Abstract
Umbilical cord blood (CB) constitutes a promising alternative to bone marrow for allogeneic transplantation and is increasingly used because of the reduced severity of graft-versus-host disease after CB transplantation. We have compared the T-cell receptor β chain (TCRB) diversity of CB lymphocytes with that of adult lymphocytes by analyzing the complementarity determining region 3 (CDR3) size heterogeneity. In marked contrast to adult samples, we observed bell-shaped profiles in all of the 22 functional β-chain variable (BV) subfamilies that reflect the lack of prior antigenic stimulation in CB samples. However, the mean CDR3 size and BV usage were comparable between CB and adult samples. BJ2 (65%) segments were used preferentially to BJ1 (35%), especially BJ2S7, BJ2S5, BJ2S3, and BJ2S1, in both CB and in adult lymphocytes. We therefore conclude that although naive as reflected by the heterogeneity of the CDR3 size, the TCRBV repertoire appears fully constituted at birth. The ability to expand TCRB subfamilies was confirmed by stimulation with staphylococcal superantigens toxic shock syndrome toxin-1 and staphylococcal enterotoxin A. This study provides the basis for future analysis of the T-cell repertoire reconstitution following umbilical CB transplantation.
UMBILICAL CORD blood (CB) has been used successfully since 1988 as a source of hematopoietic stem cells for transplantation involving sibling donors1 and, more recently, unrelated recipients.2,3 CB transplantation has been associated with a reduced risk of developing severe graft-versus-host disease (GVHD), even when cells from partially major histocompatibility complex (MHC)-mismatched donors are used.3 The maximum degree of HLA disparity that will still allow engraftment has yet to be determined, but in contrast with HLA-mismatched bone marrow transplantation, even unrelated 1 or 2 antigen mismatched CB transplants result in an acceptable grade of acute GVHD.4
The lower risk of GVHD associated with the use of CB transplants is thought to be caused by the functional immaturity of lymphocytes at birth. Phenotypically, a lower expression of the CD45RO marker of memory T cells has been shown in newborns.5 The expression of class II molecules on antigen presenting cells is decreased in CB B cells, and these molecules are predominantly empty.6Alloantigens or superantigens can induce strong initial proliferative responses of CB as well as of adult T cells, but a state of unresponsiveness is induced on restimulation in CB.7,8 Many abnormalities of lymphokine production have been described on activation, such as a decreased production of interferon-γ or tumor necrosis factor-α after phytohemagglutinin, anti-CD3, or allogeneic stimulation.9 On the whole, these data indicate a relatively unstimulated state of CB lymphocytes at least under in vitro conditions.
At birth, most of the T cells express an αβ T-cell receptor (TCR) heterodimer. γδ T cells are rare and express a diverse array of TCR.10 The TCR β-chain is produced by the combination of V, D, J, and C gene segments.11,12 In addition, this combinatorial diversity is increased by the nibbling of germline nucleotides and addition of N- and P-residues at the V-D-J junction sites. The complementarity determining region 3 (CDR3) encompassing the V-D-J junction displays the most extensive diversity and is thought to contact the antigenic peptide.13 Most T-cell repertoire studies used β-chain variable (BV)-specific monoclonal antibodies (MoAbs) or polymerase chain reaction (PCR) methods to evaluate the expression level of the various BV segments.14,15 A method called “Immunoscope” has been developed to determine the size of CDR3 regions in transcripts of whole BV families or in given BV-BJ combinations with the help of an automated DNA sequencer.16 17 This approach is particularly valuable in defining the diversity of αβ CB T cells for several reasons. First, it can give a global picture of the repertoire in all presently known functional BV subfamilies without being limited by the availability of specific MoAbs. Second, it allows a comparison of several parameters of the αβ T-cell repertoire between adults and newborns (clonality of the BV families, size of the CDR3 regions, and semiquantitative analysis of the BV and BJ usage). Herein we have defined the overall picture of the αβ T-cell repertoire in CB lymphocytes and its modification on stimulation with bacterial superantigens toxic shock syndrome toxin-1 (TSST-1) and the staphylococcal enterotoxin A (SEA) in comparison with adult donors.
MATERIALS AND METHODS
CB samples.
Umbilical CB samples were obtained at delivery from full-term healthy pregnancies after the mother's consent (Dr Brossard, Hôpital Saint-Vincent-de-Paul, Paris, France). Nine peripheral blood leucocyte (PBL) samples from healthy adult individuals were studied as controls.
RNA extraction and cDNA synthesis.
Cells were obtained by gradient density centrifugation (Ficoll/Hypaque). Pellets were frozen in liquid nitrogen before RNA extraction by lysis in guanidium thiocyanate buffer.18 The first cDNA strand was prepared starting from 5 to 10 μg total RNA and avian myeloblastosis virus (AMV) reverse transcriptase as recommended by the manufacturer (cDNA cycle kit; Invitrogen, The Netherlands).
Oligonucleotides and PCR amplification.
The primers used have been described in Puisieux et al19with modifications for BV6 (5′-CTCTGAAGATCCAGCGCACAGAGC-3′) and BV21 (5′-TCCAGCCTGCAAAGCTTGAGGACT-3′). Fluorescent primers for BC and BJ were labeled at the 5′ end with the Fam fluorophore (Applied Biosystems, Foster City, CA). Aliquots of the cDNA synthesis reaction (corresponding to 250 ng of total RNA) were amplified in 50 μL reactions with one of the BV-specific oligonucleotides as the 5′ primer and the BC oligonucleotide as the 3′ primer. The final concentration was 0.5 mmol/L for each primer, dNTP 0.2 mmol/L, MgCl2 2 mmol/L in Taq polymerase buffer (Promega, Madison, WI) in the presence of 1 unit of Taq polymerase (Promega) on a DNA thermal cycler (Perkin Elmer 9600, Norwalk, CT). The PCR cycle profile was denaturation at 94°C for 30 seconds, annealing at 60°C for 45 seconds, primer extension at 72°C for 45 seconds for 40 cycles, and a final polymerization step of 10 minutes at 72°C. Aliquots from each BV-BC PCR product (2 μL) were copied in 4- to 6-cycle run-off reactions primed with a fluorophore-labeled BC- or BJ-specific oligonucleotide as described.19 The run-off reactions were loaded on 4.25% acrylamide sequencing gels (377A DNA sequencer, Applied Biosystems) for size and fluorescence intensity determination. Fluorescent size markers were 80, 145, 210, 270, and 350 bp long or the Genescan-500 size marker (Perkin Elmer). The raw data were analyzed with the help of the Immunoscope software.16 The CDR3 region was defined to include residues 95-106.17 Since the position of the BV and the BC primers are fixed, the length distribution observed in the PCR fluorescent BV-BC products only depends on the size of the V-D-J junctions. Statistical analysis was performed to determine whether or not a profile could be considered as gaussian; a profile was not considered to be gaussian if one peak was excluded from the 95% confidence interval of peak level intensities. TCRB subfamilies BV10 and BV19 were omitted from this analysis because they are pseudogenes in most individuals.20
Vβ and Jβ gene usage.
A competitive PCR was used to quantify the TCR transcripts in each sample. A deleted (4 bp) δ-chain plasmid of the CD3 complex and CD3 primers (CD3-3′ TGTCTGAGAGCAGTGTTCCCAC and CD3-5′ CCAGGCTGATAGTTCGGTGACC) was used.17 The cDNA sample and the deleted δ-chain plasmid were amplified together for 25 cycles in the same conditions as described above. About 3 × 106 copies of cDNA from each sample were then amplified for 30 cycles with the BV primers and an internal fluorescent BC primer. The percentage of representation of each BV family was calculated and presented as histograms. BJ usage was defined after run-off reactions of the unlabeled BV-BC amplification product and is quantitative because the fluorescent primers have comparable amplification efficiencies.17 The fluorescence intensity in each BJ family was expressed as the relative percentage of total signals from the 13 BJ subfamilies and represented as histograms. Statistical comparisons between samples from adults and newborns were done by the Mann-Whitney test.
Cell culture of CB and adult PBL with superantigens.
Umbilical CB cells and adult mononuclear cells were cultured at 2 × 106 cells/mL for 4 days in RPMI 1640 medium (Seromed, Biochrom, KG, Berlin) supplemented with 2 mmol/L glutamine, 10 U/mL penicillin, 10 mg/mL streptomycin (GIBCO, Paisley, UK), and 10% heat-inactivated fetal calf serum (FCS; Seromed). Cultures were stimulated with the lowest concentration of superantigen giving an optimal response: SEA and TSST-1 (Toxin Technology, Madison, WI) at 1 ng/mL. Culture flasks (25 cm2) were incubated at 37°C in 5% CO2. Simultaneously, proliferation assays using3H thymidine uptake were performed to check for proliferation after 4 days of culture. Quantification of the TCR complex–specific RNA and BV usage analysis were performed as described above.
Flow cytometry analysis.
Phenotypic analysis was performed by flow cytometry using a FACScan flow cytometer (Becton Dickinson, Mountain View, CA). MoAbs used were CD3-phycoerythrin conjugated (PE), CD4-PE, CD8-PE, and CD25-PE (Caltag Laboratories, San Francisco, CA). 158-4D3 and UCHL1 MoAbs, specific respectively for CD45RA and CD45RO molecules, were provided during the Vth International Workshop on Human Differentiation Antigens.21 After being washed, cells stained with unlabeled MoAbs were incubated with a 1:100 dilution of fluorescein isothiocyanate-conjugated goat anti-mouse F(ab)2′ fragment (Immunotech, Marseilles, France). Results are expressed as percentages of cells staining above background.
RESULTS
Clonality of umbilical CB lymphocytes and CDR3 size analysis.
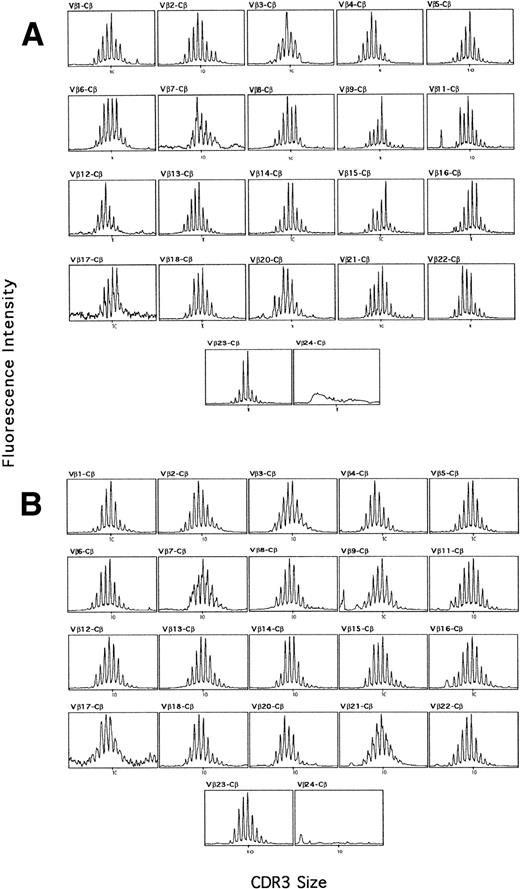
In adults, six to eight peaks spaced by three nucleotides corresponding to in-frame transcripts of the TCR β-chain CDR3 region are usually found for each BV-BC combination (Fig 1).The area under each peak is proportional to the amount of transcripts of the corresponding CDR3 size in the sample.22 Each peak corresponding to a given CDR3 length usually contains multiple distinct sequences. An increase in the height and area of a peak that modifies the bell-shaped CDR3 size distribution indicates oligoclonal or monoclonal expansions occurring on immunological stimulations. Statistical analysis allowed to define a profile as gaussian or not. Sequence analysis has previously been used to confirm the clonality of such expansions in healthy adults.23 In the samples from our healthy adult control group, oligoclonal expansions were detected in at least 5 BV families, BV3, BV7, BV9, BV15, and BV23 in Fig 1A. This was markedly different from the aspect observed in nine different umbilical CB samples showing a reproducible gaussian profile in each BV family (Fig 1B). This profile displayed a size distribution of 8 to 10 identifiable peaks spaced by 3 nucleotides corresponding to in-frame transcripts as previously described in adults.17 CDR3 lengths varied between 5 and 14 amino acids, with a mean length of 9 or 10 amino acids. Two BV families, BV20 and BV4, had a shorter mean CDR3 length of 8 amino-acids (aa) (Fig 1). The bell-shaped profile of the BV-BC amplifications was modified in only 5 out of 220 cases; therefore, we examined these cases for a possible oligoclonal expansion. An in-depth analysis of these BV families with BJ run-off revealed a polyclonal profile in each of the 13 BJ families. Because the majority of expansions occurring in young healthy adults have been described in the CD8+ lymphocyte subpopulation,24 we selected CD4+ and CD8+ lymphocytes in three different CB samples. Similar gaussian profiles and CDR3 lengths were found in both subpopulations (data not shown). We concluded from these data that the umbilical CB T-cell repertoire is definitely polyclonal.
TCR β-chain transcript CDR3 size distribution patterns from a representative adult (A) and CB (B) sample. cDNA was amplified in PCR reactions primed by one BV subfamily and the BC specific primer. The amplification products were copied in run-off reaction primed by a nested fluorescent BC specific primer, and the labeled DNA copies were analyzed on a sequencing gel in an automated DNA sequencer.
TCR β-chain transcript CDR3 size distribution patterns from a representative adult (A) and CB (B) sample. cDNA was amplified in PCR reactions primed by one BV subfamily and the BC specific primer. The amplification products were copied in run-off reaction primed by a nested fluorescent BC specific primer, and the labeled DNA copies were analyzed on a sequencing gel in an automated DNA sequencer.
Vβ and Jβ usage in umbilical CB cells.
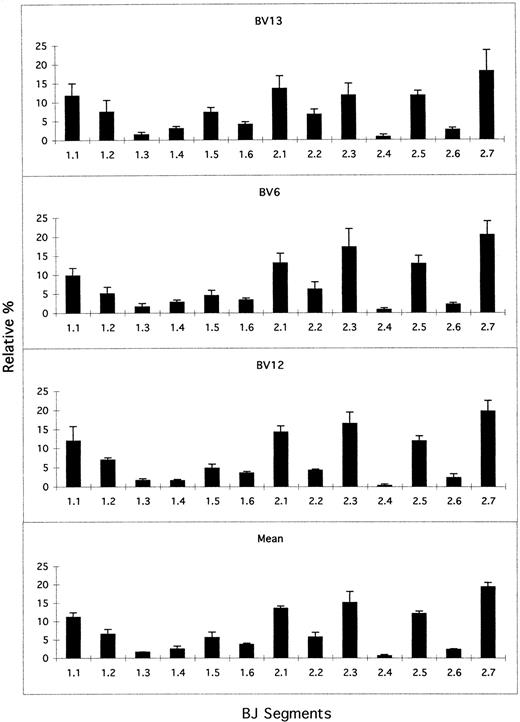
A semiquantitative analysis of the BV usage is possible after a prior quantification of the δ-chain of the CD3/TCR complex. Nine different umbilical CB samples have been analyzed, and the usage of the various BV families compared with adults is shown in Fig2A. Three groups could be distinguished arbitrarily: highly expressed BV families with more than 6% each of the total expression (BV2; BV3; BV4; BV6; BV22); poorly expressed families with less than 3% (BV7; BV11; BV12; BV14; BV15; BV16; BV17; BV18; BV20; BV23; BV24); and BV families with an intermediate expression level between 3% and 6% (BV1; BV5; BV8; BV9; BV13; BV21). In parallel, nine samples from healthy adult donors were studied under identical experimental conditions (Fig 2B). The pattern of BV expression was not markedly different between newborns and adults. There was not a significant difference between the highly expressed families of CB versus adult, taking P < .01 as a cut-off. We then focused our study on the usage of the 13 BJ segments in family BV13 (10 different CB samples), family BV6 (n = 7), and family BV12 (n = 4) previously studied in adults.25 26 The BJ profile is shown for these 3 BV families in Fig 3. The BJ2 family was always expressed more than BJ1 (65%v 35%), and three representative groups could also be distinguished (Fig 3): well-expressed families accounted for more than 8% each of the total BJ representation (BJ2S7; BJ2S1; BJ1S1; BJ2S3; BJ2S5); moderately expressed families between 3% and 8% (BJ1S2; BJ1S5; BJ2S2; BJ1S6); and poorly expressed families with less than 3% (BJ1S4; BJ2S6; BJ1S3; BJ2S4). This nonrandom use of BJ subfamilies was found also in other BV families analyzed, such as BV3, BV5, BV14, BV15, and BV16. A histogram of the mean BJ representation in all the BV families analyzed is shown in Fig 3.
BV usage in nine different CB samples (A). After quantification, cDNA was amplified for 30 cycles directly with one of the BV primers and the fluorescent BC primer. The peak values for each BV subfamily were added, and the sum was expressed as the relative percentages of the total of all the peak intensities (mean ± SD). A similar analysis was conducted for nine samples from healthy adult individuals (B).
BV usage in nine different CB samples (A). After quantification, cDNA was amplified for 30 cycles directly with one of the BV primers and the fluorescent BC primer. The peak values for each BV subfamily were added, and the sum was expressed as the relative percentages of the total of all the peak intensities (mean ± SD). A similar analysis was conducted for nine samples from healthy adult individuals (B).
BJ usage in CB T cells. Histograms were calculated as in Fig 2 and indicate the relative percentage of each BJ segment (mean ± SD) in BV subfamilies BV13, BV6, and BV12 and the mean BJ expression.
BJ usage in CB T cells. Histograms were calculated as in Fig 2 and indicate the relative percentage of each BJ segment (mean ± SD) in BV subfamilies BV13, BV6, and BV12 and the mean BJ expression.
CB T-cell repertoire under superantigenic activation.
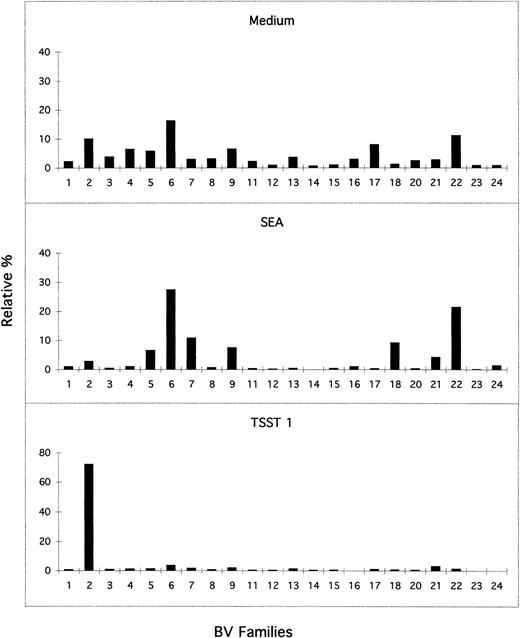
Previous studies have shown that Staphylococcus aureusenterotoxins, including TSST-1 and SEA, are powerful stimulators of T cells.27 Using this technique we analyzed the effect of toxins TSST-1 and SEA on CB and adult PBL. Cells were cultured for 4 days in the same conditions with controls, including culture in medium alone. Strong proliferation in response to superantigenic stimulations was observed for adult (n = 30) as well as CB (n = 10) samples (data not shown). T-cell activation markers CD25 and CD45RO analyzed by flow cytometry (Table 1) markedly increased on stimulation of CB with either toxin. CD45RO marker of memory T cells was very low in CB-unstimulated cells as has been previously reported.5,8,28 We have verified that a nonspecific polyclonal activation with an anti-CD3 MoAb did not change the Immunoscope profiles and BV subfamilies distribution (data not shown). A quantitative study of the BV family usage in CB (n = 4) and adult PBL (n = 4) samples was performed. One representative experiment of CB is shown in Fig 4. After TSST-1 stimulation, we observed a marked expansion of the BV2 family in CB samples, as high as 75% of the total TCRBV repertoire. The BJ usage within BV2 families of samples activated by TSST-1 from one newborn and one adult donor was not different from the representation shown in Fig3 (data not shown). Activation with SEA resulted in the expansion of several BV families in CB as well as in adult PBL. Therefore, in each type of stimulation the profile of the BV family remained strictly polyclonal. Table 2 presents data for BV families modified on SEA stimulation and for BV1, a previously reported SEA expanded family.29 Families BV6, BV18, BV22, and BV24 were expanded in almost all individuals up to eightfold in SEA-stimulated samples in comparison with cultures in medium only. BV18 and BV24, which are poorly expressed without stimulation, became notably expanded in adults and in newborns. BV5, BV7, BV9, and BV21 families were expanded in some individuals and not in others. The BV1 family was not expanded by SEA stimulation under our experimental conditions.
Activation of Adult or CB Lymphocytes With Bacterial Toxins
| Cells* . | Phenotype . | ||||||
|---|---|---|---|---|---|---|---|
| 3H-Thymidine† . | CD3 . | CD4 . | CD8 . | CD25 . | CD45RA . | CD45RO . | |
| Adult PBL (n = 4) | |||||||
| d0 | 74 ± 1.5‡ | 48 ± 4 | 33 ± 3 | 2.6 ± 2 | 68 ± 9 | 54.6 ± 1.7 | |
| d4 SEA | 63 ± 11.5 | 81 ± 7 | 53 ± 15 | 33 ± 3.2 | 36 ± 11 | 74.5 ± 5 | 53 ± 8.6 |
| d4 TSST-1 | 40 ± 15.1 | 75 ± 2.5 | 44 ± 14 | 31.2 ± 4 | 30 ± 0.4 | 68.6 ± 4 | 43 ± 10 |
| CB leucocytes (n = 4) | |||||||
| d0 | 43 ± 10.7 | 44 ± 10.8 | 21 ± 7.3 | 3 ± 1.5 | 55 ± 7.9 | 7.5 ± 3.2 | |
| d4 SEA | 60 ± 25.1 | 73 ± 2.1 | 49 ± 10 | 29 ± 6.5 | 44 ± 4.5 | 72 ± 9 | 50 ± 3 |
| d4 TSST-1 | 50 ± 16.1 | 69 ± 6 | 53 ± 12 | 22 ± 1.5 | 40 ± 6.4 | 72 ± 18 | 63 ± 8 |
| Cells* . | Phenotype . | ||||||
|---|---|---|---|---|---|---|---|
| 3H-Thymidine† . | CD3 . | CD4 . | CD8 . | CD25 . | CD45RA . | CD45RO . | |
| Adult PBL (n = 4) | |||||||
| d0 | 74 ± 1.5‡ | 48 ± 4 | 33 ± 3 | 2.6 ± 2 | 68 ± 9 | 54.6 ± 1.7 | |
| d4 SEA | 63 ± 11.5 | 81 ± 7 | 53 ± 15 | 33 ± 3.2 | 36 ± 11 | 74.5 ± 5 | 53 ± 8.6 |
| d4 TSST-1 | 40 ± 15.1 | 75 ± 2.5 | 44 ± 14 | 31.2 ± 4 | 30 ± 0.4 | 68.6 ± 4 | 43 ± 10 |
| CB leucocytes (n = 4) | |||||||
| d0 | 43 ± 10.7 | 44 ± 10.8 | 21 ± 7.3 | 3 ± 1.5 | 55 ± 7.9 | 7.5 ± 3.2 | |
| d4 SEA | 60 ± 25.1 | 73 ± 2.1 | 49 ± 10 | 29 ± 6.5 | 44 ± 4.5 | 72 ± 9 | 50 ± 3 |
| d4 TSST-1 | 50 ± 16.1 | 69 ± 6 | 53 ± 12 | 22 ± 1.5 | 40 ± 6.4 | 72 ± 18 | 63 ± 8 |
*Adult or CB leukocytes were either unstimulated (d0) or stimulated with bacterial toxins SEA or TSST-1 (1 ng/mL) during 4 days before 3H-thymidine incorporation measurement or immunophenotype analysis.
3H-thymidine incorporation (cpm × 10−3) measured at d4. 3H thymidine incorporation in unstimulated adult or CB lymphocytes at d4 was <1,000 cpm.
Percent of positive cells of the indicated specificity ± SD.
BV usage after activation by superantigens TSST-1 and SEA in a representative CB sample. Cell suspensions were cultured for 4 days in the presence of TSST-1 (1 ng/mL), SEA (1 ng/mL), or culture medium only. The semiquantitative analysis of BV family usage was performed and the results expressed as in Fig 2.
BV usage after activation by superantigens TSST-1 and SEA in a representative CB sample. Cell suspensions were cultured for 4 days in the presence of TSST-1 (1 ng/mL), SEA (1 ng/mL), or culture medium only. The semiquantitative analysis of BV family usage was performed and the results expressed as in Fig 2.
Analysis of BV Subfamily Frequencies Under SEA Stimulation of Adult and CB Lymphocytes
| BV . | Adult PBL (%) . | CB PBL (%) . | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n°1 . | n°2 . | n°3 . | n°4 . | n°1 . | n°2 . | n°3 . | n°4 . | |||||||||
| SEA . | Medium . | SEA . | Medium . | SEA . | Medium . | SEA . | Medium . | SEA . | Medium . | SEA . | Medium . | SEA . | Medium . | SEA . | Medium . | |
| 1 | 1.0 | 2.9 | 1.3 | 2.8 | 0.8 | 4.0 | 2.7 | 2.3 | 1.0 | 2.2 | 2.1 | 4.1 | 1.8 | 3.6 | 3.5 | 1.3 |
| 5 | 5.1 | 3.6 | 3.5 | 4.8 | 6.9* | 2.7 | 4.2 | 8.8 | 6.5 | 5.8 | 6.4 | 7.0 | 19.5 | 3 | 22 | 4.2 |
| 6 | 31.2 | 20.8 | 21.3 | 11.2 | 30.6 | 19.3 | 22.4 | 13.9 | 27.4 | 16.2 | 24.4 | 8.0 | 1 | 14.4 | 40.2 | 14.3 |
| 7 | 18.2 | 4.3 | 16.5 | 7.6 | 0.3 | 0.3 | <0.1 | <0.1 | 10.9 | 3.0 | <0.1 | 0.3 | <0.1 | 1.4 | <0.1 | 4.1 |
| 9 | 4.3 | 5.3 | 5.4 | 4.6 | 6.1 | 4.8 | 4.4 | 5.4 | 7.5 | 6.4 | 7.2 | 2.9 | 7.8 | <0.1 | <0.1 | <0.1 |
| 18 | 6.6 | 1.7 | 7.6 | 3.5 | 3.8 | <0.1 | 0.1 | <0.1 | 9.2 | 1.3 | 1.2 | <0.1 | 2.1 | <0.1 | 8.5 | <0.1 |
| 21 | 5.1 | 2.4 | 3.5 | 4.0 | 8.8 | 5.5 | 9.4 | <0.1 | 4.2 | 2.8 | 5.8 | 10.0 | 5.1 | 4.8 | 5.1 | 7.2 |
| 22 | 17.3 | 14.2 | 16.4 | 6.7 | 22.3 | 6.0 | 17.8 | <0.1 | 21.5 | 11.0 | 20.7 | 6.0 | 14.5 | 5.7 | 17.8 | 9.5 |
| 24 | 2.2 | <0.1 | 2.2 | <0.1 | 0.3 | <0.1 | 3.0 | <0.1 | 1.4 | 0.8 | 0.2 | <0.1 | <0.1 | <0.1 | <0.1 | <0.1 |
| BV . | Adult PBL (%) . | CB PBL (%) . | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n°1 . | n°2 . | n°3 . | n°4 . | n°1 . | n°2 . | n°3 . | n°4 . | |||||||||
| SEA . | Medium . | SEA . | Medium . | SEA . | Medium . | SEA . | Medium . | SEA . | Medium . | SEA . | Medium . | SEA . | Medium . | SEA . | Medium . | |
| 1 | 1.0 | 2.9 | 1.3 | 2.8 | 0.8 | 4.0 | 2.7 | 2.3 | 1.0 | 2.2 | 2.1 | 4.1 | 1.8 | 3.6 | 3.5 | 1.3 |
| 5 | 5.1 | 3.6 | 3.5 | 4.8 | 6.9* | 2.7 | 4.2 | 8.8 | 6.5 | 5.8 | 6.4 | 7.0 | 19.5 | 3 | 22 | 4.2 |
| 6 | 31.2 | 20.8 | 21.3 | 11.2 | 30.6 | 19.3 | 22.4 | 13.9 | 27.4 | 16.2 | 24.4 | 8.0 | 1 | 14.4 | 40.2 | 14.3 |
| 7 | 18.2 | 4.3 | 16.5 | 7.6 | 0.3 | 0.3 | <0.1 | <0.1 | 10.9 | 3.0 | <0.1 | 0.3 | <0.1 | 1.4 | <0.1 | 4.1 |
| 9 | 4.3 | 5.3 | 5.4 | 4.6 | 6.1 | 4.8 | 4.4 | 5.4 | 7.5 | 6.4 | 7.2 | 2.9 | 7.8 | <0.1 | <0.1 | <0.1 |
| 18 | 6.6 | 1.7 | 7.6 | 3.5 | 3.8 | <0.1 | 0.1 | <0.1 | 9.2 | 1.3 | 1.2 | <0.1 | 2.1 | <0.1 | 8.5 | <0.1 |
| 21 | 5.1 | 2.4 | 3.5 | 4.0 | 8.8 | 5.5 | 9.4 | <0.1 | 4.2 | 2.8 | 5.8 | 10.0 | 5.1 | 4.8 | 5.1 | 7.2 |
| 22 | 17.3 | 14.2 | 16.4 | 6.7 | 22.3 | 6.0 | 17.8 | <0.1 | 21.5 | 11.0 | 20.7 | 6.0 | 14.5 | 5.7 | 17.8 | 9.5 |
| 24 | 2.2 | <0.1 | 2.2 | <0.1 | 0.3 | <0.1 | 3.0 | <0.1 | 1.4 | 0.8 | 0.2 | <0.1 | <0.1 | <0.1 | <0.1 | <0.1 |
*Underlined percentages are families showing an increase of expression of at least 50% in SEA-stimulated samples in comparison with cells cultured in medium only. Results for BV expanded subfamilies and for BV1 are shown.
DISCUSSION
This is the first detailed characterization of the TCR β-chain diversity in CB and its capacity to undergo expansion, although a previous study provided some insight into the TCR β-chain CDR3 length distribution in CD4 and CD8 lymphocyte subpopulations.24Some characteristics of the CB T-cell repertoire are close to those described in healthy adults using either the same or other methodologies.15,17,30,31 All BV subfamilies were expressed in newborns and adults with comparable differences in the BV and BJ usage. This nonrandom usage is not a function of the number or location of genes,32 and its significance remains unknown. Genetic factors could be responsible for the variations in the BV expression levels as shown in the case of the BV3 subfamily33 and could be reflected by the standard deviation values of BV expression seen in Fig 2. Other studies have concerned the diversity of the early fetal TCRBV repertoire.25,34,35 The preferential usage of BJ2 elements has been shown in human fetal tissue as early as after 13 weeks of gestation.34 Mean CDR3 length in the different BV families was similar to that reported in adults, which suggests that the variability of the CDR3 length is not notably modified after birth in humans in contrast to the early human fetal repertoire25,33 and to observations made in mice.16 This is also in agreement with previous β-chain CDR3 sequencing data obtained from one CB sample.36 We could therefore conclude that the T-cell repertoire is fully constituted at birth. This also appears to be the case for CB B-cell repertoire.37
As an example of in vitro-induced modifications of this repertoire, we have looked at stimulation with superantigens. We did not observe significant differences in the levels of proliferative responses to stimulation between the adult and the CB lymphocytes. Staphylococcal toxin TSST-1 is known to specifically induce the expansion of BV2-expressing T cells.27 The heterogeneity of the CDR3 length in the BV2 family on TSST-1 stimulation has been recently shown in adults by TCR spectratyping.38 The global Immunoscope analysis was especially appropriate in the case of SEA, because several different BV families could be stimulated. MoAbs are presently not available for all of the BV families; BV24 and poorly expressed families would otherwise have been difficult to evaluate. Some BV families expanded by SEA stimulation in adults have been reported29,39; BV6, BV7, BV9, BV18, and BV22 specificities were confirmed in this study. BV24 is one of the most poorly expressed families in adults and in newborns, but its expansion was clearly detected in this study. BV6, BV18, BV22, and BV24 were consistently expanded on stimulation, whereas other families (BV5, BV7, BV9, BV21) were expanded more variably. We did not observe notable differences in the SEA-induced modification of the TCRBV repertoire between adults and newborns.
The major difference between the adult and the CB T-cell repertoire lies in the distribution of the CDR3 size, which reflects the clonality of the T-cell population (polyclonal v oligoclonal or monoclonal profiles). The polyclonal profiles observed in CB support the notion that these cells have not been previously exposed to antigenic stimulation. In healthy adults, oligoclonal expansions can be seen in different BV subfamilies and are the hallmark of the antigenic stimuli received throughout life. These have been reported to occur mainly in the memory CD8+CD45RO+ lymphocyte subpopulations,24 but they have also been reported in CD4+CD45RO+ T cells in elderly humans.23 Alterations of the repertoire have been observed in pathological conditions,17 such as autoimmune diseases, tumors,19 or GVHD.40,41 Mature T cells constitute the main reservoir for T-cell repopulation during the first year after bone marrow engraftment.42 Expanded populations of antigen-specific mature T lymphocytes cotransfused with adult stem cells could participate in the triggering of GVHD caused by cross-reactive recognition of alloantigens in the recipient. By using spectratyping, another method based on the CDR3 size analysis, modifications of the T-cell repertoire during the immune reconstitution after bone marrow transplantation have been documented42-44and linked to GVHD or infectious complications. The absence of clonal expansions of alloreactive cells in CB could have a crucial role in the lower incidence of GVHD observed after CB transplantation. The evaluation of the T-cell repertoire reconstitution after graft from CB hematopoietic stem cells in parallel with the clinical status, GVHD, and the quality of immune responses will be needed to assess these points. Furthermore, the polyclonal profile of the CB T-cell repertoire constantly observed here will facilitate detection of T-cell expansions during follow-up of these grafts in comparison with grafts from adult donors.
ACKNOWLEDGMENT
The authors thank Dr M. Busson for help in the statistical analysis and Drs P. Kourilsky and D. Przepiorka for critical reading of the manuscript.
L.G. and N.D. contributed equally to this work.
Supported in part by the Etablissement Français des Greffes (EFG), the Fondation de France contre la Leucémie, and EUROCORD Concerted Action Biomed II.
Address reprint requests to Antoine Toubert, MD, Laboratoire d'Immunogénétique Humaine, INSERM U.396, Centre G. Hayem, Hôpital Saint-Louis, 1, Av Claude Vellefaux, 75475, Paris Cedex 10, France.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal