Abstract
Members of the large Eph family of receptor tyrosine kinases (RTKs) display temporally and spatially restricted expression patterns during embryogenesis, suggesting a role in various developmental processes. We have begun to investigate the expression of members of this receptor family during human hematopoiesis, in particular B lymphopoiesis. Expression of Eph RTKs in cells of the B-lymphoid lineage was assessed by using degenerate oligonucleotide primers based on stretches of conserved nucleic acid sequences in members of the Eph family. First, the content of Eph-family RTKs was assessed in freshly sorted fetal bone marrow pro–B cells. This population was found to harbor transcripts of the Hek8 and Hek11 members of this gene family. Subsequent analysis of expression of these genes in B cells representing various differentiation and ontogenic stages showed that the Hek8 transcript is constitutively present in all fetal and adult B-lineage cells, with high levels of expression in peripheral blood B cells. In contrast, the Hek11 transcript was exclusively found in fetal bone marrow pro–B cells and pre–B cells, but not in more mature fetal B-lineage cells. All adult B-lineage cells, from early pro–B cells to end-stage plasma cells, lacked Hek11 transcripts. The developmentally regulated expression of Hek11 during fetal B lymphopoiesis suggests a role for this gene in pre/pro–B cell expansion and/or differentiation and defines a difference in progenitor B cell populations isolated from fetal versus adult human bone marrow.
IN THE BONE MARROW (BM) of vertebrates, cellular interactions and soluble or cell-associated growth factors promote the expansion and differentiation of pluripotent stem cells into a diversity of hematopoietic cell types, including B-lineage cells. Here, discrete stages of B lymphopoiesis can be discerned by the ordered loss and acquisition of cell surface markers, the expression of transcription factors, the state of rearrangement and expression of Ig genes, and the susceptibility to cytokines that contribute to the genesis of mature surface(s) Ig+ B lymphocytes.1-4 The analysis of the complexity of these developmental events has been significantly facilitated by the establishment of short-term and long-term stromal cell–dependent cultures of B-lymphoid cells.5-15 In humans, various stages of early B lymphopoiesis have been recapitulated using both human and murine stromal cells.9,10 Although a number of growth and differentiation factors have been identified that partake in this process, additional as yet unknown factors are expected to play a role.7,9 10
Receptor tyrosine kinases (RTKs) are type 1 membrane-bound proteins composed of a glycosylated extracellular region, a hydrophobic transmembrane region, and a cytoplasmic region that catalyzes the phosphorylation of tyrosine residues following ligand binding. RTKs and their ligands play a key role in many biological processes including cellular differentiation and proliferation, embryonic morphogenesis, and neoplastic growth.16 On the basis of predicted structural homologies, sequence conservation, and similarity of ligands, RTKs have been assigned to several subclasses. One of these, the Eph-related family of RTKs, constitutes the largest known family of RTKs, comprised of at least 13 distinct members highly conserved between different species including man, mouse, rat, chicken, and xenopus.17-22 Members of the human Eph family of RTKs are often denoted human Eph-related kinases (Hek).
Recently, seven distinct membrane-bound ligands binding to Eph-related receptors have been identified, denoted ligands for Eph-related kinases (LERKs) or Eph-family ligands (EFL's).23-26 Members of the LERK family share a number of characteristics including nucleotide sequence homology and promiscuous binding to different Eph-related receptors in vitro. The functional role of Eph RTKs and their cognate ligands is beginning to be elucidated. Recent evidence indicates a role in angiogenesis27 and in the formation of neuronal pathways.28-30 To date, Eph-related RTKs and LERKs have been sporadically found in cells of the human hematopoietic system. The RTK Hek (also denoted Hek4) was originally cloned from a pre–B cell leukemia, and expression was subsequently detected in two leukemic T cell lines but not in wide range of other cell lines of lymphoid or myeloid lineage or normal (BM) cells.31 The RTK Htk is expressed in a wide variety of tissues, including CD34+ hematopoietic progenitor cells from bone marrow and cell lines of the myeloid but not lymphoid lineage.20 A ligand for the Htk receptor has been isolated and displays a broad expression pattern, including a bone marrow stroma cell line.32 The Eph-related RTK Eck has been localized in the thymus by in situ hybridization.33 In addition, Hek5 and Esk expression have been detected in rat thymus by Northern analysis.17 34
We have begun a systematic search for expression of Eph-related RTKs in cells of the hematopoietic system, in particular B-lineage cells. By degenerate reverse transcription polymerase chain reaction (RT-PCR), we have identified three members of the Eph RTK family expressed in freshly isolated pro–B cells from human fetal BM. The expression pattern of two of these, Hek8 and Hek11, was assessed during B lymphopoiesis in fetal and adult BM, in adult peripheral B-lineage cells, in malignant early B-lineage cells, and in fetal tissues. The data suggest that Hek8 is ubiquitously expressed during human fetal B-cell differentiation, mirroring the broad tissue distribution of these transcripts. In constrast, Hek11 expression is restricted to early fetal B-cell differentiation stages and, in addition, distinguishes fetal from adult BM early B-cell differentiation.
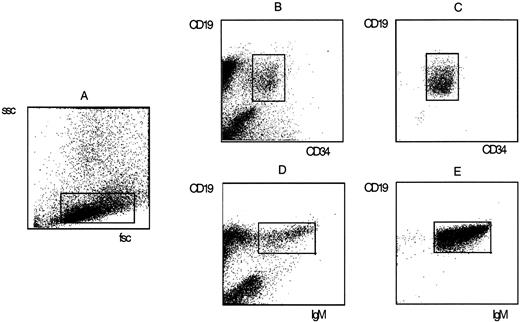
Staining of fetal BM mononuclear cells with fluorochrome-labeled monoclonal antibodies and sort criteria and purity of pro–B cells and sIgM+ B cells. Pro–B cells were defined by forward scatter (fsc) and sidescatter (ssc) properties (A) and the simultaneous expression of CD34 and CD19 (B). Mature B cells were defined by fsc and ssc properties (A) and the coexpression of CD19 and surface IgM (D). The purity of sorted pro–B (C) and B (E) populations was determined by rerun analysis on a FACSscan.
Staining of fetal BM mononuclear cells with fluorochrome-labeled monoclonal antibodies and sort criteria and purity of pro–B cells and sIgM+ B cells. Pro–B cells were defined by forward scatter (fsc) and sidescatter (ssc) properties (A) and the simultaneous expression of CD34 and CD19 (B). Mature B cells were defined by fsc and ssc properties (A) and the coexpression of CD19 and surface IgM (D). The purity of sorted pro–B (C) and B (E) populations was determined by rerun analysis on a FACSscan.
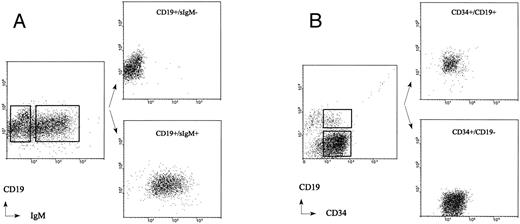
Sort criteria and cell purity analysis of adult BM CD19+/sIgM+ B cells and CD19+/sIgM− pre–B cells obtained from the immunomagnetically separated CD34− fraction (A) and CD34+/CD19+ pro–B cells and CD34+/CD19− non–B-lineage progenitor cells obtained from the immunomagnetically separated CD34+ fraction (B).
Sort criteria and cell purity analysis of adult BM CD19+/sIgM+ B cells and CD19+/sIgM− pre–B cells obtained from the immunomagnetically separated CD34− fraction (A) and CD34+/CD19+ pro–B cells and CD34+/CD19− non–B-lineage progenitor cells obtained from the immunomagnetically separated CD34+ fraction (B).
MATERIALS AND METHODS
Tissues and cell lines. Fetal tissues were obtained from aborted human fetuses (16 to 20 weeks of gestation) according to the guidelines set forth by the University Hospital Utrecht Committee on the use of Human Subjects in Scientific Research. BM cells were obtained by flushing intramedullary cavities of the femural and humoral bones with RPMI 1640 medium containing 5% fetal calf serum (FCS; GIBCO, Gaithersburg, MD). The cell suspension was washed and separated by Ficoll-Paque density gradient centrifugation. Other fetal tissues were directly snap-frozen in liquid nitrogen and processed for RNA extraction. Adult BM cells were obtained by iliac crest aspiration from volunteers with informed consent and the approval of the Ethics Committee of The Norwegian Radium Hospital. Tonsils were obtained from children undergoing routine tonsillectomy and minced, and mononuclear cells (MNC) were purified through Ficoll-Paque density gradient centrifugation. Tonsillar MNC were depleted of T cells using 2-aminoethyl-isothiouronium-bromide–treated sheep red blood cells as described.35 The non–T cell fraction was subjected to a second round of T-cell depletion using the same procedure.
Oligoprimers Used for PCR Amplification
| Primer . | Nucleotide Sequence . |
|---|---|
| MGQF | 5′-CGG ATC CAT GGG NCA STT Y |
| WT(A/S)PE | 5′-CGG ATC CGC YTC NGG NGM NGT CCA |
| Actin-5′ | 5′-GGC GAC GAG GCC CAG AGC AAG AGA G |
| Actin-3′ | 5′-TGC CCA GGA AGG AAG GCT GGA AGA G |
| HSRPTK-5′ | 5′-CAA AGC TGA CCA AGA AGG CG |
| HPTK275′ | 5′-GGG ATA TGT TCA CAG GGA CCT TGC |
| HPTK273′ | 5′-TTT CCG GTA CTG GAT GGC TTC G |
| HEK113′ | 5′-TCA TCC TGG CTA CTG ATT CAA GGG |
| HEK1125′ | 5′-GCG GAT CCA AGC TAA ACG TGG AGC AGC CG |
| HEK11LO | 5′-CGA ATT CTC TAG TGT AGC AAC ATC AAG TCT GG |
| HEK8a | 5′-GTG ATT AAA GCC ATT GAG GAA G |
| HEK8b | 5′-ACA AAC TCA TCC GCA ACC C |
| HEK8c | 5′-GCT GTG ATA CCA ATT CTT GCC |
| TdTF2 | 5′-GAC TAT TCA GAT AGC ACC AAC |
| TdTrev | 5′-CTC CAT CTT CAA TAA TCT CC |
| Primer . | Nucleotide Sequence . |
|---|---|
| MGQF | 5′-CGG ATC CAT GGG NCA STT Y |
| WT(A/S)PE | 5′-CGG ATC CGC YTC NGG NGM NGT CCA |
| Actin-5′ | 5′-GGC GAC GAG GCC CAG AGC AAG AGA G |
| Actin-3′ | 5′-TGC CCA GGA AGG AAG GCT GGA AGA G |
| HSRPTK-5′ | 5′-CAA AGC TGA CCA AGA AGG CG |
| HPTK275′ | 5′-GGG ATA TGT TCA CAG GGA CCT TGC |
| HPTK273′ | 5′-TTT CCG GTA CTG GAT GGC TTC G |
| HEK113′ | 5′-TCA TCC TGG CTA CTG ATT CAA GGG |
| HEK1125′ | 5′-GCG GAT CCA AGC TAA ACG TGG AGC AGC CG |
| HEK11LO | 5′-CGA ATT CTC TAG TGT AGC AAC ATC AAG TCT GG |
| HEK8a | 5′-GTG ATT AAA GCC ATT GAG GAA G |
| HEK8b | 5′-ACA AAC TCA TCC GCA ACC C |
| HEK8c | 5′-GCT GTG ATA CCA ATT CTT GCC |
| TdTF2 | 5′-GAC TAT TCA GAT AGC ACC AAC |
| TdTrev | 5′-CTC CAT CTT CAA TAA TCT CC |
Short-term cultures of human fetal BM stromal cells were established by allowing 1 × 107 BM cells to adhere for 2 hours at 37°C to the plastic of an 80 cm2 tissue culture flask in 20 mL RPMI 1640 medium containing 5% FCS and 5% horse serum. Nonadherent cells were removed by washing with cold PBS/1% FCS, and the adherent cells were cultured for 3 to 5 days in 20 mL RPMI containing 5% FCS, 5% horse serum, penicilline, and streptomycine.
The following human cell lines were used in this study: the pro–B cell lines TOM-136 and BV17337; the pre–B cell lines Reh (ATCC CRL-8286) and Nalm 638; the mature B cell lines Bjab (kind gift from Dr G. Moldenhauer) and Daudi (ATCC CCL-213); the plasmacytoid cell lines U266 (ATCC TIB 196), ARH-77 (CRL-1621), and IM9 (CCL-159); the T cell lines CEM (CCL-119), Jurkat (ATCC TIB-152), and H9 (HTB-176); the myeloid cell lines KG1-A (ATCC CCL-246), HL-60 (ATCC CCL-240), and U937 (ATCC CRL-1593); the erythroid precursor cell line K562 (ATCC CCL-243); the cervical carcinoma cell line HeLa (ATCC CCL-2); and the hepatoma cell line HepG2 (ATCC HB-8065). All cell lines were grown in RPMI 1640 medium supplemented with 5% FCS at 37°C in a humidified atmosphere with 5% CO2 .
Flow cytometry and cell sorting. The criteria used for identification and flow-cytometric separation of human B cell subpopulations in fetal BM have been described.39 40 Fetal BM MNC were stained with combinations of fluoresceine isothiocyanate (FITC) or allophycocyanine (APC)-labeled CD19, phycoerythrin (PE)-labeled CD34 monoclonal antibodies (Becton Dickinson, San Jose, CA), and a FITC-labeled goat anti-human IgM polyclonal antibody (Southern Biotechnology Associates, Birmingham, AL). The CD19+/CD34+ pro–B cells, the CD19+/CD34−/IgM-pre–B cells, and the CD19+/IgM+ B cells were sorted. The pro–B cell and B cell fractions were used for fluorescent-activated cell sorter (FACS) rerun analysis to assess the purity of sorted cells (Fig 1). Both fractions contained more than 99% pro–B and B cells, respectively.
For positive selection of adult BM CD34+ cells, BM MNC were rosetted with Dynabeads M-450 (Dynal, Oslo, Norway) directly coated with the CD34 monoclonal antibody 561 for 45 minutes at 4°C on an apparatus that provided tilting and gentle rotation. The bead to total cell ratio was 1:1. Rosetted cells were attracted to a samarium cobalt magnet and nonrosetting cells were removed by pipetting. Rosetted cells were washed four times. Detachment of beads from positively selected cells was performed by incubation with anti-Fab antiserum (DETACHaBEAD; Dynal) at a concentration of 35 μg/mL for 1 hour at room temperature. Isolated cells, free of beads, were washed and counted. The purity of CD34+ cells isolated by this method was reproducibly greater than 95% as determined by flow cytometric analysis (not shown).
The immunomagnetically separated CD34+ cells were used to obtain CD34+/CD19− non–B-lineage progenitor cells, CD34+/CD19+ pro–B cells, and CD19+/IgM− pre–B cells by double-staining with FITC-conjugated anti-CD34 (MCA, Groningen, The Netherlands) and PE-conjugated CD19 (Becton Dickinson) monoclonal antibodies or double-staining with PE-conjugated CD19 (HD37, DAKO, Denmark) and FITC-conjugated anti-IgM antibodies (DAKO, Denmark). To obtain mature adult BM B cells, the CD34− fraction of the immunomagnetic separation was stained with PE-conjugated anti-CD19 and FITC-conjugated anti-IgM antibodies. Flow cytometric cell sorting was performed to isolate various subpopulations of the isolated CD34+ cells and CD19+ BM cells (Fig 2). The purity of sorted adult BM pro–B-, pre–B-, and non–B-lineage progenitor cells was analyzed by FACS rerun analysis and found to be more than 99%. Representative results from experiments with three adult and five fetal BM samples are shown in Figs 1 and 2.
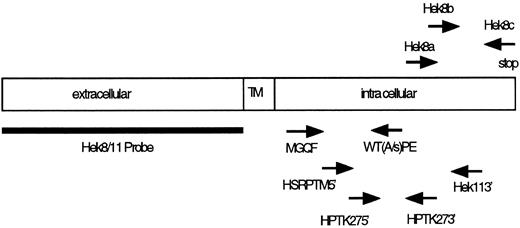
Location of PCR primers and probes used for cloning and detection of Eph-related RTKs. TM, transmembrane region.
Location of PCR primers and probes used for cloning and detection of Eph-related RTKs. TM, transmembrane region.
Plasma cells were sorted from the BM of patients with multiple myeloma based on high levels of CD38 expression and forward and side scatter properties as described in detail elsewhere.41 Immunostained cells were sorted using a FACSVantage flow cytometer (Becton Dickinson) equipped with an automated cell deposition unit. FITC and PE fluorescence was excited by a 488-nm argon ion laser and APC fluorescence by a 90-mW spectrum laser tuned at 647 nm. Doublets and cell aggregates were eliminated using a pulse processor module on the forward light scatter signal. In some experiments cell sorting was performed on a FACStar Plus (Becton Dickinson) equipped with an argon laser.
RNA isolation, Northern blot analysis, and first-strand cDNA synthesis. Total RNA was extracted from cells or tissues by the guanidine thiocyanate method essentially as described.42 Ten micrograms of total RNA from different cell types and tissues was size fractionated on a 1%–agarose formaldehyde denaturating gel, transferred to nitrocellulose membranes (Schleicher and Schull, Optitran), and crosslinked by baking for 2 hours at 80°C. Alternatively, RNA was isolated as described and spotted directly on nitrocellulose filter (dot blot) and crosslinked by baking for 2 hours at 80°C. Prehybridization (1 hour) and hybridization were performed in hybridization buffer (5× sodium chloride/sodium phosphate/EDTA buffer [SSPE], 10% dextran sulphate, 0.1% sodium dodecyl sulfate [SDS], 50% formamide, 100 μg/mL sheared salmon sperm DNA) at 42°C, and the membranes were hybridized overnight with a 32-P(dCTP)–labeled cDNA probe representing the extracellular part of Hek11 (see below) or with a control actin probe. After hybridization, the membranes were washed under high stringency in 0.2 sodium chloride/sodium citrate buffer [SSC]/0.1% SDS at 65°C. PolyA+ RNA was isolated from cells or tissues using oligo-dT Dynabeads (Dynal) as previous described.43 Briefly, 0.5 to 2 × 105 sorted cells were lysed in 100 μL phosphate-buffered saline (PBS)/1% NP-40 at 4°C for 2 minutes on ice before spinning down the nuclei at 10,000g for 2 minutes. The supernatant was added to a solution of 125 μg Dynabeads oligo(dT)25 solved in 100 μL binding buffer and incubated on a rotating wheel at 4°C for 5 minutes. The beads were subsequently attracted to a magnet (MPC-E, Dynal) and washed twice with 200 μL wash buffer 1 (10 mmol/L Tris-HCl pH 7.5, 0.15 mol/L LiCl, 1 mmol/L EDTA, 0.1% SDS) followed by two washes in buffer 2 (10 mmol/L Tris pH 7.5, 0.15 mol/L NaCl) including a change in microcentrifuge tubes between the last two washing steps.
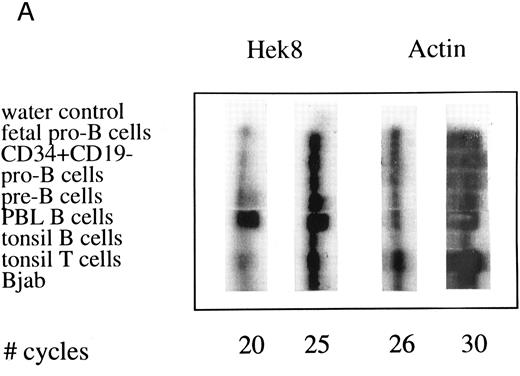
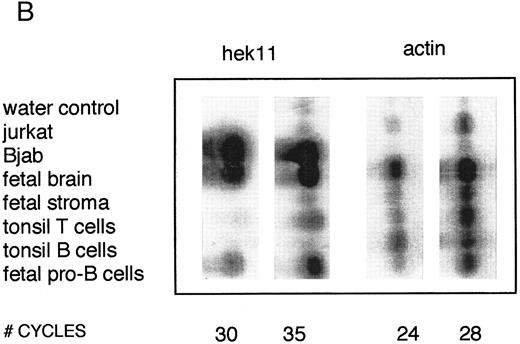
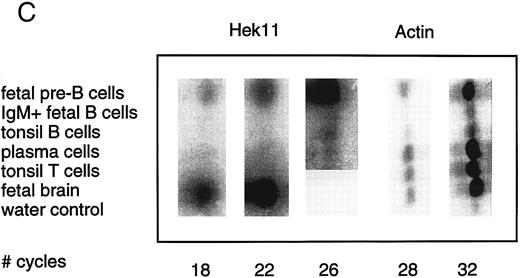
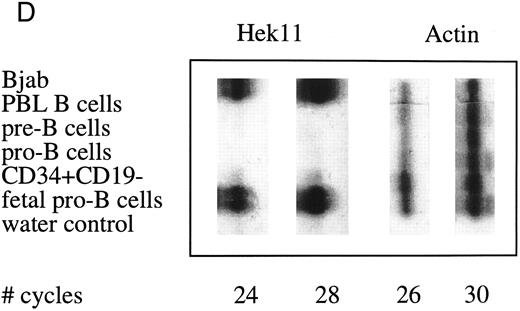
Semiquantative PCR analysis of levels of Hek8, Hek11, and β-actin expression in purified lymphoid cell subpopulations. Positive controls consisted of PCR reactions on cDNA from fetal brain and the Bjab cell line, both containing high levels of Hek11 transcripts (see figs 7 and 8). Negative controls contained all components of the PCR reaction except for the target cDNA (water control). At the indicated cycle number, 5-μL aliquots were removed from the PCR reaction mixture, separated on an agarose gel, and processed for hybridization with the Hek8, Hek11, or actin probe.
Semiquantative PCR analysis of levels of Hek8, Hek11, and β-actin expression in purified lymphoid cell subpopulations. Positive controls consisted of PCR reactions on cDNA from fetal brain and the Bjab cell line, both containing high levels of Hek11 transcripts (see figs 7 and 8). Negative controls contained all components of the PCR reaction except for the target cDNA (water control). At the indicated cycle number, 5-μL aliquots were removed from the PCR reaction mixture, separated on an agarose gel, and processed for hybridization with the Hek8, Hek11, or actin probe.
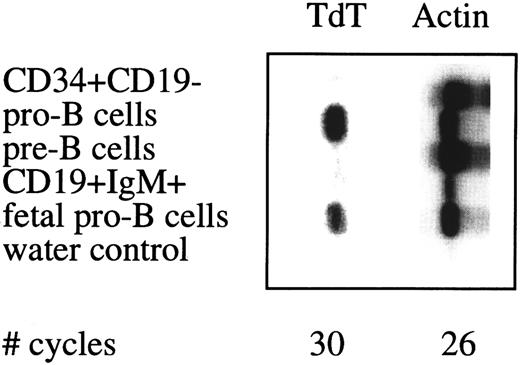
Semiquantative PCR analysis of TdT expression in sorted cell populations. See legend to Fig 4.
Semiquantative PCR analysis of TdT expression in sorted cell populations. See legend to Fig 4.
First-strand cDNA was synthesized directly on mRNA bound to Dynabeads oligo(dT). Briefly, a 25-μL cDNA synthesis mix was added to mRNA bound to beads, 5 μL 5× concentrated first-strand cDNA buffer (Boehringer Mannheim, Germany), 2.5 μL 10 mmol/L dNTP, 1 μL RNAguard (Pharmacia, Uppsala, Sweden), 12.5 U reverse transcriptase (Boehringer Mannheim), and incubated on a rotating wheel for 1 hour at 42°C. After 1 hour, 1.5 μL 10 mmol/L MnCl2 and 2.5 U rTth polymerase (Perkin Elmer) were added to the reaction mix and incubated for 10 minutes at 68°C on a rotating wheel. Finally the beads were washed twice in 100 μL Tris/EDTA (TE) buffer, resuspended in 25 μL TE, and stored at −20°C.
Identification of Eph-related RTK. To identify Eph-related RTKs from sorted B-cell populations, we designed a set of degenerate oligonucleotide primers specific for the conserved amino acid motifs MGQF (sense) and WT(A/S)PE (antisense; Table 1 and Fig 3) localized in the intracellular kinase domain of Eph-related kinases. Typically, PCR amplification was performed on immobilized cDNA obtained from 104 sorted cells using 0.75 μg of each primer and 50 cycles of 1 minute at 94°C, 2 minutes at 37°C, and 3 minutes at 63°C. Amplified products with the expected size (approximately 370 bp) were isolated from agarose gels, ligated into T-vector (Promega, Madison, WI), and used for dideoxy sequencing using the sequenase system (Stratagene, La Jolla, CA).
Semiquantitative PCR analysis of Hek8 and Hek11 expression. cDNA generated from approximately 2 × 103 or 2 × 104 cells was used in semiquantitative PCR assays employing a set of four nested oligonucleotides primers specific for the Hek11 RTK or, as a control, a pair of two actin oligonucleotide primers (Table 1 and Fig 3). The amount of cDNA generated from different cell populations was first normalized using the actin PCR and adjusted to the amount of cDNA obtained from 2 × 103 freshly isolated fetal BM pro–B cells. To that end, first-strand cDNA from 2 × 103 cells sorted fetal BM pro–B cells was amplified (2 minutes preincubation at 94°C followed by 30 cycles of 40 seconds at 94°C, 40 seconds at 56°C, and 40 seconds at 72°C) and 5-μL aliquots of the PCR reactions were removed at the various cycle intervals. The intensity of actin fragments in ethidium-stained agarose gels from the linear phase of the PCR reaction was used as a measure of amounts of cDNA present in the different samples. Approximately equal amounts of cDNA were subsequently used in the Hek11-specific PCR.
The Hek11 PCR was performed using five times the amount of cDNA used as starting material for the actin PCR. The first PCR was performed with the primers HSRPTK5′ and Hek113′, which bind to region 2367-2391 and 3037-3061, respectively17 (Table 1 and Fig 3). The PCR conditions were as follows: 2 minutes 94°C then 35 cycles of 40 seconds 94°C, 40 seconds 56°C, and 40 seconds 72°C. The second nested PCR was performed with the primer set HPTK27-5′ and HPTK27-3′, which bind to region 2441-2464 and region 2600-2621, respectively.17 A 1-μL aliquot from the first PCR was amplified in the second PCR reaction. The PCR conditions were as follows: 2 minutes preincubation at 94°C followed by 35 cycles of 30 seconds at 94°C, 30 seconds at 56°C, and 30 seconds at 72°C. Five microliters aliquot of the PCR reaction mixtures were removed during amplification reactions at indicated cycle numbers and seperated on 2% agarose gels for analysis. The agarose gel were then subjected to Southern blotting.
The Hek8 PCR was performed essentially as described for the Hek11 PCR. The first PCR was performed with the primers Hek8a and Hek8c, which bind to region 2530-2551 and 2886-2906, respectively17 (Table 1 and Fig 3). A 1-μL aliquot from the first PCR was amplified in the second PCR with the primer Hek8b (region 2669-2687) and Hek8c. The first PCR consisted of 30 cycles, and the conditions were the same as described for the Hek11 PCR. Five microliters aliquot of the PCR reaction mixtures were removed during amplification reactions at indicated cycle numbers and seperated on 2% agarose gels for analysis. The agarose gels were then subjected to Southern blotting.
Analysis of TdT expression in sorted B cells. TdT gene expression was determined by converting RNA from 5,000 freshly sorted B- and non–B-lineage cells to first-strand cDNA followed by a PCR reaction using the primer set TdTF2 and TdTrev (Table 1), yielding a fragment of 309 bp. PCR-amplified material was run on an agarose gel, blotted to nitrocellulose paper, and probed with a 417-bp 32P-labeled probe corresponding to position 608-1025 of the TdT cDNA (European Molecular Biology Laboratories [EMBL] database accession number M11722.em_hum2).
Isolation and nucleotide sequence analysis of Hek11 cDNAs. To obtain a probe for Northern blot analysis of PCR-amplified products, the entire predicted extracellular region of the published Hek11 sequence was PCR amplified from 200 ng total RNA extracted from the Bjab cell line and from fetal brain using oligonucleotide primer set Hek1125′ and Hek11LO, corresponding to nucleotide positions 39-58 and 1768-1792, respectively17 (Table 1 and Fig 2). Samples were preincubated for 2 minutes at 94°C and subsequently subjected to 32 amplification cycles consisting of 40 seconds at 94°C, 40 seconds at 56°C, and 2 minutes at 72°C. The 1,753-bp PCR fragments were digested with BamHI and EcoRI restriction enzymes, cloned into pBluescript vector, and used for nucleotide sequence analysis.
RESULTS
Identification of Eph-related RTKs expressed in fetal BM pro–B cells. PolyA+ mRNA from sorted CD34+/CD19+ fetal BM pro–B cells was converted to first-strand cDNA and amplified in PCR using degenerate oligonucleotide primers specific for the amino acid motifs MGQF and WT(A/S)PE. Amplified fragments were size-fractionated on 2% agarose gel and products of the expected size (370 bp) were excised, purified, and ligated into T-vector. Inserts were used for nucleotide sequence analysis and compared for homology to sequences in the EMBL database using the BlastN program. Of 12 sequences from the pro–B cells sorted from 2 different fetal BM samples, 11 sequences related to previously published Eph-related RTKs, 5 showed 100% identity to the human Hek11 RTK, and 5 showed 100% identity to the human Hek8 RTK. A single sequence displayed 100% identity at the amino acid level to the previously published rat Eph-related RTK sequence Ehk-2,44 except that a 42–amino acid insertion in Ehk-2 was missing in our sequence. The next best scores were Cek4/Hek4/Mek4 displaying 91% to 92% identity to our sequence of 112 amino acids in the region between the degenerate primers. The human homologue of Ehk-2 has not been described, although it has been predicted to exist and termed Hek12.17 Although the initial PCR data obtained with degenerate primers suggested the presence of Hek12 transcripts in fetal pro–B cells, we were not able to confirm these data by RT-PCR using primers designed to specifically amplify Hek12 RNA. Because these primers allowed amplification of Hek12 mRNA from total fetal BM samples, we anticipate that Hek12 is expressed by non–pro-B cells (data not shown).
Semiquantitative analysis of Hek8 and Hek11 expression during B lymphopoiesis. Levels of Hek8 and Hek11 expression in purified B cell populations were determined by semiquantitative PCR. Based on the available sequence data of Eph-related RTKs, we designed a set of four nested primers to amplify Hek11 and a set of three primers to amplify Hek8 from oligo-dT–primed first-strand cDNA. The amount of cDNA used for amplification reactions was standardized for each sample in a PCR with limited amounts of cycles using a primer pair specific for actin (results not shown). Standardized amounts of cDNA were subsequently amplified in 35 cycles of a primary Hek8 or Hek11 PCR. The amplified products were 247 bp (Hek8) or 184 bp (Hek11) in size. Aliquots of 5 μL from the second PCR were removed at various cycle intervals, separated on an agarose gel, transfered to nitrocellulose, and probed with a 32P-labeled Hek8 or Hek11 probe. The same cDNA samples were amplified with the actin-specific primer pair in a primary PCR. Aliquots from these reactions were removed at various cycle intervals, processed as described for aliquots of the Hek8 and Hek11 PCR, and probed with a 32P-labeled actin probe. The results of these experiments are depicted in Fig 4.
Hek8 transcripts were detectable in all fetal and adult BM, blood, and tonsil B cell populations analyzed, as well as in CD34+/CD19− non–B-lineage precursor cells from adult BM. The highest levels of Hek8 transcripts were reproducibly found in adult peripheral blood B cells (Fig 4A). Hek11 transcripts were found in purified CD34+/CD19+ fetal BM pro–B and pre–B cells, in the cell line Bjab, and in fetal brain (positive control), whereas a background signal at the level of the water control or no signal was detectable in fetal BM sIgM+ B cells, tonsillar B cells, plasma cells, fetal stroma cells, and the jurkat cell line (negative control). Tonsillar T cells showed a signal just above background in the first experiments, but this could not be reproduced in the second experiment (Fig 4B and C). These results were confirmed and extended in a subsequent experiment (Fig 4D): high levels of Hek11 expression were found in fetal BM pro–B cells and not in adult BM pro–B or pre–B cells or adult peripheral blood sIgM+ B cells.
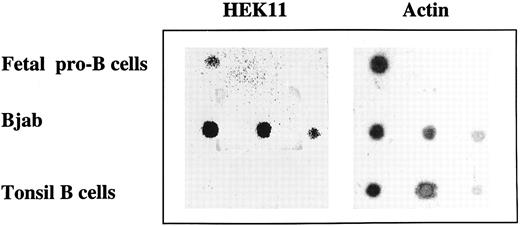
Dot-blot analysis of Hek11 expression in total RNA isolated from fetal pro–B cells, Bjab cell line, and tonsil B cells. Total RNA isolated from fetal 3 × 106 pro–B cells, and a dilution of total RNA (1 μg, 0.5 μg, and 0.1 μg from left to right) isolated from Bjab cell line or tonsil B cells was spotted on a filter and hybridized with Hek11 and actin probes.
Dot-blot analysis of Hek11 expression in total RNA isolated from fetal pro–B cells, Bjab cell line, and tonsil B cells. Total RNA isolated from fetal 3 × 106 pro–B cells, and a dilution of total RNA (1 μg, 0.5 μg, and 0.1 μg from left to right) isolated from Bjab cell line or tonsil B cells was spotted on a filter and hybridized with Hek11 and actin probes.
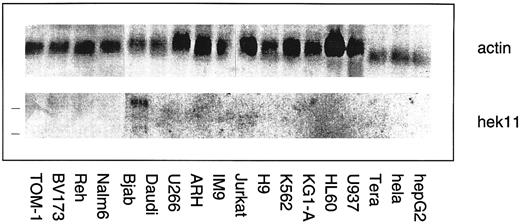
Northern blot analysis of Hek11 gene expression in a panel of human hematopoietic and epithelial cell lines. Each lane contains 10 μg of total RNA. The position of 28s and 18s rRNA bands is indicated with a line (left). Hek11 expression was solely detected in the Bjab lymphoma cell line.
Northern blot analysis of Hek11 gene expression in a panel of human hematopoietic and epithelial cell lines. Each lane contains 10 μg of total RNA. The position of 28s and 18s rRNA bands is indicated with a line (left). Hek11 expression was solely detected in the Bjab lymphoma cell line.
In humans, expression of the TdT gene during B cell differentiation is restricted to the pro–B cell stage and may be used to assess the purity of sorted B cell populations. TdT expression was found in sorted pro–B cell from adult and fetal BM, whereas TdT expression was below detection level in adult BM non–B-lineage progenitor cells, pre–B cells, and mature B cells (Fig 5).
Detection of Hek11 expression by dot-blot analysis. The extracellular portion of Hek11 was cloned by PCR amplification of first-strand cDNA synthesized from mRNA extracted from both the Bjab cell line and fetal brain. Nucleotide sequence analysis of the cloned fragments showed 100% identity with the published Hek11 sequence with the exception that two Gs are lacking at position 349 and 350 in our cDNAs cloned from both fetal brain and the Bjab cell line. As a consequence, the postulated open reading frame starting with an ATG at position 186 terminates in an stop codon at position 358. The next ATG is positioned at nucleotide 404 and yields an open reading frame that is identical to that of the published sequence. Overall, our Hek11 sequences from fetal brain and the Bjab cell line encode a protein that is shorter by 76 amino acids in the extracellular domain.
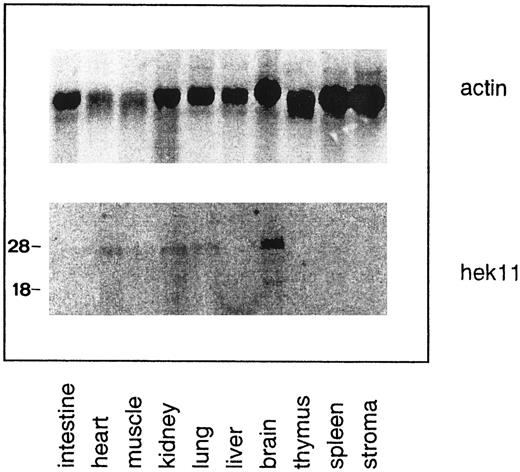
Northern blot analysis of Hek11 expression in fetal human tissues. Each lane contains 10 μg of total RNA. Stroma is defined as adherent fetal BM cells cultured for 3 to 5 days. Multiple Hek11 transcripts of approximately 6.0 kb, 4.4 kb, and 2.4 kb were detectable in brain, heart, kidney, and lung.
Northern blot analysis of Hek11 expression in fetal human tissues. Each lane contains 10 μg of total RNA. Stroma is defined as adherent fetal BM cells cultured for 3 to 5 days. Multiple Hek11 transcripts of approximately 6.0 kb, 4.4 kb, and 2.4 kb were detectable in brain, heart, kidney, and lung.
Independent evidence for Hek11 expression in fetal BM pro–B cells was obtained by dot-blot analysis of total RNA extracted from 3 × 106 sorted CD34+/CD19+ cells. Serial dilutions of total RNA from the Bjab cell line (positive control) and tonsil B cells (negative control) were spotted on the same blot as fetal pro–B cell RNA. The blot was hybridized with the cDNA spanning the extracellular domain of Hek11, analyzed, stripped, and reprobed with an actin probe. A Hek11 signal was obtained in fetal pro–B cells and in the Bjab cell line, but not in tonsillar B cells (Fig 6). The Hek11 signal was approximately 10 times weaker in pro–B cells compared with the Hek11 signal in Bjab.
Northern blot analysis of Hek11 expression in fetal tissues and cell lines of hematopoietic origin. The Hek11 cDNA was used as a probe in Northern blot analysis of mRNA extracted from fetal tissues, a collection of cell lines of hematopoietic origin, the cervical carcinoma cell line Hela, and the Hepatoma line HepG2. No Hek11 transcripts were detected in the pro–B cell lines TOM-1 and BV173; the pre–B cell lines Reh and Nalm-6; the mature B cell line Daudi (Burkit's lymphoma); or the plasmacytoid cell lines U266, ARH-77, and IM9. In addition, cell lines from the myeloid lineage (KG1-A, HL-60, and U937), the erythroid lineage (K562), or epithelial origin (HeLa and HepG2) did not express Hek11. In the mature B cell line Bjab, three Hek11 transcripts were detected with the most prominent at 6.0 kb, then 4.4 and 2.0 kb (Fig 7). In Northern blot analyses of fetal tissues, Hek11 transcripts were detectable in brain, heart, lung, and kidney, whereas the message was not detectable in intestine, liver, thymus, spleen, and short-term (3 to 5 days) cultured fetal BM stromal cells (Fig 8). Three messages were detectable in fetal brain with the most prominent at 6.0 kb, then 4.4 and 2.4 kb.
DISCUSSION
The Eph-related RTKs constitute a growing family of receptors with at least 13 members.45 These receptors bind to a family of cell-bound ligands of which at least seven distinct members have been unveiled. Members of the Eph-family receptors display temporally and spatially restricted expression patterns during embryogenesis, suggesting a role in various developmental processes.21,25,34,46 Recent evidence suggests that interactions between Eph-related RTKs and their cognate ligands play a role in angiogenesis27 and in controlling the formation of neuronal pathways.28,29 In this study, we investigated the expression of Eph RTKs in cells of the hematopoietic system, in particular B-lineage cells. By using degenerate primers specific for conserved amino acid motifs in the kinase domain of Eph-related RTKs, we identified transcripts of Hek8 and Hek11 in freshly isolated human fetal BM pro–B cells. The nucleotide sequence of a third transcript originally cloned from the pro–B cell fraction displayed most homology to the rat Ehk-2 receptor and likely represents the human homologue of this gene. This transcript, predicted to exist and denoted Hek12,17 could in subsequent experiments not be retrieved from sorted fetal B-lineage cells using a set of Hek12-specific primers. Because this primer set permitted amplification of Hek12 transcripts from unseparated fetal BM MNC populations, we anticipate that the Hek12 transcript in the original pro–B cell sample originated from contaminating non–B-lineage cells.
Hek8 transcripts were found in all B cell populations derived from fetal and adult BM, peripheral blood, and tonsils, and in addition in tonsil T cells. In adult humans, Hek8 transcripts have been found in many tissues, including brain, heart, lung, muscle, kidney, placenta, and pancreas. The expression pattern in human B-lineage cells appears to recapitulate the ubiquitous expression of Hek8 in many tissues and cell types.
In semiquantitative PCR analysis, we observed abundant expression of Hek11 in fetal BM pro–B and pre–B cells but no or very low levels of expression in more mature, sIgM+ fetal BM B-lineage cells, adult BM pro–B and pre–B cells, tonsil B cells, and plasma cells. In addition, no or very low levels of Hek11 transcripts were detected in T-lineage cells from tonsils and in short-time cultured fetal BM stroma cells. Expression of the Hek11 gene in fetal pro–B cells was confirmed by dot-blot analysis. Northern blot analysis of fetal tissues containing large populations of lymphocytes at various stages of differentiation confirmed the absence of Hek11 transcripts in fetal spleen (rich in mature T and B lymphocytes), thymus (rich in T-lineage cells), and short-term–cultured fetal BM stroma cells. In Northern blot analysis of mRNA from a panel of cell lines, Hek11 expression was solely detected in the Burkit's lymphoma cell line Bjab and not in cell lines representing the pro– and pre–B cell–differentiation stages. Of note, the leukemia and lymphoma cell lines are of adult origin and do not reflect fetal B-cell development.
The tissue distribution of Hek11 transcripts in fetal tissues (brain, heart, kidney, and lung) corresponds to that reported for adult tissues,17 whereas the number of transcripts at these two ontogenic stages differed. In adult brain, five Hek11 transcripts are detectable with major transcripts of 7.5 kb, 6.0 kb, and 3.0 kb and minor transcripts of 4.4 kb and 2.4 kb. In the corresponding organs in the fetus only three transcripts with mRNAs of length 6.0, 4.4, and 2.4 kb were detectable. A difference was also observed between fetal brain and the Bjab cell line. In Bjab the 2.4-kb transcript was missing and instead a 3.0-kb transcript was detectable. A common feature of the Eph-receptor family is the occurrence of multiple splice variants. Sequence data have shown that splice variants of other members of the Eph RTK family can result in soluble forms of the receptor, receptors with shorter or longer extracellular domains, or truncated transmembrane proteins with a disrupted kinase domain.44,46 47 Further characterization of the different Hek11 splice variants will provide more information on the differential regulation and functional relevance in the fetus and adult.
Although high levels of Hek11 mRNA were found in fetal BM pro–B and pre–B cells, no Hek11 transcripts were detectable in the equivalent populations isolated from adult BM. To our best knowledge, in only one other instance human fetal and adult BM progenitor B cells have been reported to differ in the expression of a cell surface molecule, notably the CD105 (endoglin) molecule.48 A developmental switch in B lymphopoiesis has been reported in mice, where fetal liver-derived pre–B cells can be discerned from BM-derived pre–B cells by the differential expression of a regulatory myosin light chain and class II molecules.49,50 In addition, fetal omentum contains precursors for CD5+ B cells but not conventional B cells, whereas mouse fetal liver contains progenitors for both lineages. No CD5+ progenitors are found in adult mice BM.51 Differences in the expression of as yet unknown growth and differention factor receptors between fetal and adult human progenitor B cells may be expected to exist, based on the observation that multiple stages of B-cell developments can be recapitulated in vitro using fetal cells.10,12 52 Similar in vitro cultures for growth and differentiation of human adult BM-derived B-lineage cells are cumbersome.
Establishment of the role of Hek8 and Hek11 in early B-cell differentiation/proliferation depends on the identification of their cognate ligands and a detailed understanding of the spatial and temporal expression pattern of receptors and ligands in the BM microenvironment. To date, no ligands have been described for these receptors. The notion that ligands of this family of receptors are structurally related and that a single receptor may functionally interact with more than one ligand and vice versa adds another level of complexity.23-26 53 The well-defined stages of human B lymphopoiesis and the availability of dynamic, cytokine, and/or stromal cell supported in vitro culture systems that mimick particular steps in this process provide a suitable model to address these issues.
ACKNOWLEDGMENT
We thank Drs G. Moldenhauer and M. Okabe for the Bjab and TOM-1 cell lines, respectively, and Ruth Solem and Kirsti Solberg for expert help with the cell sorting experiments.
H.-C.A. is a post-doctoral fellow of the Norwegian Research Council for Science.
Address reprint requests to Ton Logtenberg, MD, Department of Immunology, University Hospital Utrecht, 3584 CX Utrecht, The Netherlands.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal