Abstract
Fanconi anemia (FA) is an autosomal recessive genetic disorder characterized by a variety of physical anomalies, bone marrow failure, and an increased risk for malignancy. FA cells exhibit chromosomal instability and are hypersensitive to DNA cross-linking agents such as mitomycin C (MMC). FA is a clinically heterogeneous disorder and can be functionally divided into at least five different complementation groups (A-E). We previously described the use of a retroviral vector expressing the FAC cDNA in the complementation of mutant hematopoietic cells from FA-C patients. This vector is currently being tested in a clinical trial of ex vivo hematopoietic progenitor cell transduction. The FA-A group accounts for over 65% of all FA cases, and the FAA cDNA was recently identified by both expression and positional cloning techniques. We report here the transduction and phenotypic correction of lymphoblastoid cell lines from four unrelated FA-A patients, using two amphotropic FAA retroviral vectors. Expression of the FAA transgene was adequate to normalize cell growth, cell-cycle kinetics, and chromosomal breakage in the presence of MMC. We then analyzed the effect of retroviral vector transduction on hematopoietic progenitor cell growth. After FAA transduction of mutant progenitor cells, either colony number or colony size increased in the presence of MMC. In addition, FAA but not FAC retroviral transduction markedly improved colony growth of progenitor cells derived from an unclassified FA patient. FAA retroviral vectors should be useful for both complementation studies and clinical trials of gene transduction.
FANCONI ANEMIA (FA) is an autosomal recessive disorder clinically manifested by bone marrow (BM) failure, diverse developmental anomalies, and cancer susceptibility. Although FA patients are phenotypically heterogeneous, the hematologic abnormalities are the most clinically significant.1 The hallmark of the disease is the hypersensitivity of FA cells to DNA bifunctional cross-linking agents such as mitomycin C (MMC) and diepoxybutane (DEB).2 FA cells exposed to these clastogenic agents in vitro reproducibly develop chromosomal aberrations and die by apoptosis.
Somatic cell hybridization studies have established at least five complementation subtypes (A-E). The FA-A group is the most prevalent, comprising 60% to 70% of FA cases studied.3 The identification of the FAA cDNA was recently described using a positional4 and an expression cloning strategy5 previously used to identify the FAC cDNA. FAA is located on chromosome 16q24.3 and encodes a predicted protein of 163 kD. The predicted FAA and FAC proteins have no homologies to each other or to other known proteins, suggesting a novel cellular pathway for maintenance of chromosomal stability.
Cell fusion studies can reliably assign FA patients to complementation groups, because patients functionally classified as FA-C turned out to have pathogenic mutations in FAC.3 Mutation screening of FAC has shown at least 8 pathogenic mutations,6 IVS4 + 4 A to T and 322delG being the most prevalent.7 8 Four mutations have so far been reported in FAA and work is in progress in several laboratories to determine the mutation spectrum in FA-A patients.
Therapy for FA is directed at the hematologic manifestations, typically the most life-threatening complications. BM aplasia is usually treated with androgens and supportive care. Allogeneic BM transplantation (BMT) from a histocompatible sibling donor is the treatment of choice for patients with severe disease who have a suitable donor. FA patients who undergo HLA-identical sibling BMT have a 2-year survival probability of 66%, but only 29% after alternative donor transplants.9 Although BMT is potentially curative of the hematologic pathology, patients may go on to develop secondary malignancies, often solid tumors of the head and neck. Umbilical cord blood transplantation has also been successfully applied for FA patients.10 For FA patients lacking a suitable stem cell donor, other approaches to treatment are needed.
Gene augmentation therapy entails the insertion of a normal gene into an appropriate target cell to correct the function of a defective gene. Viral vectors are currently the primary vehicles due to their efficiency in entering and transferring genetic material into cells. Using two different viral vectors, we previously described the genetic complementation of FA-C lymphoblasts.11,12 Recombinant virus transduction of the FAC cDNA into FA-C CD34+ hematopoietic progenitor cells improved their survival and clonogenic growth. This in vitro survival advantage of corrected FA-C cells suggested that FAC-transduced hematopoietic progenitor cells might have a selective advantage in vivo and led to the first FA clinical gene therapy trial.13 In this report, we describe the development of two retroviral vectors for the complementation of group A FA hematopoietic cells.
MATERIALS AND METHODS
Viruses and cells. All lymphoblast cell lines were grown in RPMI containing 15% heat-inactivated fetal calf serum and glutamine as previously described.12 PA31714 and GP + E8615 cell lines were grown as previously described.
FA-A patients and cell lines. Patients 1 (43-year-old woman), and 2 (6-year-old girl) were assigned to complementation group A by somatic cell hybridization and/or mutational analysis (H. Joenje, A. Auerbach, unpublished observations). Patient 3 (8-year-old girl) was previously unclassified by FA complementation group. The following lymphoblastoid cell lines were used, characterized as FA-A by cell fusion experiments: HSC72, which is the original reference group A patient reported,16 VU5,3 EUFA471,5 and EUFA528 (unpublished observations). All patients were diagnosed as having FA on the basis of clinical symptoms and a positive chromosomal breakage test. Confirmation of the group assignment by mutation screening is currently in progress and will be reported elsewhere.
Generation of amphotropic producer clone. The 5.5-kb FAA cDNA was subcloned into the Not I site of the pG1 retroviral vector to create pG1-FAA5.5. The pG1 vector is based on the Moloney murine retrovirus and does not contain eukaryotic selection markers. The FAA cDNA is driven by the Moloney murine retrovirus long terminal repeat (LTR) promoter. The FAA termination and polyadenylation signals are maintained in this construct. A truncated 4.3-kb FAA cDNA version (pG1-FAA4.3), lacking the 3′ untranslated region, was generated using the Transformer Site-Directed Mutagenesis Kit (Clontech, Palo Alto, CA) and a mutagenic primer (5′GTG-GCAGAGCTGC TG GC TG AA AG CT AG CT AGCGGCCGCTCGA-GGC 3′) according to the manufacturer's protocol. This produces a truncation at nt 4302 leading to a loss of 94 bp in the FAA open reading frame (ORF ). The vector/insert junctions were verified by sequencing (Sequenase; US Biochemicals, Cleveland, OH). The termination and polyadenylation signals reside in the 3′ LTR.
The pG1-FAA4.3 plasmid was cotransfected with a plasmid carrying the neomycin resistance (NeoR) gene by calcium phosphate coprecipitation into the PA317 amphotropic packaging cell line. Two days later, the cells were trypsinized, replated, and placed under G418 (Geneticin; GIBCO/BRL, Gaithersburg, MD) selection (500 μg/mL, active). Individual resistant colonies were isolated and analyzed for the presence of viral RNA by dot-blot hybridization. High-titer producer clones were expanded and genomic DNA isolated. Alternatively, the pG1-FAA4.3 plasmid was transfected into the GP + E86 ecotropic packaging cell line grown in media containing 5 mmol/L sodium butyrate. After 2 days, the cell supernatant was removed, filtered, admixed with polybrene (4 μg/mL), and incubated with PA317 cells. Sequential supernatant infection was repeated twice before PA317 cells were diluted and individual colonies obtained. Producer clone 6.9 was identified as a high-titer unrearranged producer clone after dot-blot analysis for viral RNA and Southern blot analysis of the proviral genome. Genomic DNA was digested with Nhe I and Southern blot hybridization performed using ammonium acetate buffer17 and nylon transfer (Hybond N+; Amersham, Arlington Heights, IL). The blot was probed with a BamHI fragment P32 random primer-labeled fragment of the FAA cDNA.
The plasmid pG1FAA-5.5 was cotransfected into GP + E86 cells with the NeoR plasmid by calcium-phosphate coprecipitation. Neomycin-resistant colonies grown in G418 (500 μg/mL, active) were screened for viral RNA production. Producer clones were identified by viral RNA dot-blot analysis; Southern blot analyses confirmed unrearranged proviral genomes in these cell lines. High-titer ecotropic producer lines 1.16 and 2.16 were identified. Supernatant from these cell lines was used to infect PA317 cells twice daily for three days. Individual colonies were established by limiting dilution and analyzed for the presence of viral RNA and intact proviral genome. Two high-titer producer clones 17 and 29 were identified and used in our studies.
Viral titer of the producer lines was assessed by semiquantitative Southern blot analysis (unpublished results, method provided by Dr E. Vanin, St Jude Children's Research Hospital, Memphis, TN). 5 × 105 NIH 3T3 cells were incubated with various amounts of retroviral supernatant and allowed to incubate overnight. Genomic DNA was isolated and 10 μg DNA digested with Nhe I. A 300-bp Not I/Pst I probe (gift from Dr E. Vanin) from the pG1 backbone was used for hybridization. Supernatant from the previously defined FA-C complementing retroviral producer cell line no. 52:19,18 with a functional titer of 5 × 106 neomycin-resistant colonies/mL, was used a positive control. Both FAC and FAA retroviral vectors were generated from the same pG1 vector backbone to which the probe hybridizes. Extended marker rescue assay for detection of helper virus was performed as previously described.19
Retroviral gene transfer of Epstein-Barr–transformed lymphoblasts. Lymphoblasts (1 × 105 cells/mL) were incubated with an equal volume of viral supernatant containing protamine sulfate (5 μg/mL). Cells were grown in RPMI supplemented with 15% fetal calf serum for 2 days before 10 nmol/L mitomycin C (MMC) (Calbiochem, La Jolla, CA) was added. Cells were maintained in media containing 10 nmol/L MMC. Non–virus-infected lymphoblasts yielded no viable cells after 5 weeks following drug selection.
MMC sensitivity assay. Cell sensitivity to MMC was assayed as previously described.12 Lymphoblasts were plated at 1 × 105 cells/mL in 24-well plates. Increasing concentrations of MMC were added and after 4 days, cellular viability assayed by trypan blue exclusion. Samples were performed in duplicate.
Flow cytometry. Lymphoblasts were incubated at 2 × 105 cells/mL with 100 nmol/L MMC for 48 hours or 10 nmol/L MMC for 5 to 7 days. Cell nuclei were stained with propidium iodine using CycleTest+ (Becton Dickinson, Franklin Lakes, NJ) as described by the manufacturer. Stained cell nuclei were analyzed by FACSCAN (Becton Dickinson, Franklin Lakes, NJ) using Cytomation Cicero Software (Fort Collins, CO). Cell-cycle analysis was performed using MODFIT LX 2.0 (Verity Software House, Topsham, ME).
Cytogenetic breakage. Lymphoblast cultures were analyzed for cytogenetic breakage and exchange figures (radial formation) by exposure to MMC (40 ng/mL final) for 2 days in the dark. Cultures were obtained after a 90-minute exposure to 0.50 μg/mL colcemid and 10 μg/mL ethidium bromide. After a 10-minute treatment with 0.075 mol/L KCl, the cells were fixed with a 3:1 mixture of methanol:acetic acid. Slides were prepared using wet slides, air dried, and stained with Wright's stain. Fifteen or 50 metaphase figures from each culture were scored for obvious breaks, gaps larger than a chromatid width, and exchange figures.
Transduction of CD34 progenitors. BM samples were obtained after receiving informed consent on a protocol approved by the Institutional Review Board of the University of North Carolina Medical School. Somatic cell hybridization studies indicated that patient 1 belonged to the FA-A complementation group (H. Joenje, unpublished). Genomic analysis of DNA from patient 2 showed compound heterozygous deletions in the FAA coding sequence (A. Auerbach, unpublished). Patient 3 had not previously been assigned to a FA complementation group. Mononuclear cells were isolated from the marrow by lymphocyte separation medium (Organon Teknika, Durham, NC) and CD34-enriched cells immunoselected using a CellPro column (Bothell, WA) as previously described.12 CD34+ cells were incubated overnight in Dulbecco's modified essential media (DMEM) containing 10% fetal calf serum, 100 ng/mL stem cell factor (SCF; Amgen, Thousand Oaks, CA), 25 ng/mL interleukin-3 (IL-3; Sandoz, East Hanover, NJ), and 50 ng/mL interleukin-6 (IL-6; Sandoz). Cells were then incubated with retroviral supernatant from clone 17 and 5 μg/mL protamine. Incubations were repeated twice daily for 2 days. Cells were plated at 20,000/plate and then analyzed for colony formation as previously described.12 In separate experiments using CD34+ cells from patient 3, both FAA and FAC12 retroviral transduction studies were performed. CD34-immunoselected cells isolated from the umbilical cord blood of a normal newborn were used as control.
RESULTS
Generation of FAA amphotropic producer clones. The FAA vector plasmids are shown in Fig 1. pG1FAA5.5 was derived by inserting the 5.5-kb cDNA, isolated from expression cloning, into the plasmid pG1.20 This cDNA contains an open reading frame of 4,368 bp, a 31-bp 5′ untranslated region (UTR) and a 1.1-kb 3′ UTR. The sequence upstream of the first ATG has poor homology to a Kozak consensus sequence, and a polyadenylation signal was not found. The plasmid pG1FAA4.3 contains a truncated cDNA lacking the 3′ region. This truncated FAA vector was generated to determine if the 3′ UTR affected FAA retroviral expression. Amphotropic producer clones for each vector were derived from PA317 cell lines. Genomic DNA subjected to restriction enzyme digestion with Nhe I, which cuts once within each LTR, was analyzed by Southern blot hybridization (data not shown). The predicted 6.3- and 5.3-kb fragments were detected, indicating unrearranged provirus from FAA5.5 clone 17 and FAA4.3 clone 6.9. Proviral copy number was determined by comparisons with dilutions of plasmid pG1FAA estimated to give 0.5 to 2 copies/cell. Southern blot analysis (data not shown) indicated that producer clones 17 and 6.9 had 10 and 3 copy number equivalents, respectively. Titers of each clone were determined by comparison with a producer clone of known copy number (FAC producer clone 52:19).12 Viral supernatants were incubated overnight on NIH 3T3 cells, genomic DNA isolated and digested with Nhe I, and Southern blot hybridization performed. The probe used hybridizes to all pG1-based vectors. The titers of FAA 5.5/17, 5.5/29, and FAA4.3/6.9 were estimated at 5 × 106, 3 × 106, and 1 × 106, respectively, particles/mL (data not shown). Supernatant from each clone was analyzed for replication-competent retrovirus using an extended rescue assay.19 No replication-competent retrovirus (RCR) was detected from any of the producer clones (data not shown).
Schematic diagrams of FAA retroviral vector constructs. A general schema for the two retroviral constructs used is shown with the FAA coding region subcloned as a Not I fragment into plasmid pG1 (top). (Bottom) The FAA cDNA is driven by the 5′ hybrid long terminal repeat (LTR) of murine moloney sarcoma virus; psi (ψ) retroviral packaging signal; TAG represents a mutation in authentic start codon in the molony murine leukemia virus gag.
Schematic diagrams of FAA retroviral vector constructs. A general schema for the two retroviral constructs used is shown with the FAA coding region subcloned as a Not I fragment into plasmid pG1 (top). (Bottom) The FAA cDNA is driven by the 5′ hybrid long terminal repeat (LTR) of murine moloney sarcoma virus; psi (ψ) retroviral packaging signal; TAG represents a mutation in authentic start codon in the molony murine leukemia virus gag.
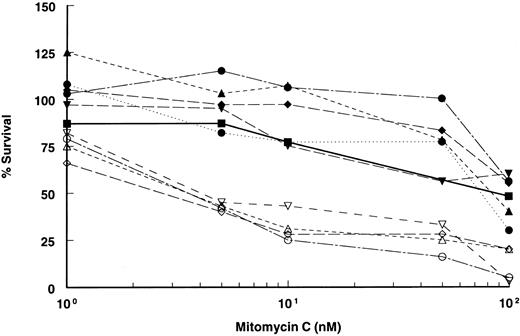
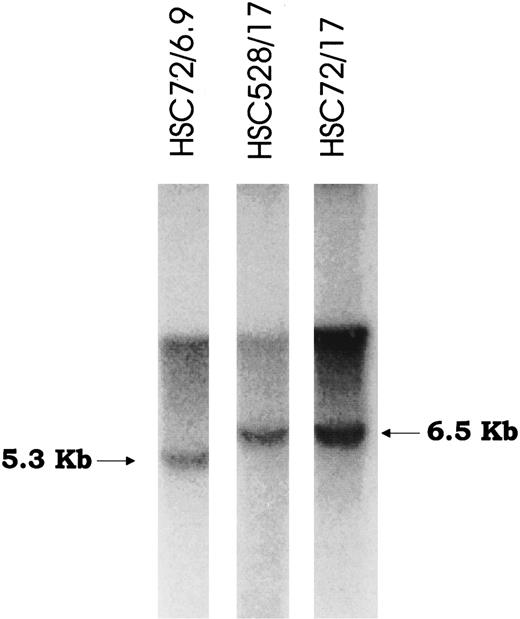
Retroviral-mediated gene transfer and phenotypic correction of FA-A lymphoblasts. Epstein-Barr virus immortalized lymphoblast lines established from FA patients were confirmed to be FA-A subtype by somatic cell hybrid complementation and mutational analyses. After viral supernatant infections, each cell line was incubated with low doses of (10 nmol/L) MMC for at least 4 to 6 weeks to select for complemented cells. MMC resistance was then measured by incubating cells at various concentrations of MMC to determine drug sensitivity. All the mutant (parental) lines were markedly sensitive to MMC, with an effective drug concentration yielding 50% reduction in cell viability (EC50 ) of 2 to 5 nmol/L MMC (Fig 2). However, in marked contrast, retroviral-transduced lymphoblasts were significantly less sensitive to MMC and equivalent in resistance to lymphoblasts established from normal donors. The EC50 of both normal and corrected lymphoblasts was greater than 100 nmol/L. Correction to normal was observed in all cell lines tested and with both retroviral constructs. Confirmation of integrated provirus in FAA gene-corrected cells was performed by Southern hybridization of genomic DNA digested with Nhe I (Fig 3).
MMC sensitivity of FAA-retroviral transduced lymphoblasts. Plot of cell survival of FAA parental and retroviral-transduced cell lines after incubation with varying concentrations of MMC. MMC sensitivity of lymphoblasts derived from a normal individual is shown. All data points are the results of quadruplicate determinations (SEM < 5%). Open symbols represent FAA mutant parental cell lines, closed symbols represent retroviral-transduced cell lines, (⋅⋅•⋅⋅) indicates retroviral-transduced cell line with vector FAA/4.3 (clone 6.9). Normal lymphoblasts are represented by the solid line (─). ( — ○ — ), HSC 72; (– –▵– –), EUFA 528; ( — • — ), HSC 72/Retro17; (⋅⋅⋅•⋅⋅⋅), HSC 72/Retro6.9; (– –▴– –), EUFA 528/Retro29; ( — ▪ — ), normal; (– –▿– –), EUFA 471; ( — ⋄ — ), EUFA 005; ( — □ — ), EUFA 471/Retro17; ( — ▾ — ), EUFA 005/Retro17.
MMC sensitivity of FAA-retroviral transduced lymphoblasts. Plot of cell survival of FAA parental and retroviral-transduced cell lines after incubation with varying concentrations of MMC. MMC sensitivity of lymphoblasts derived from a normal individual is shown. All data points are the results of quadruplicate determinations (SEM < 5%). Open symbols represent FAA mutant parental cell lines, closed symbols represent retroviral-transduced cell lines, (⋅⋅•⋅⋅) indicates retroviral-transduced cell line with vector FAA/4.3 (clone 6.9). Normal lymphoblasts are represented by the solid line (─). ( — ○ — ), HSC 72; (– –▵– –), EUFA 528; ( — • — ), HSC 72/Retro17; (⋅⋅⋅•⋅⋅⋅), HSC 72/Retro6.9; (– –▴– –), EUFA 528/Retro29; ( — ▪ — ), normal; (– –▿– –), EUFA 471; ( — ⋄ — ), EUFA 005; ( — □ — ), EUFA 471/Retro17; ( — ▾ — ), EUFA 005/Retro17.
Southern blot analysis of FAA-transduced lymphoblast cell lines. Genomic DNA was isolated from two cell lines transduced with either vector FAA 5.5 or FAA 4.3, digested with Nhe I and hybridized with probe to FAA. The expected bands (6.5 kb and 5.3 kb) represent unrearranged proviral form in each cell line.
Southern blot analysis of FAA-transduced lymphoblast cell lines. Genomic DNA was isolated from two cell lines transduced with either vector FAA 5.5 or FAA 4.3, digested with Nhe I and hybridized with probe to FAA. The expected bands (6.5 kb and 5.3 kb) represent unrearranged proviral form in each cell line.
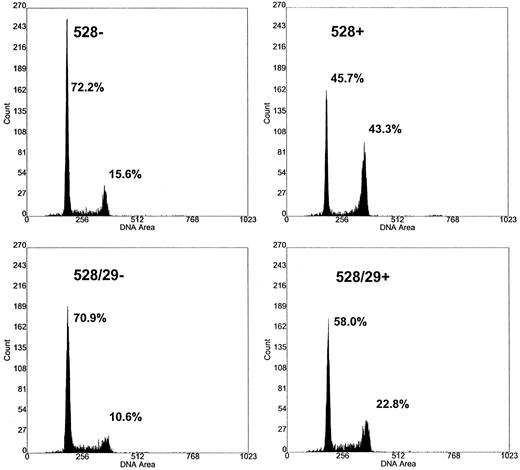
Restoration of cell-cycle progression. Cell-cycle arrest and loss of cellular proliferative capacity are the hallmarks of FA. FA cells typically respond to DNA damage induced by MMC with a delay in the G2 phase of the cell cycle. We tested FAA gene-corrected cells to determine if complementation resulted in relief of the G2 checkpoint delay. FA-A cells in each phase of the cell cycle were analyzed by propidium iodide staining and flow cytometry (Fig 4). The percentage of FA-A cells in the G2 phase was increased when compared with normal individuals. Incubation of uncorrected mutant FA-A cells with either low-dose MMC (10 nmol/L) for 1 week or 100 nmol/L for 2 days had the expected effect of increasing the percentage of cells in the G2 phase (Fig 4). Table 1 summarizes the cell-cycle results from five separate experiments. Three different mutant FAA lymphoblast cell lines and three different normal lymphoblasts were studied. Statistical analysis of the percentage of cells in the G2/M cell fraction showed a significant difference (.02 > P > .01, Student's t-test) when mutant FAA lymphoblasts (35%) were compared with either normal (14.8%) or virus-transduced cells (18.6%) in the presence of MMC. The corrected FA-A cells showed normalized cell-cycle kinetics in both the presence and absence of MMC (Table 1). No significant change in the number of cells in G1/S was observed following the addition of MMC.
Cell-cycle kinetics of corrected FAA cells. The effect of mitomycin C on DNA flow cytometry histograms of retroviral-transduced lymphoblasts. The transduced and parental FAA cell line EUFA 528 were incubated in the presence and absence of MMC. Cells were harvested, stained with propidium iodide, and DNA histograms obtained 48 hours after cells were exposed to 100 nmol/L MMC. EUFA 528, parental FAA line; EUFA 528/29, retroviral-transduced cell line. −/+ refers to the presence and absence of MMC. The percentage of cells in the G1 and G2/M fractions are indicated.
Cell-cycle kinetics of corrected FAA cells. The effect of mitomycin C on DNA flow cytometry histograms of retroviral-transduced lymphoblasts. The transduced and parental FAA cell line EUFA 528 were incubated in the presence and absence of MMC. Cells were harvested, stained with propidium iodide, and DNA histograms obtained 48 hours after cells were exposed to 100 nmol/L MMC. EUFA 528, parental FAA line; EUFA 528/29, retroviral-transduced cell line. −/+ refers to the presence and absence of MMC. The percentage of cells in the G1 and G2/M fractions are indicated.
Cell-Cycle Analysis
| Cell Type . | Condition . | % Cells (mean ± SD)* . | ||
|---|---|---|---|---|
| . | . | G1/G0 . | S . | G2/M . |
| FAA | −MMC | 69.9 (4.0) | 17.7 (6.3) | 12.2 (2.8) |
| +MMC | 52.9 (13.0) | 11.7 (6.4) | 35.0 (7.8) | |
| FAA/Retro | −MMC | 65.8 (5.3) | 22.1 (6.2) | 12.0 (3.4) |
| +MMC | 60.0 (10.8) | 20.5 (7.9) | 18.6 (3.8)† | |
| Normal | −MMC | 63.0 (3.6) | 27.9 (4.5) | 8.8 (0.7) |
| +MMC | 59.0 (11.4) | 26.2 (9.1) | 14.8 (3.9)† | |
| Cell Type . | Condition . | % Cells (mean ± SD)* . | ||
|---|---|---|---|---|
| . | . | G1/G0 . | S . | G2/M . |
| FAA | −MMC | 69.9 (4.0) | 17.7 (6.3) | 12.2 (2.8) |
| +MMC | 52.9 (13.0) | 11.7 (6.4) | 35.0 (7.8) | |
| FAA/Retro | −MMC | 65.8 (5.3) | 22.1 (6.2) | 12.0 (3.4) |
| +MMC | 60.0 (10.8) | 20.5 (7.9) | 18.6 (3.8)† | |
| Normal | −MMC | 63.0 (3.6) | 27.9 (4.5) | 8.8 (0.7) |
| +MMC | 59.0 (11.4) | 26.2 (9.1) | 14.8 (3.9)† | |
Results of 5 separate experiments. Three different FAA mutant cell lines and 3 different normal lymphoblast cell lines were used.
Significant at .02 > P > .01 using Student's t-test.
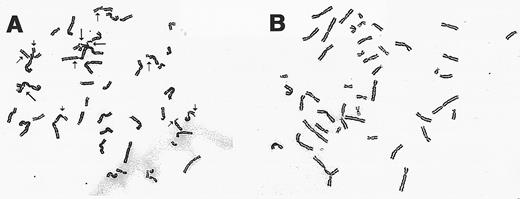
Chromosomal breakage analysis of gene-corrected FAA cells. Exposure of all FA cell types to DNA cross-linking agents such as MMC results in a characteristic increased frequency of chromosomal breakage and aberrations. Retroviral-transduced FA-A lymphoblasts were analyzed for chromosomal breakage following incubation with MMC. As shown in Table 2 and Fig 5, the mutant HSC 72 and EUFA 528 lines have a spontaneous breakage rate comparable with normal cells. After drug exposure, the percentage of mutant cells with chromosomal breaks increased dramatically as expected. Complementation with the FAA cDNA led to decreased chromosomal breakage rates for both lines tested.
Cytogenetic Analysis of FAA Lymphoblast Cell Lines
| Cell Line . | Condition* . | Radials†‡ . | Breakageρ . |
|---|---|---|---|
| HSC 72 | −MMC | 0/15 | 3/15 |
| HSC 72 | +MMC | 7/15 | 15/15 |
| HSC 72/Retro | −MMC | 0/50 | 0/50 |
| HSC 72/Retro | +MMC | 4/50 | 15/50 |
| EUFA 528 | −MMC | 0/45 | 1/45 |
| EUFA 528 | +MMC | 23/45 | 39/45 |
| EUFA 528/Retro | −MMC | 0/45 | 2/45 |
| EUFA 528/Retro | +MMC | 0/45 | 2/45 |
| Lymphoblasts-NML2-155 | −MMC | 0/50 | 0/50 |
| Lymphoblasts-NML | +MMC | 0/50 | 0/50 |
| Cell Line . | Condition* . | Radials†‡ . | Breakageρ . |
|---|---|---|---|
| HSC 72 | −MMC | 0/15 | 3/15 |
| HSC 72 | +MMC | 7/15 | 15/15 |
| HSC 72/Retro | −MMC | 0/50 | 0/50 |
| HSC 72/Retro | +MMC | 4/50 | 15/50 |
| EUFA 528 | −MMC | 0/45 | 1/45 |
| EUFA 528 | +MMC | 23/45 | 39/45 |
| EUFA 528/Retro | −MMC | 0/45 | 2/45 |
| EUFA 528/Retro | +MMC | 0/45 | 2/45 |
| Lymphoblasts-NML2-155 | −MMC | 0/50 | 0/50 |
| Lymphoblasts-NML | +MMC | 0/50 | 0/50 |
Cells were incubated in the absence or presence of 40 ng/mL MMC for 36 hours.
The number of exchange figures (radials)/number of metaphases analyzed.
Greater than 20% radial formation is diagnostic for FA.
ρ Number of chromosome/chromatid gaps, fragments, and radials/number of metaphases analyzed.
Normal lymphoblasts.
Cytogentic analysis of retroviral-transduced FAA lymphoblasts. (A) A representative metaphase spread from the cell line EUFA 528 following incubation with MMC (40 ng/mL). Chromatid breaks (→), and exchanges (➞) are indicated. (B) Metaphase preparation of retroviral-transduced EUFA 528 cells following exposure to MMC.
Cytogentic analysis of retroviral-transduced FAA lymphoblasts. (A) A representative metaphase spread from the cell line EUFA 528 following incubation with MMC (40 ng/mL). Chromatid breaks (→), and exchanges (➞) are indicated. (B) Metaphase preparation of retroviral-transduced EUFA 528 cells following exposure to MMC.
Transduction of primary hematopoietic cells. BM hematopoietic progenitor cells from three FA patients were assayed for colony growth after transduction with our FAA retroviral vector. All three patients were pancytopenic and had not received any treatment before donating BM cells. Recent BM morphology showed hypoplasia without blasts and normal karyotypes. BM aspiration, mononuclear cell collection, and CD34+ cell enrichment were performed. Transduction was done over a 3-day period with resuspension in fresh media and viral supernatant each day. After infection, cells were plated in methylcellulose in both the absence and presence of MMC. Colony growth increased slightly following FAA virus transduction of cells from patient 1 (see Table 3A). More dramatic was the morphologic difference in the size and type of colonies present following transduction. Mock-transduced colonies were small and granulocytic in appearance, whereas transduced colonies were more robust (larger) and BFU-E, CFU-GM, and CFU-GEMM were observed. As expected, when mock-transduced cells were incubated with low concentrations of MMC, the colony numbers decreased. In contrast, transduced colonies (that were significantly larger in size) grew in the presence of low concentrations of MMC.
Colony Growth After Retroviral FAA Transduction of CD34 Cells
| Condition (virus/MMC) . | BFU-E . | Colony No.3-150 . | CFU-GEMM . |
|---|---|---|---|
| . | . | CFU-GM . | . |
| A. Patient 1 | |||
| −Virus/−MMC | 1 | 15 | 1 |
| +Virus/−MMC | 0 | 30 | 3 |
| −Virus/+MMC | 0 | 8 | 0 |
| +Virus/+MMC | 0 | 25 | 2 |
| B. Patient 2 | |||
| −Virus/−MMC | 0 | 0 | 0 |
| +Virus/−MMC | 4 | 24 | 16 |
| C. Patient 3 | |||
| −Virus/−MMC | 10 | 40 | 7 |
| −Virus/+MMC | 0 | 6 | 0 |
| +FAA virus/−MMC | 26 | 115 | 12 |
| +FAA virus/+MMC | 14 | 50 | 8 |
| +FAC virus/−MMC | 26 | 46 | 6 |
| +FAC virus/+MMC | 1 | 10 | 0 |
| D. Umbilical cord blood | |||
| −Virus/−MMC | 20 | 52 | 16 |
| −Virus/+MMC | 22 | 48 | 14 |
| Condition (virus/MMC) . | BFU-E . | Colony No.3-150 . | CFU-GEMM . |
|---|---|---|---|
| . | . | CFU-GM . | . |
| A. Patient 1 | |||
| −Virus/−MMC | 1 | 15 | 1 |
| +Virus/−MMC | 0 | 30 | 3 |
| −Virus/+MMC | 0 | 8 | 0 |
| +Virus/+MMC | 0 | 25 | 2 |
| B. Patient 2 | |||
| −Virus/−MMC | 0 | 0 | 0 |
| +Virus/−MMC | 4 | 24 | 16 |
| C. Patient 3 | |||
| −Virus/−MMC | 10 | 40 | 7 |
| −Virus/+MMC | 0 | 6 | 0 |
| +FAA virus/−MMC | 26 | 115 | 12 |
| +FAA virus/+MMC | 14 | 50 | 8 |
| +FAC virus/−MMC | 26 | 46 | 6 |
| +FAC virus/+MMC | 1 | 10 | 0 |
| D. Umbilical cord blood | |||
| −Virus/−MMC | 20 | 52 | 16 |
| −Virus/+MMC | 22 | 48 | 14 |
The number of progenitor colonies (>50 cells/colony) measured at day 15 in methylcellulose culture. Results are expressed as colony no./2 × 104 CD34+ cells plated. Samples were performed in duplicate and results are expressed as mean colony number. The MMC concentration was 1.0 nmol/L in all experiments.
Due to limited number of CD34+ cells obtained from patient 2, cells were transduced but not incubated with MMC (Table 3B). In the absence of virus transduction no progenitor colonies were observed. In marked contrast, a significant increase in colony number was observed following virus transduction.
Transduction of CD34+ progenitor cells from an untyped (unknown complementation group) FA patient (no. 3) was also performed (Table 3C) using both FAA and the FAC retroviral infection. Baseline colony growth was inhibited in the presence of 1.0 nmol/L MMC. In the absence of MMC, colony growth increased after incubation of cells with FAA but not the FAC retroviral supernatant. More impressively, in the presence of MMC, colonies resistant to MMC grew only after FAA transduction. Results of colony growth using CD34 cells from umbilical cord blood from a normal newborn are shown for comparison (Table 3D).
Finally, CD34-enriched progenitor cells from a known FA-C patient were transduced with the FAA retroviral vector and plated in colony culture. No growth was observed either with or without MMC (data not shown).
DISCUSSION
We report here the functional complementation of FA-A hematopoietic cells using two amphotropic retroviral vector constructs. Phenotypic correction was determined by three different functional assays, namely, the resistance to MMC-induced cell death, insusceptibility to chromosomal breakage, and the restoration of normal cell-cycle kinetics. Complementation and correction occurred in four FA-A lymphoblastoid cell lines. Stable maintenance of the provirus was determined in all cases. Complementation of FA-A cells with these viral vectors has implications for the diagnosis, treatment, and pathophysiology of FA.
The FAA cDNA is 5.5 kb in length, making it too large for small viral vectors such as the adeno-associated virus (AAV). Consequently, we tested truncation mutants of FAA for the ability to complement. The 4.3-kb FAA fragment leads to an incomplete open reading frame that lacks 32 amino acids from the carboxyl-terminus. Unexpectedly, this fragment complemented HSC 72 and two other FA-A lymphoblastoid cell lines (K. Fu, unpublished data), suggesting that this shorter fragment of FAA may be functional.
Phenotypic complementation using recombinant viral vectors may be useful in assigning patients to either the FA-A or FA-C complementation group (collectively constitute nearly 80% of FA cases in the European and North American registries). For example, a D195V polymorphism in FAC was identified in the cell line EUFA 123.21 We were able to demonstrate retroviral complementation using an FAA vector but not an FAC vector, confirming that the line belongs to the FA-A group (K.L. Fu et al, submitted). Group assignment using this approach was applied here to an unclassified patient (no. 3). Progenitor growth improved dramatically only following FAA transduction. This result not only validates our FAA vector but suggests that retroviral gene transfer can assign patients to a FA complementation group.
Recently, immune reactivity to the neomycin phosphotransferase gene product has been described in patients who underwent cell transduction with viral vectors containing NeoR.22 Removal of the NeoR gene from amphotropic retroviral vectors therefore may be desirable. In this study we used the unique characteristics of the FAA gene to generate complemented cells by selection solely on the basis of resistance to MMC-induced death. In contrast to our previous work (which used neomycin-selected stable transfectants), we used the inherent selection property of the FAA gene as the determinant of retroviral transduction.
The rescue and growth of transformed lymphoblasts and primary hematopoietic cells further support our hypothesis that restoration of corrected FA genes provides a selective advantage over mutant cells. Improved FA cell viability after transduction may result from resistance to apoptosis. Recent reports indicate that FA-C mutant cells are hypersensitive to apoptosis, and that complementation with the normal FAC cDNA confers resistance to apoptosis.23,24 Hematopoietic progenitor cells from fac knock-out mice are hypersensitive to interferon-γ (IFN-γ), possibly through apoptotic mechanisms. Conversely, overexpression of FAC cDNA in hematopoietic cells of transgenic mice may confer resistance to Fas-mediated apoptosis.25
Rare patients with Fanconi anemia are phenotypically mosaic, meaning that one population of cells from such a patient is hypersensitive to DNA cross-linking agents while another subpopulation is normally resistant.26 Analysis of mosaicism in FA patients indicates that spontaneous intragenic mitotic recombination and/or gene conversion occurred, resulting in the loss of one pathogenic mutation; this genetic reversion may lead to an improved phenotype.27 Spontaneous reversion to normal from an inherited mutation has been observed in vivo in FA patients, with apparently improved hematopoiesis. Clonal hematopoiesis in these patients suggests that reversion occurred in single hematopoietic stem cell. Phenotypic reversion is not unique to FA but has also been reported in patients with adenosine deaminase deficiency28 and ataxia telangiectasia.29 This observation argues that FA gene-corrected cells may have a selective advantage in vivo.
Currently, a trial of hematopoietic progenitor cell transduction is being conducted at the Clinical Center of the National Institutes of Health for FA-C patients lacking a compatible BMT donor. The study thus far has confirmed that transfer of the FAC cDNA to multipotential progenitor cells is possible. Function of the wild-type FAC cDNA is suggested by the marked increase in progenitor numbers and MMC-resistant colonies with successive cycles of gene transduction in the three patients studied thus far. Coincident with this expansion in progenitor growth, two of the patients also had transient improvement in BM cellularity. These findings are encouraging regarding the potential utility of FA gene transfer in amelioration of the hematopoietic pathology. The FAA retroviral vectors described here should be evaluable in clinical trials designed on the basis of information learned from the FA-C trial.
ACKNOWLEDGMENT
We thank Dr Larry Arnold of the UNC Flow Cytometry and Janice Coleman of the UNC Cytogenetics Lab for their valued technical assistance. We thank Dr James Granfortuna (Moses Cone Hospital), Dr Sherri Zimmerman (Duke University Medical Center), and Dr John Bergsagel (Scottish Rite Children's Medical Center) for their assistance. We also thank Dr R. Jude Samulski and the UNC Gene Therapy Center for technical and material assistance. We thank the editorial assistance of Lee Hodge in preparing this paper.
Supported in part through the Leukemia Society Translational Research Grant No. 6229-96. C.E.W. is a recipient of the Lucille B. Markey Charitable Trust.
Address reprint requests to Christopher E. Walsh, MD, PhD, Room 7101 Thurston-Bowles Bldg, CB#7352, Chapel Hill, NC 27599.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal