sequence. Elution of the receptor from porcine IF-cobalamin-Sepharose column loaded with Triton-X100–solubilized human renal cortex membranes was performed by changing the pH of the running buffer to 4.0 and adding 5 mmol/L EDTA to the buffer. The determined amino-terminal sequence of the 460-kD protein is indicated. Ten microliters of each fraction was loaded on a 4% to 16% polyacrylamide gel and run under reducing conditions.
SDS-gel electrophoresis of ligand affinity-purified human IF-cobalamin receptor/cubilin and determination of its amino-terminal sequence. Elution of the receptor from porcine IF-cobalamin-Sepharose column loaded with Triton-X100–solubilized human renal cortex membranes was performed by changing the pH of the running buffer to 4.0 and adding 5 mmol/L EDTA to the buffer. The determined amino-terminal sequence of the 460-kD protein is indicated. Ten microliters of each fraction was loaded on a 4% to 16% polyacrylamide gel and run under reducing conditions.
Receptor-ligand binding analysis.
Surface plasmon resonance measurements were performed on a BIAcore 2000 instrument (Biosensor, Uppsala, Sweden). This technology relies on the phenomenon of the surface plasmon resonance (SPR) that occurs when surface plasmon waves are excited at a metal/liquid interphase. Light is directed at, and reflected from, the side of the surface not in contact with sample, and SPR causes a reduction in the reflected light intensity at a specific combination of angle and wavelength. Biomolecular binding events cause changes in the refractive index at the surface layer, which are detected as changes in the SPR signal. In general, the refractive index change for a given change of mass concentration at the surface layer is the same for all proteins and peptides.16 The BIAcore sensor chips (type CM5; Biosensor) were activated with 1:1 mixture of 0.2 mol/L N-ethyl-N′-(3-dimethylaminopropyl) carbdiimide and 0.05 mol/L N-hydroxysuccimide in water. Human cubilin was immobilized at a concentration of 40 μg/mL in 10 mmol/L sodium acetate, pH 4.5, and the remaining binding sites were blocked with 1 mol/L ethanolamine, pH 8.5. The surface plasmon resonance signal from immobilized receptor generated BIAcore response units (RU) equivalent to 33 fmol ligand/mm2, respectively. The flow cells were regenerated with 6 mol/L guanidine-HCl. The flow buffer was 10 mmol/L HEPES, 150 mmol/L NaCl, and 1.5 mmol/L CaCl2, 1 mmol/L EGTA, pH 7.4. The binding data were analyzed using the BIAevaluation program.
cDNA cloning and sequencing.
Initial cDNA clones encoding bp −26 to 1633, 1452 to 3612, and 3624 to 8038 of human cubilin were identified by screening of a λgt11 library of human fetal kidney cDNA (Clontech, San Diego, CA) using digoxygenin (Boehringer Mannheim, Mannheim, Germany) -labeled rat cubilin cDNA probes (bp 1659-2178 and bp 6362-6621). In addition, a clone encoding bp 7144-9276 was identified in the same way using a digoxygenin-labeled human cubilin cDNA probe (bp 7534-7872). The cloned inserts of λgt11were amplified by polymerase chain reaction (PCR) with primers designed to match vector sequence before and after the EcoRI site into which the clones were inserted. The 5′ end of the coding sequence (bp −26 to 495) was generated by rapid amplification of cDNA ends (RACE) of Marathon Ready cDNA (Clontech) kit of human fetal kidney cDNA using a 5′ end adaptor primer and a 3′ end gene-specific primer (bp 468-495). The coding sequence from bp 3612-3624 and the 3′ end of the coding region of the human cubilin cDNA were generated by PCR of the same Marathon Ready cDNA using gene-specific primers (bp 3243-3261 and bp 3855-3874) for the first PCR product and a 5′ end gene-specific primer bp 8727-8750 together with a 3′ end primer bp 10908-10931 designed from an expressed sequence tag (EST accession no. AA341460) sharing high homology with the rat cubilin cDNA (accession no. AF022247). The 3′ end noncoding region including the poly A tail of the human cubilin cDNA was generated by RACE using a 5′ end gene-specific primer bp 10392-10415 and a 3′ end adaptor primer. PWO polymerase and Expand High Fidelity PCR System (both Boehringer Mannheim) were used in the PCR reactions. Sequencing of isolated cDNA clones and PCR products was performed by cycle sequencing in both directions with IRD-41–labeled primers and the sequence reactions were analyzed on a LICOR 4000 automatic sequencer (LI-COR, Lincoln, NE). All PCR products were sequenced directly to avoid PCR-generated sequence artefacts.
Fluorescence in situ hybridization (FISH).
FISH of methaphase chromosomes with corresponding 4′,6-diamidine-phenylindole dihrydrochloride (DAPI)-banding and contour length measurement of the Flpter-value (fractional length relative to the fixed reference point pter in the short arm telomer) were performed essentially as described previously17 using 100 ng of a biotin-labeled human cubilin cDNA clone (bp −26 to 1633). The metaphases were visualized on a Leica DMRB epifluorescence microscope equipped with a Sensys 1400 CCD camera (Photometrics, München, Germany) and an IPLab Spectrum imaging software (Vysis, Stuttgart, Germany).
Radiation hybrid mapping.
The 86 GeneBridge4 clones for radiation hybrid (RH) mapping18 were obtained from the UK Human Genome Mapping Project Resource Center (http://www.hgmp.mrc.ac.uk/homepage.html). The primers used for RH-mapping were CTACAGAATCAACAGGGACC-3′ and 5′-CCACAGTATCTTCCAAGGGA-3′, which amplify a 210-bp fragment located within the area encoding the fifth CUB domain of human cubilin. PCR was performed on a PTC-225 DNA Engine Tetrad thermocycler (MJ Research, Watertown, MA) for 40 cycles, each consisting of 3 steps of 30 seconds at 95°C, 59°C, and 72°C. Before thermocycling, samples were predenatured at 95°C for 5 minutes. Thermocycling was followed by a final extension at 72°C for 5 minutes. PCR were performed in standard PCR buffer (Perkin Elmer, Norwalk, CT) with 10 pmol of each primer, 10 nmol dNTP (Pharmacia), and 0.5 U AmpliTaq (Perkin Elmer) in a total volume of 15 μL. Gel-loading buffer at final concentrations of 0.2 mmol/L cresol red and 12% sucrose were included in the PCR reaction buffer.19 Fragments were electrophoresed on a 2% agarose gel (FMC, Rockland, ME) and photographed. By visual inspection of the gel, PCR products of correct length were scored as positives, whereas PCR products of incorrect length or absent were scored as ambiguities or negatives. Faint bands of correct length were likewise scored as ambiguities. Mapping was performed using the UK MRC HGMP-RC interface at the Whitehead Institute/MIT Center for Genome Research (WICGR;http://www-genome.wi.mit.edu/cgi-bin/contig/rhmapper.pl). After mapping, markers neighboring or coinciding with the radiation hybrid data vector were looked up using the ENTREZ genome database browser at the National Center for Biotechnology Information (NCBI;http://www.ncbi.nlm.nih.gov/Entrez/Genome/org.html), and the chromosomal location was documented.
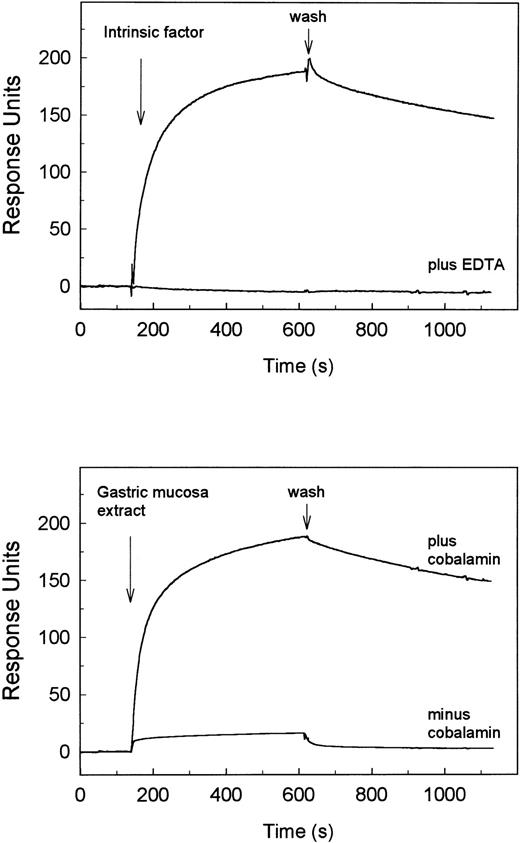
SPR analysis of the binding of IF-cobalamin to human cubilin. Human cubilin was immobilized to a sensor chip, and the on and off rates for ligand binding were recorded on a BIAcore 2000. The arrows indicate the start of flow with ligand (association phase) and the start of wash with buffer alone (dissociation phase). The recorded sensorgrams in the upper panel show binding of 400 nmol/L porcine intrinsic factor in normal flow buffer with 0.5 mmol/L free Ca2+ and the absence of binding when 5 mmol/L EDTA was added to the flow buffer. The lower panel shows binding activity of diluted porcine gastric mucosa extract (114 μg protein/mL) with and without saturation with cobalamin. The sensorgrams display the values after subtraction of the values from the same recordings on a blank chip.
SPR analysis of the binding of IF-cobalamin to human cubilin. Human cubilin was immobilized to a sensor chip, and the on and off rates for ligand binding were recorded on a BIAcore 2000. The arrows indicate the start of flow with ligand (association phase) and the start of wash with buffer alone (dissociation phase). The recorded sensorgrams in the upper panel show binding of 400 nmol/L porcine intrinsic factor in normal flow buffer with 0.5 mmol/L free Ca2+ and the absence of binding when 5 mmol/L EDTA was added to the flow buffer. The lower panel shows binding activity of diluted porcine gastric mucosa extract (114 μg protein/mL) with and without saturation with cobalamin. The sensorgrams display the values after subtraction of the values from the same recordings on a blank chip.
The primary structure of human cubilin as translated from cDNA clones. The predicted signal sequence is shown in italic type. The amino-terminal defined by protein sequencing is underlined. Potential N-glycosylation sites are shown in bold type. The figure on the right side represents the organization of extracellular modules. Their positions correspond to the sequence to the left. The mass of the peptide of the precursor is 396.280 kD and 395.041 kD after cleavage at the furin recognition site, thus suggesting that carbohydrate accounts for approximately 14% of the processed 460-kD glycoprotein.
The primary structure of human cubilin as translated from cDNA clones. The predicted signal sequence is shown in italic type. The amino-terminal defined by protein sequencing is underlined. Potential N-glycosylation sites are shown in bold type. The figure on the right side represents the organization of extracellular modules. Their positions correspond to the sequence to the left. The mass of the peptide of the precursor is 396.280 kD and 395.041 kD after cleavage at the furin recognition site, thus suggesting that carbohydrate accounts for approximately 14% of the processed 460-kD glycoprotein.
Screening of yeast artificial clones (YACs).
A minimum set of 11 YACs covering the 7-cM region between D10S191 and D10S548 on the Whitehead WC10.1 YAC contig (http://www.genome.wi.mit.edu/cgi-bin/contig/sts_info/300) were obtained from Fondation Jean Dausset-CEPH (Paris, France). YACs were directly screened with PCR (Hansen et al, manuscript submitted). Primers and PCR conditions were as described for radiation hybrid mapping above. PCR fragments were electrophoresed on a 2% agarose gel in 1× TBE buffer for 40 minutes at 200 V and photographed.
RESULTS
Characterization of the human-purified IF-cobalamin receptor/cubilin.
IF-cobalamin affinity chromatography of human renal cortex membranes yielded high amounts of receptor protein (Fig 1). The electrophoretic mobility was identical to that of the previously characterized 460-kD rodent receptor7 (not shown). The 460-kD band was absolutely predominant compared with some inconsistent minor bands, which are probably accounted for by partial degradation in the postmortem tissue and a high molecular weight band with a size corresponding to the position of 600-kD megalin. Microsequencing of the electroblotted 460-kD band showed the amino-terminal sequence Ser-Iso-Asn-Leu-Glu-Glu.
The binding of IF-cobalamin to the ligand affinity-purified receptor preparation was verified and characterized by the surface plasmon resonance technique described in detail in the Materials and Methods. This analysis demonstrated a high-affinity Ca2+-dependent binding of porcine IF-cobalamin (Fig 2). The prompt increase in response units when the sensor chip with immobilized purified cubilin was exposed to a flow with the ligand reflects the association phase of ligand-receptor interaction. The bound ligand dissociated slowly as seen by the weak decrease in the response units after wash with buffer alone. The binding curves fitted according to simple one binding site kinetics (Kd = 5 nmol/L). A similar curve with similar binding kinetics was obtained with human IF-cobalamin (Kd = 7 nmol/L; data not shown). Analysis of porcine gastric mucosa extract, which is rich in apoIF, showed only binding when the cobalamin binding activity was saturated (Fig 2B), thus indicating that apoIF does not bind to the receptor. Surface plasmon resonance analysis showed also binding of human RAP and rabbit megalin (data not shown) with sensorgrams similar to those reported for the binding of the two molecules to the rabbit receptor.7 9
Determination of the primary structure of the human IF-cobalamin receptor/cubilin.
An 11.4-kb human cubilin cDNA was obtained from several pieces by human kidney cDNA library screening as described in the Materials and Methods. Figure 3 shows the primary structure of the receptor as deduced from the 10,864-bp encoding cDNA sequence.*
A hydrophobic signal peptide of 24 amino acids precedes a 3597 amino acid protein with 47 potential N-glycosylation sites. The encoded protein has 69% sequence identity to rat cubilin.9 The protein sequence predicts, as for the rat receptor, a peripheral membrane protein with eight EGF repeats followed by 27 CUB domains. The receptor is rather acidic (pK = 5.14) and has 153 cysteines. The amino terminal located cysteine is not included in a known extracellular module and is therefore suggested to be a free cysteine, whereas the remaining cysteines confined to EGF and CUB domains are suggested to form 76 disulfide links. The same disulfide bridge pattern has been predicted in the rat receptor, except that one of the two sugested disulfide bridges is missing in the sixth CUB domain of the human receptor. The N-terminal sequence of the purified human receptor is identical to the sequence succeeding Arg10. Interestingly, this sequence follows the sequence Arg7-Glu8-Lys9-Arg10that abides by the consensus sequence Arg-X-Arg/Lys-Arg for cleavage by the trans-Golgi proteinase furin. Consequently, these data indicate that the receptor is synthesized as a precursor undergoing further proteolytic processing in the trans-Golgi causing a 10 residue propeptide to be cleaved off.
Chromosomal mapping by FISH, radiation hybrid, and YAC screening.
By using a biotin-labeled probe encoding the eight EGF repeats and the first CUB domain, specific FISH signals were only observed on the midportion of the short arm of chromosome 10, with 49 of 50 analyzed metaphases (98%) displaying at least one specific signal. In total, 147 of the 200 chromatids (73.5%) were labeled, with 4 cells with one, 12 cells with two, 9 cells with three, and 23 cells with four labeled chromatids. The unusual high FISH signal might be accounted for by the presence of the 27 homologous regions encoding the CUB domains. The FLpter value was 0.14 ± 0.04 corresponding to a localization at 10p12-p14.20 The localization of the FISH signals on elongated metaphase chromosomes suggested a fine localization to 10p12.33-p13 (Fig 4). Using primers specific for the fifth CUB domain of cubilin for RH mapping, PCR products from the 86 clones of the hybrid panel were detected. The data vector, consisting of 0's, 1's, and 2's (negative, positive, and ambiguous results, respectively) from each of the clones, was submitted to and compared at the WIGCR radiation hybrid mapping facility. The results of this comparison showed that the cubilin gene maps to chromosome 10, 18.3 cR* from marker NIB1436 (lod>3) and 3.2 cR from marker D10S203.21,22 Searching the chromosome 10 integrated map from NCBI shows that NIB1436 and D10S203 are located at a position corresponding to 10p12-10p14.20
Partial metaphases hybridized with biotin-labeled cubilin cDNA, showing (a) the specific FISH signals on the homologous chromosome 10p-regions (arrows) superimposed on the DAPI-stained chromosomes, (b and c) FISH signals on more elongated chromosomes, and (d) a chromosome ideogram31 showing the localization of the intrinsic factor receptor gene and the mean FLpter value (arrow). The box indicates the variation in Flpter values on individual chromosomes.
Partial metaphases hybridized with biotin-labeled cubilin cDNA, showing (a) the specific FISH signals on the homologous chromosome 10p-regions (arrows) superimposed on the DAPI-stained chromosomes, (b and c) FISH signals on more elongated chromosomes, and (d) a chromosome ideogram31 showing the localization of the intrinsic factor receptor gene and the mean FLpter value (arrow). The box indicates the variation in Flpter values on individual chromosomes.
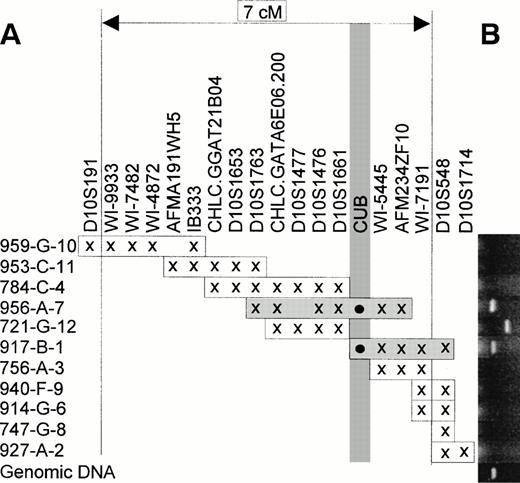
Correctly sized PCR products were obtained by PCR screening of YACs 956a7 and 917b1 (Fig 5B) using the same primer as used for radiation hybrid maping. Besides correctly sized PCR products, a larger fragment was seen corresponding to YAC 721g12 (Fig 5B). Although this fragment in theory could represent a rearrangedCUB-sequence, the same oversized fragment was also obtained from a few YACs from other chromosomes as well (data not shown). Combined with the negative YACs 784c4 and 756a3, the two positive YACs indicate that the cubilin gene is located between markers D10S1661 and WI-5445 (Fig 5A).
PCR-screening of YAC contig between markers D10S191 and D10S548. (A) The inferred position of the cubilin gene (designatedCUB) between D10S1661 and WI-5445. The individual YACs and their genetic marker content (x) are arranged essentially as in the Whitehead WC10.1 contig. (B) Agarose gels aligned to the individual YAC clones, with CUB-specific PCR fragments corresponding to YACs 956a7 and 917b1. The bottom lane is genomic test DNA. The incorrectly sized PCR fragment in lane 721g12 is scored as negative (see text).
PCR-screening of YAC contig between markers D10S191 and D10S548. (A) The inferred position of the cubilin gene (designatedCUB) between D10S1661 and WI-5445. The individual YACs and their genetic marker content (x) are arranged essentially as in the Whitehead WC10.1 contig. (B) Agarose gels aligned to the individual YAC clones, with CUB-specific PCR fragments corresponding to YACs 956a7 and 917b1. The bottom lane is genomic test DNA. The incorrectly sized PCR fragment in lane 721g12 is scored as negative (see text).
DISCUSSION
The present data describe the molecular characterization, including the determination of the complete primary structure, of the processed human IF-cobalamin receptor/cubilin and the chromosomal localization of the receptor gene. The human receptor shows 69% identity with the rat receptor. The organization of eight EGF repeats and 27 CUB domains is conserved among the two species as well as the lack of any potential hydrophobic transmembrane and cytoplasmic domains. Furthermore, both the human and the rat receptor show a similar calcium-dependent binding of IF-cobalamin, RAP, and megalin. ApoIF has only a weak affinity, thus suggesting that the conformational change IF undergoes upon binding of cobalamin23 is essential for the human receptor to bind or that cobalamin is directly involved in receptor binding. Amino-terminal sequencing of affinity-purified human receptor showed that it is truncated at a consensus recognition cleavage site for furin, an endoproteinase located in the trans-Golgi.24 In view of several reports indicating that furin-mediated cleavage is essential for the transformation of inactive precursors to bioactive proteins, such as the insulin receptor25 and transforming growth factor β,26 cubilin might also be synthesized as an inactive precursor. A similar amino-terminal trimming may occur in the rat receptor that has a similar sequence (Arg-Gly-Glu-Lys-Arg)9 at the same position. However, in contrast to the human receptor, the amino-terminal of the purified rat receptor has not been accessible for amino-terminal sequencing9 in our hands.
Previous pulse-chase experiments of 35S-labeled cells have shown that cubilin appears to have an unusual processing pathway.27 Although the bulk of the plasma membrane cubilin is endoglycosidase H resistant, the protein is initially targeted to the plasma membrane in an endoglycosidase H-sensitive form, indicating that it has not been processed through the early Golgi stacks. This implies that processing of cubilin takes place by recycling from the plasma membrane to the Golgi apparatus. A similar recycling process from the plasma membrane to the Golgi has been suggested for a number of membrane proteins, including Thy1 and the transferrin and LDL receptors.28 This apparent processing pathway of cubilin, the amino-terminal trimming demonstrated here, and the fact that furin and furin-like enzymes are present in the trans-Golgi may also suggest that cubilin initially can be expressed on the plasma membrane as a noncleaved precursor. From a hematological point of view, the complex processing of cubilin is intriguing, because a recent study of a dog phenotype with hereditary cobalamin malabsorption and proteinuria showed that the IF-cobalamin receptor is concentrated in the Golgi stacks and is inappropriately glycosylated.29 30
The chromosomal mapping of the human cubilin gene localized the gene to the same region as the gene locus of autosomal recessive megaloblastic anemia (MGA1) in 38 well-characterized Norwegian and Finnish Imerslund-Gräsbeck patients with severe malabsorption of cobalamin6 and proteinuria. The phenotype of these patients is similar to that of the dog phenotype characterized by Fyfe et al.29 The linkage analysis of the multiplex Imerslund-Gräsbeck families from Norway and Finland by Aminoff et al6 assigned the MGA1 gene to a 6-cM region on chromosome 10p flanked by the markers D10S548 and D10S466, with a multipoint maximum lod score of 5.36 near marker D10S1477. The screening of a YAC-contig between D10S548 and D10S191 (which should include D10S466) with CUB-specific primers identified two positive YACs (956a7 and 917b1), placing the cubilin gene within the 6-cM region very close to D10S1477. The present FLpter value of 0.14 ± 0.04 corresponds to a localization at 10p12-p14,20 and our FISH signals on elongated metaphase chromosomes indicated a fine localization to 10p12.33-p13 (Fig 4b and c). In conclusion, the present refinement of the integrated cytogenetic, genetic, and physical map of the MGA1 region has precisely mapped the cubilin gene in this part of the human genome.
A cubilin gene defect might either directly affect the synthesis of receptor, its processing (eg, maturation in Golgi or megalin-mediated transport), or the binding of IF-cobalamin. Any of these possibilities might explain a recent observation5 that four French Imerslund-Gräsbeck patients with similar symptoms have decreased IF-cobalamin binding activity in ileal homogenates.
We have now initiated studies to further define the ligand binding region of the receptor and to identify the likely cubilin gene defect and its functional implications in Imerslund-Gräsbeck patients.
ACKNOWLEDGMENT
Technical assistance by Kirsten Lassen is gratefully acknowledged. Dr Ebba Nexø is thanked for providing human IF-cobalamin and porcine gastric mucosa extract.
The 10.9-kb nucleotide sequence of the encoding region of human cubilin cDNA has been submitted to the GenBank/EBI Data Bank with accession no. AF034611.
The unit of genetic map distance that corresponds to an interval in which there is a 1% probability of x-ray–induced breakage.
The name CUB introduced by Bork and Beckmann32 is an abbreviation of Complement subcomponents C1r/C1s, Uegf, and Bone morphogenic protein-1.
R.K. and M.K. have contributed equally to this report.
Supported by the Novo Nordisk Foundation, the Danish Medical Research Council, the Danish Biomembrane Center, the Aarhus University Research Foundation, the Velux Foundation, the Fondation Vaincre les Maladies Lysosomales, le Ministère des Affaires Etrangères, Danish Biotechnological Research and Development Programme 1996-1998, the Danish Cancer Society, the Danish Research Center for Growth and Regulation, the Danish Research Council, Åge Bangs Foundation, the German Genome Programme/Deutsche Forschungsanstalt für Luft- und Raumfahrt e.V. (Grant No. 4763), and the EU-Commission (BMH4-CT97-2268).
Address reprint requests to Søren K. Moestrup, MD, Department of Medical Biochemistry, University of Aarhus, 8000 Aarhus C, Denmark; e-mail: skm@biobase.dk.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal