Abstract
Cyclin E is one of the G1 cyclins that play an important role in cell proliferation. Overexpression of cyclin E protein has been reported in several solid tumors, but little is known about the involvement of cyclin E in leukemia. In this study, we analyzed the expression of cyclin E gene product in 85 patients with acute myelogenous leukemia (AML) by Western blot analysis. In 23 of 85 AML samples (27%), cyclin E expression was enhanced in blasts. Among the French-American-British classification of AML, the ratio of the samples with enhanced cyclin E expression was high in M5 and low in M2 and M3. No rearrangements were observed by Southern blot analysis in these AML blasts with enhanced cyclin E expression. Flow cytometric analysis showed no correlation between overexpression of cyclin E and cell cycle distribution. Immunoblot analysis of cyclin D1 showed no correlation between overexpression of cyclin E and that of cyclin D1. Interestingly, p27 expression detected by Western blotting was apparently enhanced in 18 of 23 AML cells with enhanced cyclin E expression but none of 14 AML cells without enhanced cyclin E exhibited enhanced p27 expression. The rates of complete remission and of disease-free survival of the patients with M4 or M5 leukemia blasts with overexpressed cyclin E seemed to be low. Therefore, we suggest the necessity of a larger-scale study to elucidate the contribution of cyclin E overexpression to the phenotype and the prognosis of certain AML.
CYCLINS PLAY important roles in regulation of the cell cycle.1-6 Five classes of cyclins have been identified in mammalian cells.7,8 Cyclin E was discovered by screening human complementary DNA for rescue of a deficiency of G1 cyclin function in budding yeast.9,10 Synthesis of cyclin E gene product starts in mid-G1 phase, and its expression peaks at the G1-S transition, then decreases as cells proceed through S phase.1,2,4,9,11 Cyclin E associates with cdk2 and activates its serine-threonine kinase activity shortly before entry into S phase.1-6,12 Overexpression of cyclin E in fibroblasts shortened the duration of G1, decreased cell size, and diminished the serum requirement for the transition from G1 to S phase.13 Induction of cyclin E in asynchronous Rat-1 fibroblasts decreased the length of the G1 interval.14 Constitutively expressed cyclin E overcame retinoblastoma protein-mediated suppression of proliferation.15
Recently, several authors have shown overexpression of cyclin E in solid tumors.7,8,16-20 Leach et al17 reported amplification of the cyclin E gene and high levels of cyclin E protein in colorectal carcinoma cell line. Keyomarsi et al18 19 showed that altered expression of cyclin E protein occurred in most breast tumors, and the incidence of alterations increased with increasing grade and stage of the tumor. However, the expression of cyclin E has not been reported in acute myelogenous leukemia (AML). In this study, we analyzed the expression of cyclin E at the protein level in 85 AML samples.
MATERIALS AND METHODS
Cells. We used 85 bone marrow (BM) samples containing more than 90% leukemia blasts. Twenty-nine fresh heparinized leukemia samples were separated by Ficoll-Hypaque (Pharmacia Biotech, Uppsala, Sweden) and washed twice with phosphate-buffered saline (PBS), followed by extraction of whole cell lysates. The other 56 samples, previously frozen by a controlled-rate freezer to −80°C and stored in liquid nitrogen, were subjected to quick thawing and extraction of whole cell lysate. Viability was assessed by the Trypan-blue exclusion test. Every sample contained over 90% viable cells. The 85 AML specimens included 18 M1, 32 M2, 13 M3, 8 M4, and 14 M5 (French-American-British classification). Whole cell lysates were extracted as reported previously.21 Concisely, the cells were lysed in RIPA buffer (50 mmol/L Tris-HCl, pH 7.5, 0.3 mol/L NaCl, 1% Nonidet P-40, 0.1% sodium deoxycholate, 0.1% sodium lauryl sulfate [SDS], 1 mmol/L EDTA) containing 20 μg/mL aprotinin, 20 μg/mL leupeptin, 10 mmol/L sodium pyrophosphate, 50 mmol/L sodium fluoride, and 1 mmol/L sodium vanadate. After incubating for 1 hour at 4°C, lysates were centrifuged at 17,000g for 20 minutes. The protein concentration of the lysates was examined with a commercial kit (Bio-Rad Protein Assay; Bio-Rad Laboratories, Hercules, CA). Equal amounts of protein were used for Western blotting.
CD34+ cells were separated from BM specimen of volunteer donor by magnetic separation kit (MiniMACS Separation Columns; Miltenyi Biotec GmbH, Bergisch Gladbach, Germany). Purity of separated cells was measured by a flow cytometer (EPICS XL; Coulter Corp, Hialeah, FL). Normal monocytes were collected as follows. Mononuclear cells were separated by Ficoll-Hypaque and incubated for 1 hour at 37°C in CO2 incubator. After incubation, nonadherent cells were removed and adherent cells were collected by adding 0.2% EDTA. These cells were confirmed as monocytes by nonspecific esterase staining.22
Western blot analysis. Equal amounts of whole cell lysates (10 μg) were separated by electrophoresis through an 8% SDS polyacrylamide gel. Proteins were electroblotted onto polyvinylidene difluoride (PVDF) membranes and probed with monoclonal antibody to cyclin E and p27 (PharMingen, San Diego, CA), actin (Boehringer Mannheim, Mannheim, Germany), and cyclin D1 (Oncogene Science, Cambridge, MA). The quality and quantity of the lysates were confirmed by Coomassie blue staining of the gels. Immunoreactive proteins were detected using an antimouse secondary antibody conjugated to horseradish peroxidase followed by ECL (Amersham, Buckinghamshire, UK). The same sets of membranes were subjected to SuperSignal ULTRA Chemiluminescent Substrate (Pierce, Rockford, IL) followed by image analyzer (Molecular Imager System GS-525, Bio-Rad Laboratories) to calculate the relative intensity to the control.
Southern blot analysis. Ten micrograms of genomic DNAs were digested with either HindIII or Pst I, separated by electrophoresis through a 0.8% agarose gel, and transferred by capillary action to a nylon filter as described previously.23 The immobilized DNA was hybridized to a purified radiolabeled probe of cyclin E. The cyclin E cDNA was kindly provided by Dr M. Funk (Phillipps-Universität Marburg).
Flow cytometric analysis. Cryopreserved cells were thawed and washed quickly. The cell pellets were suspended and incubated with standard propidium iodide (PI) buffer (0.1% sodium citrate, 0.2% Nonidet P-40, 50 μg/mL PI) at 4°C for 1 hour. DNA content was measured on a flow cytometer, EPICS XL.
RESULTS
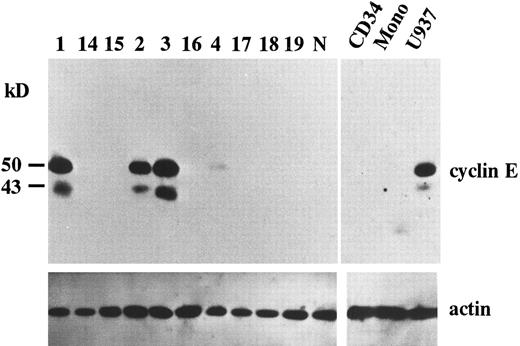
Overexpression of cyclin E in AML. To search for potential roles of cyclin E in human leukemia as an “oncogene,” we examined the expression of cyclin E in acute leukemia cells by immunoblotting. We found that two gene products were reactive with cyclin E antibody. One was the normal cyclin E protein of 50 kD, the other was a 43 kD splicing-variant of human cyclin E, which had lost its ability to complex with cdk2.24 Representative data are shown in Fig 1. Ten samples exhibited very strong bands (Fig 1, patients no. 1, 2, and 3), and 13 samples had faint bands (Fig 1, no. 4). No bands were detected in other samples, including samples derived from normal BM cells, even by a longer exposure after ECL procedure. In cultured human AML cell lines, HL60 and U937 exhibited strong bands. But other cell lines such as KG-1, NB-4, Kasumi-1, and K562 exhibited only faint bands (data not shown). The 43-kD product was observed in all samples with enhanced 50-kD–cyclin E. To measure the intensity of each band, we used an image analyzer (Molecular Imager) after the procedure of SuperSignal ULTRA following the manufacturer's recommendation. This detection system has linearity from an almost invisible signal to a very strong one. Each blot contained U937 sample as a positive control to calculate relative intensity. Intensity of normal BM cells and cyclin E− samples were almost similar to background. Faint bands exhibiting less than 10% intensity of U937 and strong bands exhibiting more than 20% of U937 were designated as intermediate and high expression, respectively. The rate of the samples with enhanced (intermediate and high) expression was significantly high in M4 and M5 blasts (chi square test, P = .002) and low in M2 and M3 blasts (P < .0001: Table 1). This result obtained from clincal samples seems to be consistent with the data from cell lines.
The expression of cyclin E gene products. Equal amounts of whole cell lysates (10 μg) from AML blasts were applied in each lane. Full-length cyclin E is defined as the band of approximately 50 kD. Lanes number 1 to 4 correspond to the patient number in Table 2. The other 6 samples (lanes number 14 to 19) represent cyclin E− AML blasts. “N” represents the lysate from mononuclear cells in normal BM. “CD34” and “Mono” represents the lysate from enriched CD34+ cells and monocytic cells separated from normal BM by the method as described in Materials and Methods. “U937” represents the lysate from U937 cells. The blots were reprobed with anti-actin antibody to show that each sample was equally loaded.
The expression of cyclin E gene products. Equal amounts of whole cell lysates (10 μg) from AML blasts were applied in each lane. Full-length cyclin E is defined as the band of approximately 50 kD. Lanes number 1 to 4 correspond to the patient number in Table 2. The other 6 samples (lanes number 14 to 19) represent cyclin E− AML blasts. “N” represents the lysate from mononuclear cells in normal BM. “CD34” and “Mono” represents the lysate from enriched CD34+ cells and monocytic cells separated from normal BM by the method as described in Materials and Methods. “U937” represents the lysate from U937 cells. The blots were reprobed with anti-actin antibody to show that each sample was equally loaded.
Summary of Patient Characteristics and Enhanced Expression of Cyclin E
| Phenotype . | No. of Samples . | Enhanced Expression of Cyclin E . | |
|---|---|---|---|
| . | . | No. . | % . |
| AML M1 | 18 | 8 (2)* | 44 |
| M2 | 32 | 3 | 9 |
| M3 | 13 | 0 | 0 |
| M4 | 8 | 3 (2) | 38 |
| M5 | 14 | 9 (6) | 64 |
| Total | 85 | 23 (10) | 27 |
| Phenotype . | No. of Samples . | Enhanced Expression of Cyclin E . | |
|---|---|---|---|
| . | . | No. . | % . |
| AML M1 | 18 | 8 (2)* | 44 |
| M2 | 32 | 3 | 9 |
| M3 | 13 | 0 | 0 |
| M4 | 8 | 3 (2) | 38 |
| M5 | 14 | 9 (6) | 64 |
| Total | 85 | 23 (10) | 27 |
Numbers in parentheses represent the number of samples with very high cyclin E expression.
Because the expression level of cyclin E seemed to be altered in a differentiation-specific manner, we analyzed the expression of cyclin E in enriched CD34+ cells as a normal counterpart of stem cell leukemia and in normal monocytes as a normal counterpart of monocytoid leukemia. Purity of CD34+ cells was 95%. Neither CD34+ cells nor monocytes exhibited any bands of cyclin E (Fig 1).
We next analyzed the expression of cyclin D1 at the protein level by Western blotting to find any possible correlation with the overexpression of cyclin E. In 28 samples (13 with positive cyclin E and 15 without), four samples exhibited the enhanced band of cyclin D1 protein while the other 24 samples showed basal level of cyclin D1 (data not shown). Statistic analysis (chi square test) indicated no significant correlation between the positivity of cyclin E and cyclin D1.
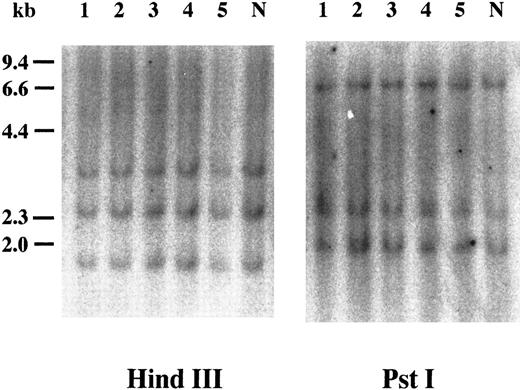
Southern blot analysis of cyclin E+ samples. To examine whether overexpression of cyclin E occurred by rearrangements or gross mutations of the gene, we performed Southern blot analysis. Genomic DNAs extracted from three highly positive samples (Fig 2, no. 1 to no. 3) and two intermediately positive samples (no. 4 and no. 5) as well as normal lymphocytes (lane “N”) were digested with HindIII and Pst I. Band patterns from normal lymphocytes were similar to those reported previously17 and were identical to those from leukemia samples.
Southern blot analysis of the cyclin E gene. Genomic DNAs were digested with either HindIII or Pst I, and the separated fragments were probed with human cyclin E cDNA. Each number over the lane corresponds to the patient number in Table 2. The applied amount of DNA of lane 5 in HindIII panel was small revealed by etidium bromide staining of the gel. “N” represents DNA from mononuclear cells in normal BM.
Southern blot analysis of the cyclin E gene. Genomic DNAs were digested with either HindIII or Pst I, and the separated fragments were probed with human cyclin E cDNA. Each number over the lane corresponds to the patient number in Table 2. The applied amount of DNA of lane 5 in HindIII panel was small revealed by etidium bromide staining of the gel. “N” represents DNA from mononuclear cells in normal BM.
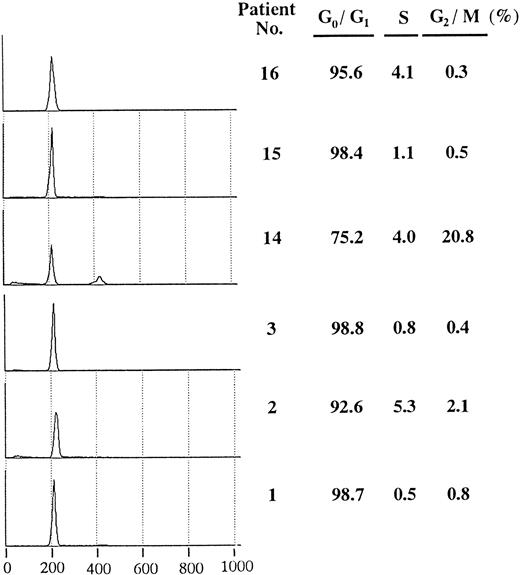
Flow cytometric analysis of positive samples. To assess cell cycle distribution in leukemia samples with overexpression of cyclin E, we examined DNA content of samples with positive or negative expression (Fig 3). Each sample had a high G0/G1 peak and a low percentage of S phase compared with cultured leukemia cell lines (data not shown). Except for case no.14, which showed a small G2/M peak, no significant differences in cell cycle distribution were observed among the leukemia samples.
Cell cycle distribution of AML blasts with or without overexpressed cyclin E. In each sample, a total of 10,000 cells were analyzed, and the half peak CV was between 1.5 and 2.6. Histograms show PI-stained cellular DNA. DNA-staining fluorescence intensity on a linear scale (X-axis) and cell number (Y-axis) are represented. The right hand table shows the percentage in G0/G1, S, and G2/M phase. Patients no. 1, no. 2, and no. 3 correspond to respective numbers in Table 2.
Cell cycle distribution of AML blasts with or without overexpressed cyclin E. In each sample, a total of 10,000 cells were analyzed, and the half peak CV was between 1.5 and 2.6. Histograms show PI-stained cellular DNA. DNA-staining fluorescence intensity on a linear scale (X-axis) and cell number (Y-axis) are represented. The right hand table shows the percentage in G0/G1, S, and G2/M phase. Patients no. 1, no. 2, and no. 3 correspond to respective numbers in Table 2.
Clinical features of patients with enhanced cyclin E expression. Clinical features of patients with positive cyclin E are presented in Table 2. Patients 1 and 43 had received BM transplantation during the first complete remission (CR), and were alive and disease-free. Other patients who are presented as “alive” were still under chemotherapy. The CR rate by intensive remission induction chemotherapy was 67% in cyclin E positive patients and 83% in negative patients (M3 patients were excluded because the principles and the protocol of therapy were completely different). Among M4 or M5 patients, while seven of eight (88%) patients with negative cyclin E achieved CR, only five of 10 (50%) patients with positive cyclin E achieved CR. However, we failed to find any statistical significance because of the small number of samples (chi square test: P = .09).
Characteristics of Cyclin E Positive AMLs
| Patient No. . | Age/Sex . | FAB . | Response to Induction Chemotherapy . | Remission Duration (mo) . | Current Status/Survival . | Cyclin E Expression . |
|---|---|---|---|---|---|---|
| 1 | 45/M | M4 | CR | 108 | Alive | High |
| 2 | 26/F | M5 | NR | 0 | Died of leukemia | High |
| 3 | 48/M | M5 | NE | 0 | Died of RF | High |
| 4 | 60/F | M5 | NE | 4 | Died of HF | Intermediate |
| 5 | 45/M | M1 | CR | 16 | Second relapse | Intermediate |
| 6 | 19/F | M1 | CR | 72 | Died of relapse | Intermediate |
| 7 | 46/M | M2 | NR | 0 | Died of leukemia | Intermediate |
| 36 | 69/M | M4 | NR | 0 | Died of leukemia | High |
| 37 | 21/F | M1 | NR | 0 | Died of leukemia | High |
| 38 | 47/M | M5 | CR | 5 | Died of relapse | High |
| 39 | 66/F | M5 | NR | 0 | Died of leukemia | High |
| 40 | 63/F | M5 | NR | 0 | Died of leukemia | High |
| 41 | 26/M | M5 | CR | 5 | Alive | High |
| 42 | 45/F | M1 | CR | 2 | Alive | High |
| 43 | 37/F | M1 | CR | 40 | Alive | Intermediate |
| 44 | 59/M | M4 | CR | 2 | Died of relapse | Intermediate |
| 45 | 48/M | M5 | NR | 0 | Alive | Intermediate |
| 46 | 54/F | M2 | CR | 3 | Alive | Intermediate |
| 47 | 53/M | M5 | NR | 0 | Alive | Intermediate |
| 48 | 32/M | M2 | CR | 10 | Alive | Intermediate |
| 49 | 48/M | M1 | CR | 6 | Died of relapse | Intermediate |
| 50 | 16/M | M1 | CR | 16 | Died of relapse | Intermediate |
| 51 | 43/F | M1 | CR | 1 | Alive | Intermediate |
| Patient No. . | Age/Sex . | FAB . | Response to Induction Chemotherapy . | Remission Duration (mo) . | Current Status/Survival . | Cyclin E Expression . |
|---|---|---|---|---|---|---|
| 1 | 45/M | M4 | CR | 108 | Alive | High |
| 2 | 26/F | M5 | NR | 0 | Died of leukemia | High |
| 3 | 48/M | M5 | NE | 0 | Died of RF | High |
| 4 | 60/F | M5 | NE | 4 | Died of HF | Intermediate |
| 5 | 45/M | M1 | CR | 16 | Second relapse | Intermediate |
| 6 | 19/F | M1 | CR | 72 | Died of relapse | Intermediate |
| 7 | 46/M | M2 | NR | 0 | Died of leukemia | Intermediate |
| 36 | 69/M | M4 | NR | 0 | Died of leukemia | High |
| 37 | 21/F | M1 | NR | 0 | Died of leukemia | High |
| 38 | 47/M | M5 | CR | 5 | Died of relapse | High |
| 39 | 66/F | M5 | NR | 0 | Died of leukemia | High |
| 40 | 63/F | M5 | NR | 0 | Died of leukemia | High |
| 41 | 26/M | M5 | CR | 5 | Alive | High |
| 42 | 45/F | M1 | CR | 2 | Alive | High |
| 43 | 37/F | M1 | CR | 40 | Alive | Intermediate |
| 44 | 59/M | M4 | CR | 2 | Died of relapse | Intermediate |
| 45 | 48/M | M5 | NR | 0 | Alive | Intermediate |
| 46 | 54/F | M2 | CR | 3 | Alive | Intermediate |
| 47 | 53/M | M5 | NR | 0 | Alive | Intermediate |
| 48 | 32/M | M2 | CR | 10 | Alive | Intermediate |
| 49 | 48/M | M1 | CR | 6 | Died of relapse | Intermediate |
| 50 | 16/M | M1 | CR | 16 | Died of relapse | Intermediate |
| 51 | 43/F | M1 | CR | 1 | Alive | Intermediate |
Abbreviations: CR, complete remission; NR, no response; NE, not evaluable; RF, respiratory failure; HF, heart failure.
Overall Kaplan-Meier curve for event-free survival from positive-cyclin E patients is always under that from negative-cyclin E patients (Fig 4). The event-free survival rate obtained by the Cox-Mantel test showed a statistically significant difference in survival between positive-cyclin E patients and negative-cyclin E patients (P = .015).
Kaplan-Meier curves for event-free survival from positive-cyclin E patients and negative-cyclin E patients. M3 patients were excluded from this analysis. Patients who underwent BM transplantation were censored at the date of it. P = .208 by g-Wilcoxon test. P = .015 by Cox-Mantel test.
Kaplan-Meier curves for event-free survival from positive-cyclin E patients and negative-cyclin E patients. M3 patients were excluded from this analysis. Patients who underwent BM transplantation were censored at the date of it. P = .208 by g-Wilcoxon test. P = .015 by Cox-Mantel test.
Correlation between enhanced expression of cyclin E and p27kip1. p27 is a member of cyclin/cdk inhibitors and inhibits kinase activity of several cyclin/cdk complexes including cyclin D/cdk4 and cyclin E/cdk2. It was reported that high levels of cyclin E and low levels of p27 were strongly predictive of increased mortality in breast cancer patients.25 We examined p27 expression in all cyclin E+ samples and 14 cyclin E− samples by Western blotting. Eighteen of 23 (78%) samples with enhanced cyclin E exhibited the bands corresponding to p27 protein, while none of the samples without enhanced cyclin E nor normal BM cells exhibited any detectable bands (Fig 5). There was a strong correlation between enhanced expression of cyclin E and enhanced expression of p27 (chi square test, P < .001). The disease-free survival rate of 18 patients with positive p27 (31% at 2 year) was worse than that of 19 patients without enhanced p27 (65%), but no statistical significance was obtained.
The expression of p27 gene product. Western blotting probed by anti-p27 antibody was performed. Equal amounts of whole cell lysates (10 μg) from AML samples with enhanced cyclin E expression (upper panel) and those without enhanced cyclin E (lower panel) were analyzed. Each number over the lane corresponds to the patient number in Table 2. “N” and “U937” represent the lysates from mononuclear cells in normal BM and U937 cells.
The expression of p27 gene product. Western blotting probed by anti-p27 antibody was performed. Equal amounts of whole cell lysates (10 μg) from AML samples with enhanced cyclin E expression (upper panel) and those without enhanced cyclin E (lower panel) were analyzed. Each number over the lane corresponds to the patient number in Table 2. “N” and “U937” represent the lysates from mononuclear cells in normal BM and U937 cells.
DISCUSSION
In this study, we found that a proportion of AML blasts overexpressed cyclin E. Normal BM samples had no visible band in our assay. Furthermore, cultured AML cell lines at exponential growth, which should have a higher percentage of cells in mid-G1 and early S phase, did not exhibit elevated cyclin E expression compared to positive AML blasts. We failed to show any difference in the duration of G1 and S phase between cyclin E-overexpressed samples and -unexpressed samples. We speculate that this overexpression of cyclin E may not simply be regulated by ordinary cell cycle events but may be implicated in the pathology of leukemia. A link between oncogenesis and cyclins has been made with the discovery of inappropriate expression of cyclins in tumors.17,26,27 Cyclin D1, a member of the G1 cyclin family, has been reported to be involved in chromosomal rearrangement, and the resulting amplification of cyclin D1 plays a crucial role in a variety of tumors including lymphoma28-30 and breast cancer.18 Overexpression of cyclin D2 has been reported in chronic B-cell malignancies31 and colorectal carcinoma.17 Although these observations emphasize the importance of cyclins in tumorigenesis, the question remains as to how the overexpressed cyclin E may be responsible for the onset or development of primary AML.
With respect to the mechanism of overexpression, few studies conducted have been successful in primary cancer cells, except in the case of amplification of cyclin D1 gene.7,8,28 Although no gross gene rearrangements, mutations or deleted fragments of cyclin E gene were observed in our study, it is still possible that small mutations of cyclin E gene itself are responsible for this overexpression. Recently, the promotor of human cyclin E gene was reported and E2F was found to be one of the transcription factors binding to the promotor.32,33 Cyclin E protein has been reported to be degraded by ubiquitin-proteasome machinery.34 35 Northern blot analysis might be helpful to see whether the overexpression occurs at the transcriptional level or posttranscriptional level in these cyclin E-enhanced AML blasts.
We found that M5 blasts frequently exhibited enhanced cyclin E expression but that M2 and M3 blasts rarely did. Interestingly, in human cell lines HL60 and U937, which are capable of differentiating towards monocytes by the addition of phorbol ester, exhibited enhanced cyclin E expression,36 although the reason still remains unclear. The enhanced cyclin E expression seemed to be highly correlated with a monocytic leukemia. The expression of cyclin E in CD34+ normal marrow cells and normal monocytes which are thought to be at the same stage of differentiation as CD34+ leukemia blasts and monocytic leukemia blasts, respectively, remained basal level. This may support our suggestion that enhanced cyclin E expression does not simply reflect the proliferation and differentiation status. That might be closely associated with leukemogenesis.
In breast cancer, cyclin E protein was highly overexpressed at advanced stages; thus, it may be used as a prognostic marker.18,19 Very recently, high cyclin E expression and low p27 expression were reported to be worse prognostic markers for breast cancer.25 In acute lymphoblastic leukemias, cyclin E level was higher in relapsed samples than in samples at initial diagnosis.30 In the present study, CR rates of cyclin E+ samples were worse than E− samples. Among M4 or M5 patients, the rates of CR of patients with enhanced cyclin E expression were lower than in patients without it. Event-free survival was longer in the cyclin E-negative group rather than in the cyclin E+ group, and only two patients in the cyclin E+ group survived without relapse over 2 years except for those who received BM transplantation. According to p27 expression, our data show that enhanced expression of p27 was highly correlated with enhanced cyclin E expression in contrast to breast cancer case25: no correlation was observed between the level of cyclin E expression and that of p27 in breast cancer. p27 has been regarded as a negative regulator of cyclin E/cdk2; overexpression of cyclin E may cause the augmentation of p27 by feedback mechanism. The reason for the discrepancy between AML and breast cancer remains to be elucidated. We failed to show statistical significance of high p27 expression as a poor prognostic marker (Cox-Mantel test, P = .07) possibly due to a small number of the sample analyzed. Although multiple factors including white blood cell count at diagnosis and age have been reported to contribute to the prognosis of AML patients, the prognosis of patients with enhanced cyclin E expression seems to be poor. The existence of overexpression of cyclin E may be helpful to predict patient outcome. A larger scale study with multivariate analyses is required to evaluate whether cyclin E overexpression has value in phenotyping and prognosis of AML.
ACKNOWLEDGMENT
The authors thank S. Suzuki, M. Suzuki, and C. Wakamatsu for their technical assistance. We are also grateful to Y. Yamakawa for his assistance of flow cytometry.
Address reprint requests to Masayuki Towatari, MD, PhD, First Department of Internal Medicine, Nagoya University School of Medicine, Tsurumai-cho 65, Showa-ku, Nagoya 466, Japan.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal