Abstract
Defects in the gene for Bruton's tyrosine kinase (Btk) result in the disorder X-linked agammaglobulinemia (XLA). Whereas XLA is characterized by a profound defect in B-cell development, Btk is expressed in both the B lymphocyte and myeloid cell lineages. We evaluated a patient with XLA who had reduced amounts of Btk transcript but no abnormalities in his coding sequence. A single base-pair substitution in the first intron of Btk was identified in this patient, suggesting that this region may contain regulatory elements. Using reporter constructs we identified two transcriptional control elements in the first 500 bp of intron 1. A strong positive regulator, active in both pre-B cells and B cells, was identified within the first 43 bp of the intron. Gel-shift assays identified two Sp1 binding sites within this element. The patient's mutation results in an altered binding specificity of the proximal Sp1 binding site. A negative regulator, active in pre-B cells only, was located between base pairs 281 and 491 of the intron. These findings indicate that regulation of Btk transcription is complex and may involve several transcriptional regulatory factors at the different stages of B-cell differentiation.
BRUTON'S TYROSINE kinase (Btk) has been identified as a key regulator of B-cell development.1-3Mutations in the Btk gene result in the human immunodeficiency X-linked agammaglobulinemia (XLA),1,2 which is characterized by low levels of circulating immunoglobulins and a severe decrease in B-cell numbers.4 In mice, a defective Btk gives rise to thexid phenotype,5 a milder defect in B-cell development in which affected mice fail to make antibody to some T-independent antigens and have about half the normal number of B cells.6 7
Btk is a cytoplasmic tyrosine kinase that is expressed throughout B-cell and myeloid development but it is not expressed in T cells or other nonhematopoietic cell lineages.8-10 Amino acid sequence identity places Btk in an emerging family of Src-related tyrosine kinases including Itk,11 Tec,12DrSrc28C,13 and Bmx.14 A unique feature of this family is the long amino terminal end, which includes a proline rich region and a pleckstrin homology domain.15 The proline rich region has been shown to bind the SH3 domains of the protein tyrosine kinases; Fyn, Lyn, and Hck.16 For Itk, in vitro studies have shown that the proline rich region is able to bind its SH3 domain in an intramolecular reaction that regulates the binding of these two domains to their respective targets.17 Current data suggest that the pleckstrin homology (PH) domain is likely to be involved in the intracellular translocation of Btk15,18-20 and perhaps in binding to critical substrates.21 Although Btk can be activated by cross-linking of surface immunoglobulin,22,23FcεR,24 interleukin-5 receptor (IL-5R),25 and IL-6,26 the mechanisms by which mutations in Btk result in a failure of B-cell development are not known.
Btk consists of 19 exons in a 37-kb stretch of DNA at Xq22.27-30 The first exon and 30 bp of the second exon constitute the 5′ untranslated region of the Btk mRNA. Expression of Btk has been shown in the CD34+ progenitor cell line KG-1 and begins before immunoglobulin heavy and light chain gene rearrangements.10 Multiple transcriptional start sites between base pairs −5 to −30 upstream of exon 1 (numbering according to the original sequence published by Vetrie et al) have been identified,30,31 which is consistent with the lack of a clear TATAA box in the promoter region. Studies on the promoter region have identified binding sites critical for lineage-specific expression within the first 200-bp upstream of exon 1. Transcription factor binding sites included a PU.1 site at −61 bp, an Sp1 site at −169 bp, and an Sp3 site at −38 bp.31,32 When a more upstream sequence was included in promoter reporter constructs activity dropped approximately twofold in B cells, suggesting that regulation of Btk expression was likely to be more complex and involve more than just the Sp1 and PU.1 family members binding at the minimal promoter.31 Both phorbol ester and retinoic acid were able to increase Btk expression in myeloid cells, whereas addition of phorbol ester and TGF-β1 to B cells decreased Btk mRNA levels.9
Over 200 different mutations in Btk have been identified.33-39 These varied mutations include single base pair substitutions (70%), small insertions or deletions (27%), and gross deletions (3%). However, no mutations affecting the regulation of the gene have been reported. In this report we describe a mutation in Btk that results in decreased amounts of Btk transcript but does not affect the integrity of the coding sequence. Analysis of this mutation led to the identification of two regulatory elements within the first 500 bp of intron 1.
MATERIALS AND METHODS
RNA and DNA analysis.
Northern blots, polymerase chain reaction (PCR), single-strand conformation polymorphism (SSCP) analyses of Btk, and cloning and sequencing of PCR fragments were performed as previously described.33 RNA was isolated from Epstein-Barr virus (EBV)–transformed cell lines or freshly isolated peripheral blood lymphocytes (PBLs) using the Qiagen RNeasy spin column (Qiagen, Santa Clarita, CA). cDNA was prepared by reverse transcription of the RNA using reverse transcriptase (Superscript; GIBCO-BRL, Gaithersburg, MD) and then PCR amplified using the appropriate primers. Forward primers used for amplifying the exon 1/exon 2 border sequence of Btk were: 5′-CAGACTGTCCTTCCTCTC-3′ and 5′-AATGCATCTGGGAAGCTA-3′, and the reverse primer was 5′-CAAGAGAAACAGGCGCTT-3′. Amplification of genomic DNA at the Xq22 polymorphic loci DXS101 and DXS178 was performed as previously described.40
Luciferase reporter constructs.
A 3.4-kb EcoRI-HindIII fragment, beginning 2.3 kb upstream of the first exon of Btk and ending 1 kb downstream of the exon, was subcloned from cosmid 237D10 into pBluescript (pBSK, Stratagene, La Jolla, CA).27 From this construct a 2.9-kbSma I to Xho I fragment was subcloned into the luciferase reporter backbone pGL2 (Promega, Madison, WI). TheSma I site is 1.8 kb upstream of exon 1, and the Xho I site is 1.0 kb downstream of the exon. Reporter pBtkpro was constructed by deleting the sequence between the Ppu10I site (within exon 1) and the HindIII site (within the pGL2 cloning cassette). All other constructs that contained a splice donor site and varying lengths of intron 1 also included a splice acceptor sequence. The intron 1 acceptor site was derived from a PCR product, which was amplified using the forward primer 5′-AAACCTGACAGATCTGGG-3′ and the reverse primer 5′-TGCGGCCAAAGCTTCTTC-3′. This product was cloned into the pGL2 vector using the BglII and HindIII sites included in the above primer sequences. The pGL2 construct, containing the 2.9-kb Sma I-Xho I fragment and the acceptor sequence was called pBtkpro+1029 because it contained 1,029 bp of 5′ intron 1 sequence. Constructs pBtkpro+491, pBtkpro+281, and pBtkpro+43 were deletion constructs of pBtkpro +1029 from the Stu I, Kpn I, and Rsa I sites, respectively.
The pBtkpro+6 reporter was also derived from a PCR-amplified product. Sense primer 5′-CAGACTGTCCTTCCTCTC-3′ and antisense primer 5′-AGATCTACCCACCTCAGTCCTGA-3′ were used to amplify exon 1 and the first 6 bp of the intron. This fragment was cloned into the pBtkpro construct to give pBtkpro+6.
Introduction of the patient mutation into the pBtkpro+ constructs was done using a PCR-based strategy. The same sense primer as used in the construction of pBtkpro+6 was used with the antisense primer 5′-AGCCAGCTCTGACCCTGG-3′ to amplify patient genomic DNA. The PCR product was cloned into pBtkpro+1029 at the Ppu10I andKpn I sites. Again, deletion constructs as described above were made using the Stu I, Kpn I, and Rsa I sites. All constructs were sequenced to check for PCR errors and correct ligation. Plasmid DNA for transient transfections was prepared by CsCl/EtBr density gradient centrifugation as described by Maniatis et al.41
Cell culture.
Human pre–B-cell line REH, B-cell lines Daudi and Raji, and T-cell line Jurkat were maintained as suspension cultures in RPMI supplemented with heat inactivated fetal bovine serum (FBS; 15%), 2-ME (2.5 × 10−5 mol/L), and gentamicin (250 ng/mL).
Transient transfections.
Transient transfection assays were done by electroporation using the BioRad Gene Pulsar (BioRad Laboratories, Richmond, CA).42 Cells were grown to early log phase, harvested, resuspended in serum-free Iscove's modified Dulbecco's media (IMDM) at 2 × 107 cells per 200 μL and placed in a cuvette (0.4 cm electrode gap) containing 20 μg of the test plasmid and 8 μg of an internal control reporter construct, secreted alkaline phosphatase driven by a PGK promoter (PGKSEAP; kindly provided by Dr L.H. Shapiro; St Jude Children's Research Hospital, Memphis, TN). After a 10-minute room temperature (RT) incubation the cells were subjected to a predetermined electrical field (REH and Jurkat 250V, Daudi 190V, and Raji 220V), with the capacitance extender set to 960 μF. After another 10 minutes at RT the cells were plated into 100-mm tissue culture plates containing 10 mL of IMDM supplemented with 15% FBS and gentamicin. The cells were left to recover for 24 hours, at which stage 300 μL was drawn off to be assayed for secreted alkaline phosphatase, and the remaining cells were harvested, lysed, and resuspended in 200 μL of assay buffer, and 100 μL was used in the assay as described elsewhere.42 Once the luciferase units had been normalized using the PGKSEAP to control for transfection efficiency, all results were expressed as a fold increase or decrease over the basic pBtkpro construct. All experiments were repeated at least three times using two different preparations for each construct tested.
Gel-shift assays.
Nuclear extracts were prepared as described elsewhere.43Two double-stranded oligonucleotide probes that covered the −5 to +32 bp region at the exon 1/intron 1 border were made. Probe sequences were as follows: −5+16 5′-ACTGAGGTGGGTCTGGGGTATG-3′ and +12+32 5′-GTATGGCAGGGGCTGGGCAGC-3′. An additional −5+16 probe including the patient's T to G mutation at position +6 was also made (−5+16M). The Sp1 consensus oligonucleotide, 5′-ATTCGATCGGGGCGGGGCGAGC-3′, was purchased from Promega (Madison, WI). One hundred micrograms of each complementary strand was annealed in the presence of 100 mmol/L NaCl by heating the mixture to 95°C for 5 minutes and then allowing the samples to cool to RT for 1 hour followed by a 1 hour incubation at 37°C. Double-stranded probes were radiolabeled with γ32P-adenosine triphosphate (ATP) using polynucleotide kinase (Pharmacia Biotech, Piscataway, NJ) and purified over a G50 Sephadex spin column (Boehringer Mannheim, Indianapolis, IN). For the gel-shift, 0.1 ng of probe was incubated with 10 to 15 μg of nuclear extract, 2 μg of poly(dI-dC; Boehringer Mannheim), and 20 μg bovine serum albumin (BSA; New England Biolabs, Beverly, MA) in a 20 μL volume with 10% glycerol, 10 mmol/L HEPES pH 7.9, 2 mmol/L Tris pH 7.9, 100 mmol/L KCl, 1 mmol/L DTT, 1 mmol/L EDTA, 1 mmol/L 2-ME, and 0.5 mmol/L phenylmethylsulfonyl fluoride (PMSF) at RT for 25 minutes. Samples were electrophoresed at 4°C on 6% polyacrylamide gels in 0.4× Tris borate EDTA (TBE) at 10 V/cm for 3 hours. Gels were dried under vacuum and exposed to x-ray film (Eastman Kodak, Rochester, NY) for 3 to 6 hours.
RESULTS
Patient 0030 has reduced amounts of normal Btk mRNA.
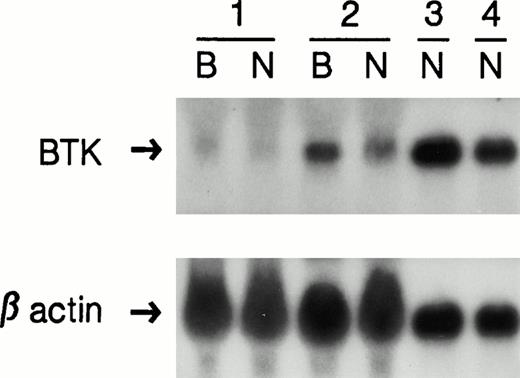
We have previously shown that most mutations in Btk that cause premature stop codons, frameshifts, or splice defects are associated with markedly reduced amounts of Btk transcript, whereas mutations resulting in amino acid substitutions are associated with normal amounts of Btk mRNA as detected by Northern blot analysis.33 A single EBV-transformed cell line from a patient with late onset XLA (UPN 0030) consistently showed decreased but easily detectable Btk transcripts. Because EBV transformation and prolonged in vitro culture can alter the phenotype of a cell line, mRNA from freshly isolated patient neutrophils was also analyzed by Northern blot. Figure 1 shows that the amount of Btk transcript was equivalently decreased in the cell line and in the freshly isolated cells.
Northern blot analysis. The top panel shows RNA from EBV-transformed cell lines (B) and neutrophils (N) from an XLA patient with a 4-bp deletion (1); the patient described in this study (2); an XLA patient with an amino acid substitution (3); and a normal control (4). To control for sample loading, the blot was stripped and rehybridized with a β-actin specific probe, shown in the lower panel.
Northern blot analysis. The top panel shows RNA from EBV-transformed cell lines (B) and neutrophils (N) from an XLA patient with a 4-bp deletion (1); the patient described in this study (2); an XLA patient with an amino acid substitution (3); and a normal control (4). To control for sample loading, the blot was stripped and rehybridized with a β-actin specific probe, shown in the lower panel.
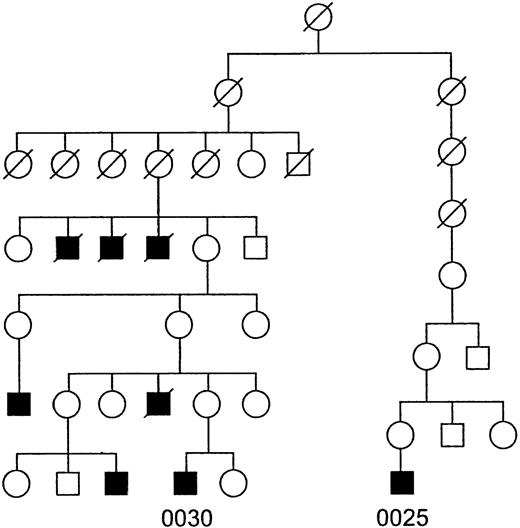
To exclude the possibility of a mutation that altered the stability of the Btk message in the patient, the coding region and 5′ and 3′ untranslated regions of the cDNA were sequenced, but no abnormalities were identified. The absence of mutations in Btk mRNA and the preservation of significant amounts of transcript raised questions about the reliability of the XLA diagnosis. However, this patient was part of a four-generation pedigree that included at least five other affected males (Fig 2). Linkage studies in this family had mapped the defect to the site of the Btk gene at Xq22 with a lod score greater than 3.0.44
Pedigree of the patient described in this study. Squares represent males and circles denote females. The shaded symbols represent patients with XLA. A slash through a symbol indicates that the individual is deceased. Patient numbers are written below their respective symbols.
Pedigree of the patient described in this study. Squares represent males and circles denote females. The shaded symbols represent patients with XLA. A slash through a symbol indicates that the individual is deceased. Patient numbers are written below their respective symbols.
Mutation detection in patient 0030.
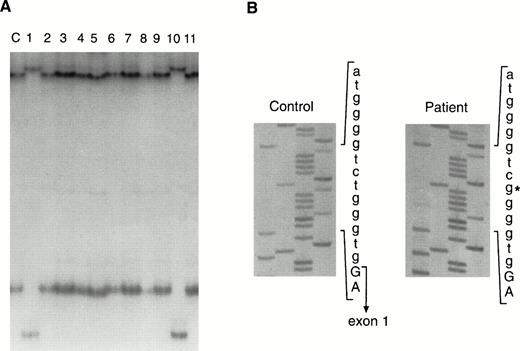
Screening for mutations in Btk outside the coding region was performed using SSCP analysis.33 Primer pairs that flanked each of the 19 exons were used to examine the genomic DNA of the patient. Analysis of exon 1, the noncoding exon, showed an abnormal migration pattern (Fig 3A); all other exons were normal. The same alteration was seen in DNA from a second XLA patient who had no known family history of immunodeficiency. Similar to Patient 0030, this patient had slightly higher concentrations of serum immunoglobulins than patients with typical XLA, and he had no other alterations in the remaining 18 exons as tested by SSCP. The altered SSCP pattern for exon 1 was not seen in 111 unrelated X chromosomes, making it unlikely that this alteration represented a polymorphism.
Mutation detection in the genomic DNA from Patient 0300. (A) SSCP analysis of exon 1 and flanking sequences from a control (lane labeled c) and patients with XLA. Patients 0030 and 0025 are shown in lanes 1 and 10, respectively. (B) DNA sequence of the 5′ part of intron 1 from a control and from Patient 0030. The 3′ end of exon 1 is indicated in the control sequence. The T to G transversion in the patient is denoted by an asterisk.
Mutation detection in the genomic DNA from Patient 0300. (A) SSCP analysis of exon 1 and flanking sequences from a control (lane labeled c) and patients with XLA. Patients 0030 and 0025 are shown in lanes 1 and 10, respectively. (B) DNA sequence of the 5′ part of intron 1 from a control and from Patient 0030. The 3′ end of exon 1 is indicated in the control sequence. The T to G transversion in the patient is denoted by an asterisk.
Exon 1 and its flanking regions were sequenced, and a T to G transversion at position +6 of the intron 1 splice donor site was identified in both patients (Fig 3B). To determine whether this alteration occurred independently in the two patients or as a result of common descent, highly polymorphic markers, DXS178 and DXS101,40 near the Btk gene at Xq22 were used to establish an XLA haplotype. Both boys had inherited the same alleles at DXS178 and DXS101, suggesting common descent. Further inquiries established that the patients were descendants of a common ancestor (Fig 2).
The proximity of the base pair substitution to a splice donor site suggested that the alteration might affect splicing, although defects at the +6 position in an intron are rarely responsible for splice defects.45 To determine if any cryptic splice sites were activated because of the +6 alteration, PCR primers were designed to amplify the cDNA region surrounding the exon 1/exon 2 cDNA border. Two 5′ primers, one beginning at the transcriptional start site position −28 (numbering according to the original cDNA sequence published by Vetrie et al) and the other closer to the 3′ end of exon 1 (position +62) were used with a 3′ primer from exon 2 (position +210). When cDNA from freshly isolated peripheral blood mononuclear cells from the patient was used as the template, both PCR reactions showed the expected products. No products using cryptic splice sites were identified. In the absence of evidence that the base-pair substitution was influencing splicing, we explored the possibility that the alteration affected transcription regulatory elements within intron 1 of Btk.
Identification of transcriptional regulators within the first 1,029 bp of intron 1.
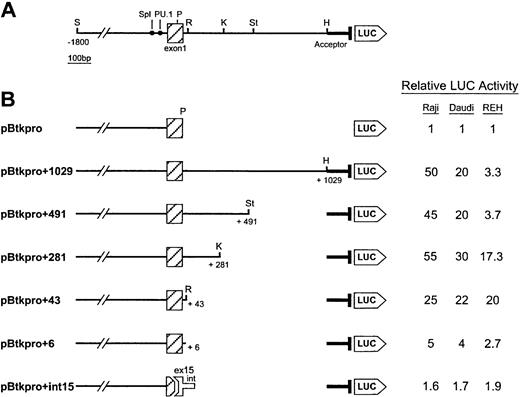
A Btk promoter reporter construct (pBtkpro, Fig4) was designed by subcloning 1.8 kb of Btk promoter DNA upstream of exon 1 plus 66 bp of this exon into the promoterless pGL2 luciferase expression vector. The activity of pBtkpro was compared with that of the empty pGL2 vector by transient transfection in the pre–B-cell line REH, the B-cell lines Raji and Daudi, and the T-cell line Jurkat (Table1). Activities were consistently above background and were in agreement with previous observations showing that the Btk promoter was more active in B cells than in T cells.31 32 To identify any cis acting elements within intron 1 that could affect Btk expression, a construct containing the same 1.8-kb upstream region plus the entire 102 bp of exon 1 and 1,029 bp of intron 1, pBtkpro+1029 (Fig 4A) was designed. Because this construct contained a splice donor site, an acceptor site was included to facilitate splicing. The acceptor site was derived from a 182-bp PCR fragment that included the last 152 bp of intron 1 and the first 30 bp of exon 2, ending 1 bp upstream of the Btk start codon. When comparing pBtkpro+1029 with pGL2, marked increases above background were observed in the two B-cell lines, a less dramatic increase was seen for the pre–B-cell line, and no change was observed for the T-cell line (Table1). The 20- to 50-fold increase in luciferase activity seen in the two B-cell lines for the intron 1 containing construct, when compared with pBtkpro, suggested that intron 1 contained a strong positive regulatory element active in B cells. In the pre–B-cell line, the approximate threefold increase suggested that this element was either nonfunctional or was balanced by negative regulatory elements.
Functional characterization of the 5′ portion of Btk intron 1. (A) The diagram shows the extent of the Btk genomic sequence flanking exon 1 that was subcloned to produce the largest reporter construct pBtkpro+1029. The Sma I site (S) 1.8 kb upstream of exon 1 and the HindIII site (H) 1.0 kb downstream indicate the 5′ and 3′ extents of the subclone, respectively. Also shown are the Sp1 and PU.1 transcription factor binding sites.31 32Restriction sites used to make the deletions included: P,Ppu10I; R, Rsa I; K, Kpn I; and St, StuI. The intron 1 acceptor sequence is shown 5′ to the start of the luciferase gene (LUC). (B) The deletion constructs with the extent of the remaining intron given in base pairs. The final construct represents the fusion between 5′ exon 1 and 3′ exon 15 and the first 119 bp of intron 15. The relative luciferase activity for each construct in the three cell lines tested is given in the table on the right. Reporter construct pBtkpro was assigned the relative activity level of 1 and all other values indicate the fold increase from this basal level. Experiments were performed in duplicate and repeated three times. Values within experiments and between experiments consistently differed by less than 10%.
Functional characterization of the 5′ portion of Btk intron 1. (A) The diagram shows the extent of the Btk genomic sequence flanking exon 1 that was subcloned to produce the largest reporter construct pBtkpro+1029. The Sma I site (S) 1.8 kb upstream of exon 1 and the HindIII site (H) 1.0 kb downstream indicate the 5′ and 3′ extents of the subclone, respectively. Also shown are the Sp1 and PU.1 transcription factor binding sites.31 32Restriction sites used to make the deletions included: P,Ppu10I; R, Rsa I; K, Kpn I; and St, StuI. The intron 1 acceptor sequence is shown 5′ to the start of the luciferase gene (LUC). (B) The deletion constructs with the extent of the remaining intron given in base pairs. The final construct represents the fusion between 5′ exon 1 and 3′ exon 15 and the first 119 bp of intron 15. The relative luciferase activity for each construct in the three cell lines tested is given in the table on the right. Reporter construct pBtkpro was assigned the relative activity level of 1 and all other values indicate the fold increase from this basal level. Experiments were performed in duplicate and repeated three times. Values within experiments and between experiments consistently differed by less than 10%.
Luciferase Activities of pGL2, pBtkpro, and pBtkpro+1029 in the Four Cell Lines Tested
| Construct . | Cell Line . | |||
|---|---|---|---|---|
| Raji . | Daudi . | REH . | Jurkat . | |
| pGL2 | 20 | 8 | 20 | 20 |
| pBtkpro | 272 | 69 | 110 | 38 |
| pBtkpro+1029 | 13,709 | 1,360 | 364 | 17 |
| Construct . | Cell Line . | |||
|---|---|---|---|---|
| Raji . | Daudi . | REH . | Jurkat . | |
| pGL2 | 20 | 8 | 20 | 20 |
| pBtkpro | 272 | 69 | 110 | 38 |
| pBtkpro+1029 | 13,709 | 1,360 | 364 | 17 |
Luciferase activities were normalized for transfection efficiency by cotransfection of a secreted alkaline phosphatase control plasmid.
Serial 3′ end deletions were made in pBtkpro+1029 (Fig 4B) to localize potential regulatory elements active in the pre-B and B cells. No differences were seen when pBtkpro+491 was compared with pBtkpro+1029 in either the B-cell or pre–B-cell lines; however, deletion of an additional 210 bp (pBtkpro+281) resulted in a fivefold increase in activity in the pre–B-cell line REH. This indicated the presence of a pre-B cell–specific negative regulatory element between base-pairs 281 and 491 of intron 1. By specifically deleting the 210 bp between theKpn I and Stu I sites in pBtkpro+1029, no change in activity was seen in the B cells, but a twofold to threefold increase was observed in the pre-B cells confirming the presence of a negative regulator within this sequence. No significant differences in activity were observed for the next serial deletion, pBtkpro+43, when compared with pBtkpro+281. This suggested that the positive regulatory element, first identified in the pBtkpro+1029 construct, was active in both B cells and pre-B cells and was localized within the first 43 bp of the intron. To confirm the presence of the positive regulatory element within the first 43 bp of intron 1, a luciferase construct containing only the first 6 bp of the intron, pBtkpro+6, was compared with pBtkpro and pBtkpro+43. pBtkpro+6 showed a fivefold to sevenfold reduction in luciferase activity in the B-cell lines Daudi and Raji and in the pre–B-cell line REH when compared with pBtkpro+43; however, the activity remained higher than that of the pBtkpro construct.
A 79-bp fragment containing the first 43 bp of intron 1 and spanning the exon 1/intron 1 border sequence was analyzed for enhancer activity by reversing the orientation and/or moving the fragment upstream of the transcriptional start site. However, this Btk sequence was only active in its original position and orientation, suggesting that it was not a “classical” enhancer element.
It has been shown that the presence of an intron in a reporter construct can increase expression levels by stabilizing the mRNA.46 To determine if the residual increase in activity of pBtkpro+6 over pBtkpro could be attributed to splicing, the last 36 bp of exon 1 and the intron 1 sequence were replaced with the last 44 bp of exon 15 and the first 119 bp of intron 15. The 182-bp acceptor site remained the same in the new construct, pBtkpro-int15. The luciferase activity seen in cells transfected with pBtkpro-int15 was marginally higher than that of pBtkpro but lower than all of the intron 1–containing constructs, indicating that splicing was making only a minor contribution to the luciferase activity of the intron 1 containing constructs.
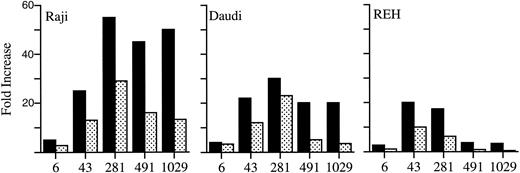
The effect of the base pair substitution found in the patient, a T to G transversion at position +6 in intron 1, was evaluated in all of the constructs containing intron 1 sequence. Luciferase activities elicited by these constructs were consistently lower when compared with their normal counterparts (Fig 5). The sixfold to eightfold reduction seen in the Daudi and REH cells when transfected with the altered pBtkpro+1029 construct, containing the most intron 1 sequence, approximated the reduction in Btk mRNA as originally seen on the Northern blots.
Relative luciferase activities of the normal constructs (▪) compared with the mutant constructs (▧). The fold increase is relative to pBtkpro, the construct with no intron 1 sequence. Construct numbers are given below their respective bars: 6, pBtkpro+6; 43, pBtkpro+43; 281, pBtkpro+281; 491, pBtkpro+491; and 1029, pBtkpro+1029. Cell lines are indicated in the top left-hand corner of each series.
Relative luciferase activities of the normal constructs (▪) compared with the mutant constructs (▧). The fold increase is relative to pBtkpro, the construct with no intron 1 sequence. Construct numbers are given below their respective bars: 6, pBtkpro+6; 43, pBtkpro+43; 281, pBtkpro+281; 491, pBtkpro+491; and 1029, pBtkpro+1029. Cell lines are indicated in the top left-hand corner of each series.
Nuclear extracts specifically bind sequences within the first 43 bp of intron 1.
DNA sequence analysis of the first 43 bp of intron 1 identified a putative Sp1 binding site47 between base pairs +20 and +28. The T to G transversion at position +6 identified in the patient's DNA created an identical Sp1 binding site at the mutant locus between base pairs +3 and +11. To determine whether these sites or additional sites within this region could bind nuclear extracts, we performed gel-shift assays.
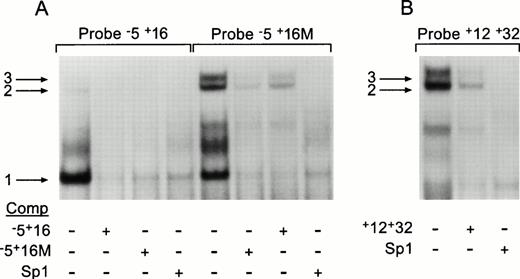
Using a sequence spanning nucleotides −5 to +16 at the exon/intron border (probe −5+16) to identify nuclear DNA binding factors, a single gel-shift was observed (Fig 6A, band 1). This band could be competed with cold wild type −5+16 probe, a probe bearing the +6 T to G transversion (−5+16M), and by an Sp1 consensus oligonucleotide. When the mutant probe, −5+16M, was used as a target for DNA binding proteins, the single band observed for the normal probe was again seen but an additional doublet (Fig 6A, bands 2 and 3) was also seen, suggesting that the mutation created additional binding sites in this region. In the competition assays, all three shifts were inhibited by the addition of cold −5+16M, −5+16, or Sp1 oligonucleotides, indicating the involvement of Sp1 transcription factors. Finally the +12+32 probe was used to establish if the putative Sp1 binding site between base pairs 20 to 28 could bind Sp1 family members. In these gel-shifts, a doublet corresponding to bands 2 and 3 seen for the mutant probe was observed (Fig 5B); however, no clear band corresponding to band 1 was seen. The doublet was competed by both the cold +12+32 and Sp1 probes.
Gel-shift analyses of the sequence at the exon 1/intron 1 border. Raji nuclear extracts were prepared and tested for their ability to shift three γ32P-labeled oligonucleotides by EMSA. (A) The normal −5+16 probe and the mutant −5+16M probe, indicated above their respective lanes, were incubated in the absence or presence of 200× excess unlabeled oligonucleotide, shown below each lane. (B) The +12+32 labeled probe, indicated above the lanes, was incubated in the absence or presence of 200× excess cold probe as shown below. The three arrows indicate the observed gel-shifts.
Gel-shift analyses of the sequence at the exon 1/intron 1 border. Raji nuclear extracts were prepared and tested for their ability to shift three γ32P-labeled oligonucleotides by EMSA. (A) The normal −5+16 probe and the mutant −5+16M probe, indicated above their respective lanes, were incubated in the absence or presence of 200× excess unlabeled oligonucleotide, shown below each lane. (B) The +12+32 labeled probe, indicated above the lanes, was incubated in the absence or presence of 200× excess cold probe as shown below. The three arrows indicate the observed gel-shifts.
As expected, because of the ubiquitous expression of Sp1, similar band shifts were identified when REH or Jurkat nuclear extracts were used, and in all cases a nonspecific unlabeled oligonucleotide was unable to compete for the specific band shifts 1, 2, and 3 (data not shown).
DISCUSSION
In this report we document the location of two transcriptional control elements within the first 1,029 bp of intron 1 of Btk. These two regulatory elements were identified through a series of 3′ end deletions of the intron 1 sequence. The dramatic increase in activity of pBtkpro+1029 compared with pBtkpro in the B-cell lines indicated the presence of a positive regulator within intron 1. The substantial loss in activity of pBtkpro+6 compared with pBtkpro+43 placed this element within the first 43 bp of the intron. This drop in activity was not caused by an unacceptably short intron as the construct still contained 162 nucleotides between the splice donor and acceptor sites, a sequence long enough for efficient splicing. Replacing the exon 1/intron 1 border sequence with that of exon/intron 15 (pBtkpro-int15, intron length 287 bp) gave similar results to that of pBtkpro+6 and not of the other constructs containing more of the intron 1 sequence. This confirmed that splicing was only playing a minor role in the activity of the reporter constructs and that intron 1 contained a positive regulator of transcription.
The positive regulator also increased luciferase activity in pre-B cells by fivefold, but its activity was reduced by the presence of a negative regulator located between base pairs 281 and 491. The potential importance of the two regulatory elements in intron 1 of Btk is underscored by the high DNA sequence conservation in this region between the murine and human genes.48
DNA sequence analysis of the positive regulator identified a putative Sp1 binding site between +20 and +28 bp. Using a +12 to +32 bp probe we were able to show specific binding of one or more proteins belonging to the Sp1 family of transcription factors to this site. When a −5 to +16 bp probe spanning the patient mutation region at +6 bp was used in an EMSA, a different gel-shift was seen; however, it was also competed by the Sp1-specific probe, suggesting that another combination of Sp1 family members was able to bind this region. The T to G alteration at position +6 in the patient created an Sp1 binding site identical to the one identified between base pairs +20 to +28. When the patient −5+16M probe was used in the EMSA, the doublet seen for the +12 to +32 bp probe was seen in addition to the single band detected for the −5 to +16 bp normal probe. These data taken together suggest that the first 32 bp of intron 1 can bind two different proteins or sets of proteins that belong to the Sp1 family of transcription factors. The patient mutation, by altering the specificity of the 5′ binding site, probably disrupts the orderly binding of these factors leading to inefficient initiation of transcription.
Transcription control elements have been identified within the introns of several lymphoid associated genes, including those found within the immunoglobulin49 and T-cell receptor50 loci. Intron 1 of the CD4,51 c-Fes,52 and adenosine deaminase genes53 and intron 2 of the IL-4 gene54 have all been shown to contain regulatory elements. Some of these elements, like the T-cell receptor α enhancer,55 the c-Fes negative regulator, and the adenosine deaminase enhancer, differ from the classical enhancer in that they are both site- and orientation-dependent for correct functioning. Other orientation-dependent transcription control elements include the interferon-β virus inducible enhancer56 and the c-fos negative regulator.57 These elements have a common mode of action that involves modifying the architecture of the surrounding chromatin to facilitate the functioning of the enhancer or repressor. Of note therefore is the fact that DNA-bound Sp1 can self-associate to bring together distant DNA segments that facilitate transcriptional enhancer function.58 It is possible therefore that the elements identified in Btk function in an analogous manner.
Studies done in both the human and the mouse suggest that expression of Btk is likely to be tightly regulated and complex. We have previously shown that intron 10 of Btk has a methylated site in pre-B cells that becomes specifically demethylated in B cells. This and the presence of several transcription regulator binding sites near the methylation site, including two E box motifs and a silencer element, imply that the intron is involved in gene regulation.59
An indication that timely expression of Btk is crucial for B-cell development comes from transgenic experiments done in the xidmouse. The xid defect was not corrected by a Btk transgene that was under the transcriptional control of an immunoglobulin promoter/enhancer.60 Although the possibility remains that this could have been caused by a dominant negative effect of the mutant Btk, the fact that the normal enzyme is expressed before immunoglobulin heavy and light chain gene rearrangement suggests that Btk may be required at the earliest stages of B-cell development to promote B-cell maturation. The presence of disease in our patient also indicates that the amount of Btk may be crucial.
The patient's mutation is unique with respect to previously reported mutations in Btk; although his cDNA sequence was normal he had lower amounts of mRNA and protein. Identification of the mutation at position +6 of the intron 1 donor site suggested that the observed phenotype could have resulted from a regulatory or splicing defect. The normal splice donor site of intron 1 closely resembles the splice donor consensus sequence (Shapiro Senapathy61 score of 85.9) and the alteration at position +6 did not significantly alter the score (81.4). In a study of 63 different donor site mutations only two were at the +6 position. In both cases, the wild type sequence corresponded poorly with the donor consensus sequence and the alterations compared even less favorably,45 making it unlikely that the T to G transversion in the patient was affecting the intron deletion reaction. A consistent drop in luciferase activity was noted when the patient mutation was substituted for the normal T at position +6 in the reporter constructs. However, the most striking difference, approximating the decrease in mRNA seen on the Northern blots, was noticed for the mutant pBtkpro+1029 containing the most intron 1 sequence. The much smaller drop in luciferase activity for mutant pBtkpro+6 suggested that the mutation was only having a negligible effect on the splicing reaction.
This study emphasizes that B-cell development will not progress appropriately unless sufficient Btk is provided at the earliest stages of B-cell development. We have identified regulatory elements within intron 1 that may account for the fine tuning of Btk expression. Examination of the intron 1 mutation shows the importance of patient mutation analyses not only with respect to structure/function studies of a protein, but also for the identification of sequences involved in the regulation of transcription.
Supported in part by grants from the National Institutes of Health (Grant No. AI 25129) and NCI CORE (Grant Nos. PO1 CA20180 and P30 CA21765); by funds from the Federal Express Chair of Excellence; by the American Lebanese and Syrian Associated Charities; and by the Assisi foundation.
Address reprint requests to Jurg Rohrer, PhD, Department of Immunology, St Jude Children's Research Hospital, 332 N Lauderdale, Memphis, TN 38101.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal