Abstract
Human CD34+ selected cells are able to reconstitute hematopoiesis in patients receiving a myeloablative treatment. Although the role of reinfused tumor cells contaminating the grafts on the determination of postautograft relapses remains unclear, the major interest of CD34+ cell selection is to reduce the tumor contamination of the graft. This can be achieved if tumor cells do not express the CD34 antigen. We previously showed that this approach was effective with bone marrow (BM) collections in patients with non-Hodgkin's lymphoma (NHL). Because peripheral blood progenitor cells (PBPC) allow faster hematologic recovery than BM and are expected to contain less tumor contamination, we have compared the results of CD34+ cell selection in 35 BM and 16 PBPC from 48 patients with NHL. The PBPC were collected after a course of chemotherapy followed by granulocyte colony-stimulating factor (G-CSF ) administration. The data showed that the final CD34+ cell purity achieved with PBPC was higher than with BM (medians, 70% v 50%; P = .02). The CD34+ cell recovery was also better for PBPC (medians, 42% v 24%; P = .001). Tumor contamination was assessed by detection of BCL2/JH rearrangement using polymerase chain reaction (PCR) in 38 of 48 patients (22 BM, 16 PBPC). In addition, immunoglobulin heavy chain gene (IgH) rearrangements were investigated using PCR with consensus IgH primers. At harvesting, 10 of 22 BM and two of 16 PBPC contained BCL2/JH+ cells, one of 22 BM and 14 of 16 PBPC contained abnormal IgH+ cells (one PBPC contained both BCL2/JH+ and abnormal IgH+ cells) at harvesting. However, because lymphoma tissue specimens from patients at diagnosis were not available, the malignant character of IgH rearrangements could not be confirmed by sequencing and probing with allele-specific nucleotides. After CD34+ cell selection, a reduction to below the level of detection of BCL2/JH+ cells of BM and PBPC was effective in seven of 12 informative selections. In contrast, a reduction to below the level of detection of abnormal IgH+ cells was effective in only three of 15 informative selections. However, the detection of cells with an abnormal IgH pattern in the context of chemotherapy plus G-CSF progenitor mobilization in patients with NHL and its correlation with actual tumor contamination needs further investigation.
IT HAS NOW BEEN clearly demonstrated that human CD34+ selected cells are able to reconstitute hematopoiesis in patients receiving myeloablative treatments.1-5 Besides the better tolerance resulting from the reduction in the number of autologous cells and consequently the total volume of dimethyl sulfoxide (DMSO) reinfused to the patient, the major interest of the CD34+ cell selection is to reduce tumor contamination of the graft in cases where tumor cells do not express the CD34 antigen.
Based on previous data showing that lymphoma cells are not CD34+,6 we recently performed a pilot trial for the separation and transplantation of CD34+ cells selected from bone marrow (BM) in patients with non-Hodgkin's lymphoma (NHL).3 In this study, CD34+ cell selection performed by immunoabsorption (Ceprate SC; Cellpro Inc, Bothell, WA) led to a substantial reduction of the amount of BCL2/JH+ lymphoma cells in eight of nine initially contaminated BM samples. Patients infused with autologous CD34+-selected marrow cells recovered a 0.5 × 109/L neutrophil count 15 days after administration of the high-dose pretransplant regimen (BEAM [BCNU, etoposide, aracytine, melphalan]).
Peripheral blood progenitor cells (PBPC) allow faster hematologic recovery than BM and may contain fewer tumor cells.7,8 Therefore, our next step was to investigate CD34+ cell selection in PBPC from NHL patients using the same purification technique and to compare the results with those obtained with BM. Although the impact of tumor cell reinfusion with the autograft in the determination of subsequent relapse is not yet clearly established, there are indications that reinfusion of tumor cells to NHL patients is an adverse prognosis factor.7,9 10 In the present study, we also assessed tumor contamination by detection of BCL2/JH rearrangement. In addition, immunoglobulin heavy chain gene (IgH) rearrangements were investigated using polymerase chain reaction (PCR) with IgH consensus primers.
We report here on the results of CD34+ cell selections in 35 BM and 16 PBPC samples from 48 patients with NHL. The PBPC were collected after a course of chemotherapy followed by granulocyte colony-stimulating factor (G-CSF ) administration. The data show that the final CD34+ cell purity achieved with PBPC was higher than with BM (70% v 50%; P = .02). The recovery of CD34+ cells was also better with PBPC (42% v 24%; P = .001). At harvesting, 10 of 22 BM and two of 16 PBPC contained BCL2/JH+ cells, one of 22 BM and 14 of 16 PBPC contained abnormal IgH+ cells (one PBPC contained both BCL2/JH+ and abnormal IgH+ cells) at harvesting. However, because lymphoma tissue specimens from patients at diagnosis were not available, the malignant character of the observed IgH rearrangements could not be confirmed by sequencing and probing with allele-specific nucleotides.
After selection of CD34+ cells, BCL2/JH+ cells were reduced to below the level of detection in seven of 10 BM, but not in the two PBPC positive products; in contrast, abnormal IgH+ cells persisted in one of one BM and in 11 of 14 PBPC samples (one with both BCL2/JH+ and abnormal IgH+ cells). The discrepancy between tumor cell depletion efficiency as evaluated by BCL2/JH+ and by abnormal IgH+ cells is a matter of concern. However, because samples of the initial lymphoma tumor were not available, it is unclear whether the detection of an abnormal IgH in the context of chemotherapy plus G-CSF progenitor mobilization in patients with NHL was due to tumor cell contamination.
MATERIALS AND METHODS
Patients.A total of 48 patients included in this study were diagnosed with NHL (31 men and 17 women with a median age of 46 years (range, 27 to 60). Forty-three patients were stage III/IV and 5 were stage I/II; 35 patients had low-grade NHL (32 follicular and 3 diffuse histology) and 13 had high- or intermediate-grade NHL. Twenty-nine had BM involvement at diagnosis by conventional histopathology. At the time of stem cell collection, 16 of the patients were in complete remission, 26 were in partial remission, and 6 had progressive disease; three still had BM involvement detected by histopathology. The BCL2/JH and IgH rearrangement status of the BM and the blood of almost all of the patients was not known at diagnosis.
The patients had either their BM or their PBPC collected for further autografting. The proportion of follicular lymphomas was similar in the two groups (21 of 32 in BM, 11 of 16 in PBPC). They were not assigned at random to either the BM or the PBPC group. We first did a series of BM collections in 32 consecutive patients. For 3 of 32, the selected fractions contained insufficient numbers of CD34+ cells to perform a transplant. Therefore, these three patients had cells collected a second time. Thus, a total of 35 BM were collected. The first 10 BM collections (10 patients) were not tested for BCL2/JH and IgH rearrangements because the techniques of detection were not available at that time in our institution. The subsequent 22 patients (nos. 11 to 32) had their BM cells tested for both rearrangements.
A second series of 16 consecutive patients had PBPC collections. Eleven had follicular lymphoma and five high-grade lymphoma. All were tested for the BCL2/JH and IgH rearrangements.
BM processing.A total of 35 BM were harvested from 32 patients under general anesthesia from the posterior iliac crests using modified 12-gauge Rosenthal aspiration needles. A buffy-coat (BC) concentrate was prepared by centrifugation of the whole marrow at 3,000 rpm for 5 minutes using a Cobe 2991 Blood Cell Processor (Cobe Laboratories, Lakewood, CO). The BC was washed on the Cobe Processor with 1 L of phosphate-buffered saline (PBS) (Braun Laboratories-92; Boulogne, France). An aliquot containing 0.5 × 108 nucleated cells per kilogram of body weight (kg bw) was removed and cryopreserved as a back-up marrow fraction to serve in case of engraftment failure of the selected cells. The remaining BC cells were incubated for 25 minutes at room temperature with 3 mg of the biotinylated anti-CD34 monoclonal antibody (MoAb) 12.8 (CellPro Inc) in 150 mL of PBS containing 1% autologous plasma or, more recently, 0.1% human serum albumin (HSA; LFB, Orsay, France). The treated cells were washed with PBS on the Cobe Processor to remove any unbound antibody. This fraction, in a volume of 300 mL, was passed through the Ceprate SC Stem Cell Concentrator (CellPro Inc), which contains a sterile column of avidin-coated polyacrylamide beads. After washing with 300 mL of PBS, CD34+ cells were removed from the beads by gentle mechanical agitation and eluted with 90 mL of PBS containing 10 U/mL of heparin and 4 mL of HSA. After centrifugation for 10 minutes at 500g, the cells were resuspended in PBS containing 7.5% dimethyl sulfoxide (Braun Laboratories) and 2% HSA to a final volume of 4.5 mL. The cells were then cryopreserved in at least two 5-mL vials (Nalgene, Redmond, WA), using a controlled-rate freezing method and stored at −196°C.
PBPC processing.PBPC were collected by two leukaphereses from 16 patients using a Cobe Spectra apheresis machine. PBPC were collected during hematopoietic recovery after treatment with ACVBP (Adriamycin:75 mg/m2, day 1; Endoxan: 1,200 mg/m2, day 1; Eldiesine: 2 mg/m2, day 1 and day 5; Bleomycin: 10 mg, day 1 and day 5; Prednisolone: 60 mg/m2, from day 1 to day 5) plus G-CSF (5 mg/kg/d). For each apheresis, approximately 2.5 blood volumes were processed for 3 hours through a dual-lumen central venous catheter resulting in approximately a 200-mL collection. The first apheresis was performed in the afternoon of the day when patients recovered 5 × 109 leucocytes/L and the second in the morning of the following day. The two apheresis products were mixed and a BC was prepared using the Cobe 2991. The PBPC CD34+ cells were isolated with the Ceprate SC device and cryopreserved using the same procedure described above for marrow.
Flow cytometry analysis.The initial BC and CD34-selected fractions were studied. MoAbs: fluorescein isothiocyanate (FITC)- and phycoerythrin (PE)-anti CD34 (8G12), IgG1, PE-anti–CD33 (P67.6) IgG1, FITC-anti–HLA-DR (L243) IgG2a, PE-anti–CD38 (HB-7) IgG1, FITC-anti–CD2 (S5.2) IgG2a, and FITC-anti–CD19 (4G7 and S125C1) IgGa1 were purchased from Becton Dickinson (BD; Le Pont de Claix, France). PE-anti–CD117 (95C3) IgG1 was purchased from Immunotech (Marseille, France). Mouse IgGa1-FITC/IgG2a–PE (simulitests, BD) were used as isotype controls.
A direct double staining procedure with the conjugated MoAbs was used to evaluate the following phenotypes: CD34+/CD33−, CD34+/CD38−, CD34+/CD117+, CD34+/HLA-DR− and CD34+/CD19+. Approximately, 5 × 105 cells from BC and CD34-selected fractions were incubated for 30 minutes at 4°C with 10 μL MoAbs (except for CD117, 20 μL), in 100 μL PBS. Erythroid cells from BC fraction were lysed in 1 mL fluorescence-activated cell sorting (FACS) lysis solution for 5 minutes. The cells from both fractions were washed twice in PBS, 1% bovine serum albumin (BSA; Sigma, Saint Quentin Fallavier, France) and then fixed in 500 μL PBS with 1% paraformaldehyde.
The flow cytometric analysis of cells was performed with a FACSort (BD) coupled with CellQuest software (BD). At least 20,000 cells were analyzed. The morphologic forward and side scatter parameters allowed exclusion of debris from analysis. The percentage of labeled cells in the two fractions was then obtained using dual parameter FL1 or FL2 staining together with side scatter gating analysis.
The initial BC could reliably be analyzed only for the percentage of CD34+ cells due to the low concentration of this specific cell population.
Hematopoietic culture assays.Two types of culture assays were used to quantitate late and early progenitors.
The assay for early colony-forming unit–granulocyte-macrophage (CFU-GM) was performed according to our previously described technique.11 Briefly, 5 × 104 mononuclear cells (MNC) were plated in 35-mm petri dishes in 1 mL aliquots of Iscove's modified Dulbecco's medium (IMDM) containing 30% fetal calf serum (FCS) (GIBCO, Cergy Pontoise, France), detoxified BSA 10 mg/mL and 0.92% methylcellulose (Flucka, Buchs, Switzerland). Cultures were stimulated with a mixture of human recombinant growth factors: interleukin-3 (IL-3, 10 ng/mL), G-CSF (10 ng/mL), granulocyte-macrophage colony-stimulating factor (GM-CSF, 10 ng/mL), and stem cell factor (SCF, 100 ng/mL). Dishes were incubated at 37°C in a humidified 5% CO2 atmosphere. Three dishes were plated for each culture. Granulocyte-macrophage colonies (CFU-GM: more than 50 cells) were scored on day 14.
The assay for late CFU-GM was performed in agar according to the technique of Pike and Robinson.12 The basic medium was McCoy's 5A medium without serum (GIBCO-Biocult) supplemented with 30% heat-inactivated FCS. Colony-stimulating activity (CSA) was supplied by 10% human placenta conditioned medium. BC and CD34+ selected cells were seeded at 5 × 104/mL and 103/mL, respectively in medium containing equal volumes of 0.6% agar and 2× McCoy's 5A medium to achieve a final serum concentration of 15%. Three 35 × 10-mm petri dishes (Greiner, Frickenhausen, Germany) were plated for each assay. The cultures were incubated for 10 days at 37°C in a humidified 5% CO2 atmosphere. The CFU-GM were scored on day 10.
Study of minimal residual disease (MRD) by PCR detection of IgH and BCL2/JH rearrangements.For molecular studies, the same number of cells, obtained from CD34+ and CD34− cell fractions and from unseparated cell fractions, was used. High molecular weight DNA was extracted according to standard methods.13
For IgH rearrangement analysis, PCR was performed in a 50 μL final volume using 1 μg of DNA, 600 nmol/L oligonucleotide primers, 200 mmol/L each of deoxyadenosine triphosphate (dATP), deoxycytidine triphosphate (dCTP), deoxyguanosine triphosphate (dGTP), 400 mmol/L of deoxyuridine triphosphate (dUTP), 2.5 U Taq polymerase (Cetus, Emeryville, CA), MgCl2 mmol/L in PCR buffer (500 mmol/L KCl, 100 mmol/L Tris HCl, pH 8.3). One unit of the enzyme Uracyl DNA glycosylase (UDG) was also added to avoid carryover contamination from previous PCR products.14 The amplification was performed for 40 cycles in a Perkin Elmer Cetus thermal cycler with the consensus oligonucleotides 5′-ACCTGAGGAGACGGTGACC-3′ (FR4) located on the 3′ end of the JH segment and 5′- ACACGGC(CT)(GC)TGTATTACTGT-3′ (FR3) located on the 3′ end of FR3 of the VH segment.15 Aliquots of the PCR together with a molecular weight marker (FX174RF/HaeIII; GIBCO-BRL, Cergy Pontoise, France) were analyzed by vertical electrophoresis in 6% polyacrylamide gel in Tris borate electrophoresis buffer. The size of the expected band was 80 to 110 bp. The sensitivity of this PCR technique, determined by serial dilutions of a IgH-positive B-acute lymphoblastic leukemia (B-ALL) cell line in normal BM mononuclear cells followed by subsequent extraction of DNA, was approximately 5 in 103 cells for a final number of 106 cells.
In 17 cases (8 BM, 9 PBPC), independently extracted DNA samples were amplified as above using a 5′ fluorescein-labeled FR3 consensus primer and a FR4 consensus primer. This technique modified from Shiach et al16 allows analysis of PCR products with a fluorescent automated laser DNA sequencer (A.L.F., Pharmacia LKB Biotechnology, Uppsala, Sweden). Electrophoresis was achieved on a 6% long ranger gel (Bioprobe, Paris, France) containing 8 mol/L urea in a 0.6× Tris-borate-EDTA buffer. Areas of the peaks were determined using the integration software Smart Manager (Pharmacia).
The amplification of the BCL2/JH rearrangement at the major breakpoint region (MBR) was performed by nested PCR according to a technique previously described by us and others.3 17 For each experiment, a positive control consisting of DNA from RL cells, a lymphoma cell line with a t(14; 18) translocation and a negative control consisting of the PCR buffer with heat-inactivated proteinase K was performed. Each sample was analyzed at least twice. The sensitivity of the nested PCR, determined by serial dilutions of BCL2/JH+ RL cells in normal BM mononuclear cells followed by subsequent extraction of DNA, was approximately 1 in 105 cells for a final number of 106 cells. No increase in sensitivity was obtained by hybridization with an internal probe.
Statistical analysis.Results of cell processing are given as medians and ranges. The statistical significance of observed differences between BM and PBPC was assessed using the Mann-Whitney U-test. Correlations between initial CD34+ cell content and final CD34+ cell purity of BM and PBPC after processing were calculated using the Spearman correlation test.
RESULTS
Cell Recovery
BM.A total of 35 BMs from 32 patients were collected (patients 18, 24, and 27 were harvested twice). After removing a back-up marrow containing 0.5 × 108 nucleated cells/kg bw, a median of 2.1 × 1010 nucleated cells (corresponding to 3.1 × 108 nucleated cells/kg bw) containing 0.9% (range, 0.2 to 7.2) CD34+ cells were incubated with 3 mg of the 12.8 anti-CD34 antibody and then processed on the Ceprate SC column (Table 1). The adsorbed CD34+ selected cell fraction contained 0.5% (range, 0.1% to 1.3%) of the original nucleated cells, with a CD34+ cell purity of 50% (range, 13 to 81). A median of 0.7 × 106 CD34+ cell/kg bw (range, 0.1 to 3.0) was present in the cryopreserved selected fractions. This corresponded to a 24% (range, 4 to 76) CD34+ cell median recovery. The median enrichment produced by the column (final/initial % CD34+) was 45-fold (range, 9 to 140). The CD34+ cell adsorbed fraction contained a median of 2.6 × 104 late CFU-GM/kg (50% median recovery; range, 4 to 150) and a median of 2.8 × 104 early CFU-GM/kg (49% median recovery; range, 6 to 135).
Characteristics of BC and CD34+ Cell Selected Fractions (medians and ranges) of BM and PBPC From NHL Patients
| . | BM (n = 35) . | PBPC (n = 16) . | ||
|---|---|---|---|---|
| . | Median . | Range . | Median . | Range . |
| BC | ||||
| NC (×1010) | 92.1 | (1-5.2) | 2.4 | (1.2-5.5) |
| % CD34+ | 0.9% | (0.2-7.2) | 1.1% | (0.4-2.8) |
| Selected fractions | ||||
| NC/kg (×106) | 1.6 | (0.3-4.1) | 3.1 | (0.7-7.5) |
| NC recovery | 0.5% | (0.1-1.3) | 0.9% | (0.3-3.1) |
| % CD34+ | 50% | (13-81) | 70% | (16-99) |
| Enrichment | ×45 | (9-140) | ×70 | (12-123) |
| CD34+ cell/kg (×106) | 0.7 | (0.1-3.0) | 2.0 | (0.1-6.6) |
| . | BM (n = 35) . | PBPC (n = 16) . | ||
|---|---|---|---|---|
| . | Median . | Range . | Median . | Range . |
| BC | ||||
| NC (×1010) | 92.1 | (1-5.2) | 2.4 | (1.2-5.5) |
| % CD34+ | 0.9% | (0.2-7.2) | 1.1% | (0.4-2.8) |
| Selected fractions | ||||
| NC/kg (×106) | 1.6 | (0.3-4.1) | 3.1 | (0.7-7.5) |
| NC recovery | 0.5% | (0.1-1.3) | 0.9% | (0.3-3.1) |
| % CD34+ | 50% | (13-81) | 70% | (16-99) |
| Enrichment | ×45 | (9-140) | ×70 | (12-123) |
| CD34+ cell/kg (×106) | 0.7 | (0.1-3.0) | 2.0 | (0.1-6.6) |
Abbreviations: NC, nucleated cells; BC, buffy coats.
PBPC.Two leukaphereses were collected on 2 consecutive days from 16 patients. They were mixed and a BC prepared and processed on the Ceprate SC column. The BC contained a median of 2.4 × 1010 nucleated cells (corresponding to 3.8 × 108 nucleated cells/kg bw) with 1.1% (range, 0.4 to 2.8) CD34+ cell content (Table 1). After Ceprate SC processing, a total of 0.9% (range, 0.3% to 3.1%) of the cells were recovered in the adsorbed fraction, with 70% (range, 16% to 99%) CD34+ cell purity. From the 16 processings, only three resulted in a final purity less than 50%. The median enrichment was 70-fold (12 to 123). The CD34+ cell recovery was 42% (range, 7 to 135). The cryopreserved grafts contained 2.0 × 106 CD34+ cell/kg bw (0.1 to 6.6) and medians of 0.6 × 104 late CFU-GM/kg bw and 11.1 × 104 early CFU-GM/kg bw. The mean recoveries post-Ceprate SC were only 14% and 28% for late and early CFU-GM, respectively.
BM versus PBPC.Taken together (BM and PBSC), the data on CD34+ cell selection showed a correlation (P = .003) between the final CD34+ cell purity and the initial CD34+ cell content of the processed BCs (Fig 1) : the higher the initial % CD34+ cells, the higher the final purity. The CD34+ cell recovery was higher in PBPC than in marrow (42% v 24%; P = .001). In contrast, the median recoveries post-Ceprate SC were lower in PBPC than in BM for late and early CFU-GM (P = .001 and P = .05, respectively). When late and early CFU-GM were plotted together (Fig 2), the slopes of the curves were different for BM and PBPC selected fractions. These data suggest that CD34 cells selected from BM and PBPC products were different.
Correlation between CD34+ cell content (%) in BC and CD34+ cell selected fractions from BM (•) and PBPC (▴) samples.
Correlation between CD34+ cell content (%) in BC and CD34+ cell selected fractions from BM (•) and PBPC (▴) samples.
Correlation between CD34+ cell recovery and early CFU-GM recovery in BM (•) and PBPC (▴) selected fractions.
Correlation between CD34+ cell recovery and early CFU-GM recovery in BM (•) and PBPC (▴) selected fractions.
Flow Cytometry Analysis
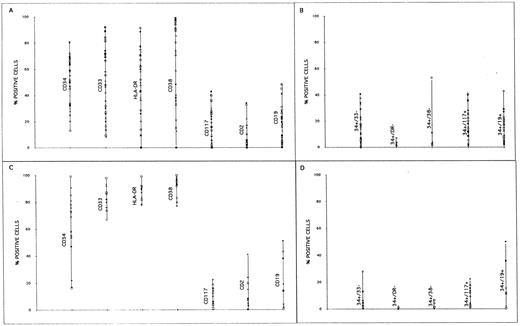
The immunophenotyping analysis of the CD34+ cell selected fractions either from BM or PBSC showed that they were highly heterogeneous for the CD33, CD38, HLA-DR, CD117, CD2, and CD19 antigens (Fig 3). The CD34+ cell selected fractions from PBPC contained a higher proportion of HLA-DR+, CD38+, and CD33+ cells than those from BM products. The median proportion of CD19+ cells was 14% (range, 1 to 43) and 2% (range, 0.4 to 51) in fractions selected from BM and PBPC, respectively, with medians of 14% (1 to 43) and 1.6% (0.3 to 5.0) of CD34+/CD19+ cells for BM and PBPC, respectively. It is noteworthy that in several instances almost all CD19+ cells were CD34+.
Immunophenotype profiles of CD34+ cell fractions selected from BM (A and B) and PBPC (C and D).
Immunophenotype profiles of CD34+ cell fractions selected from BM (A and B) and PBPC (C and D).
The double staining showed that the proportion of CD34+/CD33−, CD34+/CD38−, and CD34+/HLA-DR− cells (corresponding to the more immature cells) was low both in BM (respective medians 15%, 0.7%, 1.0%) and PBPC (respective medians, 3.3%, 0.3%, 1.3%).
Evaluation of MRD
The MRD in BM and PBPC before and after Ceprate SC selection was assessed in 38 of 48 patients by detection of cells containing the BCL2/JH rearrangement (within the major breakpoint region of the BCL2). Abnormal IgH rearrangement patterns in the FR3/FR4 region were also investigated by detection of monoclonal or oligoclonal IgH bands using electrophoresis.
Detection of MRD before CD34 Selection
BM.From the 32 patients whose BM was harvested, 22 (18 of 22 with follicular lymphoma) were tested for the presence of cells carrying BCL2/JH and/or IgH rearrangements. At harvesting, 10 of 22 had BCL2/JH+ cells in the BM and 1 of 22 had abnormal IgH+ cells in the BM (Table 2 and Figs 4 and 5). It is noteworthy that among these 11 patients with positive MRD (BCL2/JH or IgH rearrangement) in BM, 10 of 11 had a follicular lymphoma with BM involvement at diagnosis as detected by histopathology (Table 2).
MRD as Assessed by Detection of Cells Bearing a BCL2 and/or a IgH Gene Rearrangement on BM (patients 11 to 32) and PBPC (patients 34 to 48) Before (buffy coat) and After CD34 Selection (selected)
| Patient No. . | Histology . | BM Involvement at Diagnosis . | MRD . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | BCL2/JH . | IgH . | Phenotype . | . | |||||||||
| . | . | . | BC . | Selected . | BC . | Selected . | BC . | Selected . | . | . | . | . | |||
| . | . | . | . | . | . | . | CD34 . | CD19 . | CD34 . | CD19 . | CD34+/19+ . | . | . | . | . |
| . | . | . | . | . | . | . | (%) . | (%) . | (%) . | (%) . | (%) . | . | . | . | . |
| 11 | F | + | + | − | − | − | 1.8 | 1.5 | 40 | 41.2 | 14.0 | ||||
| 14 | F | + | + | + | − | − | 2.2 | 2.0 | 81 | 3.0 | 2.2 | ||||
| 15 | F | + | + | − | − | − | 0.7 | 10.0 | 32 | 7.4 | 7.0 | ||||
| 19 | F | + | + | − | − | − | 0.5 | 0.4 | 28 | 6.1 | 5.4 | ||||
| 20 | F | + | + | − | − | − | 0.7 | 1.0 | 34 | 3.0 | 2.9 | ||||
| 22 | F | + | + | − | − | − | 0.8 | 0.8 | 68 | 1.0 | ND | ||||
| 23 | DIFF | − | + | − | − | − | 0.9 | 2.3 | 59 | 23.5 | 21.1 | ||||
| 24 | F | + | + | − | − | − | 0.5 | 1.7 | 40 | 9.0 | 9.0 | ||||
| 25 | F | + | − | − | + | + | 1.4 | 4.2 | 68 | 17.9 | 15.0 | ||||
| 28 | F | − | + | + | − | − | 1.1 | ND | 51 | 48.3 | 43 | ||||
| 32 | F | + | + | + | − | − | 0.6 | 0.3 | 69 | ND | ND | ||||
| 34 | F | + | − | − | + | − | 0.8 | 6.0 | 16 | 14.0 | 11.7 | ||||
| 35 | HG | + | − | − | + | + | 2.0 | 0.9 | 81 | ND | ND | ||||
| 36 | F | + | − | − | + | − | 0.6 | 0.6 | 55 | 43.0 | 11.0 | ||||
| 37 | HG | − | − | − | + | + | 2.2 | 0.3 | 76 | 0.4 | 0.3 | ||||
| 38 | F | + | − | − | + | + | 0.4 | 1.9 | 54 | 2.0 | 1.6 | ||||
| 39 | F | + | − | − | + | + | 1.3 | 4.0 | 58 | 51.0 | 50.0 | ||||
| 40 | HG | − | − | − | + | + | 1.2 | ND | 78 | ND | 1.5 | ||||
| 41 | F | + | − | − | + | − | 1.1 | 0.9 | 85 | 1.0 | 0.6 | ||||
| 42 | F | + | − | − | + | + | 0.8 | 12.5 | 69 | 4.0 | 1.0 | ||||
| 43 | F | + | − | − | + | + | 1.1 | 0.8 | 72 | 1.4 | 1.1 | ||||
| 44 | F | + | − | − | + | + | 1.0 | 2.0 | 91 | 2.0 | 2.0 | ||||
| 45 | F | + | + | + | − | − | 1.4 | ND | 17 | 14.0 | ND | ||||
| 46 | HG | − | − | − | + | + | 0.8 | 1.3 | 73 | 2.0 | 2.0 | ||||
| 47 | F | + | + | + | + | + | 0.7 | 1.1 | 99 | 0.4 | 0.3 | ||||
| 48 | F | + | − | − | + | + | 0.3 | 8.0 | 22 | 19.0 | 15.5 | ||||
| Patient No. . | Histology . | BM Involvement at Diagnosis . | MRD . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | BCL2/JH . | IgH . | Phenotype . | . | |||||||||
| . | . | . | BC . | Selected . | BC . | Selected . | BC . | Selected . | . | . | . | . | |||
| . | . | . | . | . | . | . | CD34 . | CD19 . | CD34 . | CD19 . | CD34+/19+ . | . | . | . | . |
| . | . | . | . | . | . | . | (%) . | (%) . | (%) . | (%) . | (%) . | . | . | . | . |
| 11 | F | + | + | − | − | − | 1.8 | 1.5 | 40 | 41.2 | 14.0 | ||||
| 14 | F | + | + | + | − | − | 2.2 | 2.0 | 81 | 3.0 | 2.2 | ||||
| 15 | F | + | + | − | − | − | 0.7 | 10.0 | 32 | 7.4 | 7.0 | ||||
| 19 | F | + | + | − | − | − | 0.5 | 0.4 | 28 | 6.1 | 5.4 | ||||
| 20 | F | + | + | − | − | − | 0.7 | 1.0 | 34 | 3.0 | 2.9 | ||||
| 22 | F | + | + | − | − | − | 0.8 | 0.8 | 68 | 1.0 | ND | ||||
| 23 | DIFF | − | + | − | − | − | 0.9 | 2.3 | 59 | 23.5 | 21.1 | ||||
| 24 | F | + | + | − | − | − | 0.5 | 1.7 | 40 | 9.0 | 9.0 | ||||
| 25 | F | + | − | − | + | + | 1.4 | 4.2 | 68 | 17.9 | 15.0 | ||||
| 28 | F | − | + | + | − | − | 1.1 | ND | 51 | 48.3 | 43 | ||||
| 32 | F | + | + | + | − | − | 0.6 | 0.3 | 69 | ND | ND | ||||
| 34 | F | + | − | − | + | − | 0.8 | 6.0 | 16 | 14.0 | 11.7 | ||||
| 35 | HG | + | − | − | + | + | 2.0 | 0.9 | 81 | ND | ND | ||||
| 36 | F | + | − | − | + | − | 0.6 | 0.6 | 55 | 43.0 | 11.0 | ||||
| 37 | HG | − | − | − | + | + | 2.2 | 0.3 | 76 | 0.4 | 0.3 | ||||
| 38 | F | + | − | − | + | + | 0.4 | 1.9 | 54 | 2.0 | 1.6 | ||||
| 39 | F | + | − | − | + | + | 1.3 | 4.0 | 58 | 51.0 | 50.0 | ||||
| 40 | HG | − | − | − | + | + | 1.2 | ND | 78 | ND | 1.5 | ||||
| 41 | F | + | − | − | + | − | 1.1 | 0.9 | 85 | 1.0 | 0.6 | ||||
| 42 | F | + | − | − | + | + | 0.8 | 12.5 | 69 | 4.0 | 1.0 | ||||
| 43 | F | + | − | − | + | + | 1.1 | 0.8 | 72 | 1.4 | 1.1 | ||||
| 44 | F | + | − | − | + | + | 1.0 | 2.0 | 91 | 2.0 | 2.0 | ||||
| 45 | F | + | + | + | − | − | 1.4 | ND | 17 | 14.0 | ND | ||||
| 46 | HG | − | − | − | + | + | 0.8 | 1.3 | 73 | 2.0 | 2.0 | ||||
| 47 | F | + | + | + | + | + | 0.7 | 1.1 | 99 | 0.4 | 0.3 | ||||
| 48 | F | + | − | − | + | + | 0.3 | 8.0 | 22 | 19.0 | 15.5 | ||||
Abbreviations: MRD, minimal residual disease; F, follicular; HG, high grade; DIFF, diffuse; ND, not done.
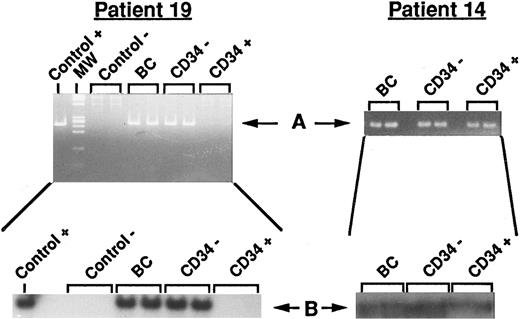
PCR detection of BCL2/JH rearrangement. Two representative results from unprocessed and processed BM are shown. Left panel: Data from patient 19 (disappearance of the BCL2/JH translocation in the CD34+ cell fraction). Right panel: Data from patient 14 (persistance of the BCL2/JH translocation in the CD34+ cell fraction). Control+, RL cells; Control−, normal PB lymphocytes; BC, DNA extracted from buffy coat (unprocessed BM); CD34−, DNA extracted from unadsorbed CD34− cell fraction; CD34+, DNA extracted from adsorbed CD34+ cell fraction; MW, molecular weight markers. Arrows show the specific BCL2/JH+ band.
PCR detection of BCL2/JH rearrangement. Two representative results from unprocessed and processed BM are shown. Left panel: Data from patient 19 (disappearance of the BCL2/JH translocation in the CD34+ cell fraction). Right panel: Data from patient 14 (persistance of the BCL2/JH translocation in the CD34+ cell fraction). Control+, RL cells; Control−, normal PB lymphocytes; BC, DNA extracted from buffy coat (unprocessed BM); CD34−, DNA extracted from unadsorbed CD34− cell fraction; CD34+, DNA extracted from adsorbed CD34+ cell fraction; MW, molecular weight markers. Arrows show the specific BCL2/JH+ band.
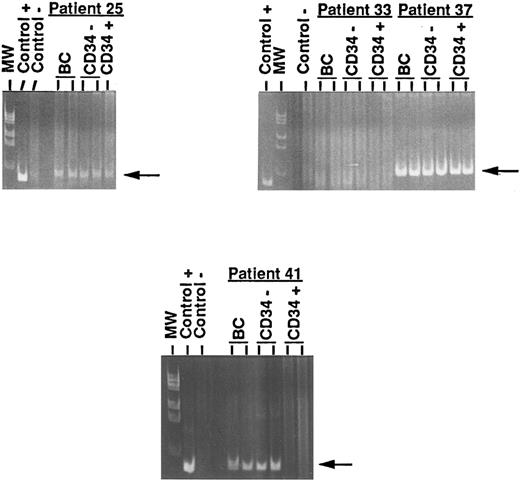
PCR detection of IgH rearrangement. Four representative results from unprocessed and processed BM and PBPC are shown. Left upper panel: Data from patient 25 (persistance of the IgH rearrangement in the CD34+ cell fraction of BM). Right upper panel: Data from patients 33 (negativity of the IgH rearrangement in the three fractions) and 37 (persistence of the IgH rearrangement in the CD34+ cell fraction of PBPC). Lower panel: Data from patient 41 (disappearance of the IgH rearrangement in the CD34+ cell fraction of PBPC). Control+, B-ALL cells; Control−, normal PB lymphocytes; BC, DNA extracted from buffy coat (unprocessed BM or unprocessed PBPC); CD34−, DNA extracted from the unadsorbed CD34− cell fraction; CD34+, DNA extracted from the adsorbed CD34+ cell fraction; MW, molecular weight markers. Arrows show the specific abnormal JH rearranged band.
PCR detection of IgH rearrangement. Four representative results from unprocessed and processed BM and PBPC are shown. Left upper panel: Data from patient 25 (persistance of the IgH rearrangement in the CD34+ cell fraction of BM). Right upper panel: Data from patients 33 (negativity of the IgH rearrangement in the three fractions) and 37 (persistence of the IgH rearrangement in the CD34+ cell fraction of PBPC). Lower panel: Data from patient 41 (disappearance of the IgH rearrangement in the CD34+ cell fraction of PBPC). Control+, B-ALL cells; Control−, normal PB lymphocytes; BC, DNA extracted from buffy coat (unprocessed BM or unprocessed PBPC); CD34−, DNA extracted from the unadsorbed CD34− cell fraction; CD34+, DNA extracted from the adsorbed CD34+ cell fraction; MW, molecular weight markers. Arrows show the specific abnormal JH rearranged band.
PBPC.All of the 16 PBPC products were tested for BCL2/JH and IgH rearrangements: at harvesting, one of 16 (patient 45) had BCL2/JH+ cells, 13 of 16 had abnormal IgH+ cells, and one of 16 (patient 47) had BCL2/JH+ and abnormal IgH+ cells (Table 2 and Figs 4 and 5) at harvesting. Only PBPC of patient 33 lacked both markers. These results indicate that a high proportion of patients had PBPC with an abnormal IgH pattern and hence potentially tumor contamination, despite the fact that the number of CD19+ cells (Table 2) was in the range as found in healthy donors.18 To exclude the possibility of false positive results due to hematopoietic disturbance related to G-CSF administration, we further analyzed, for IgH rearrangement, PBPC harvested after G-CSF mobilization from 8 patients with solid tumors (2 breast cancers, 2 Ewing sarcomas, 1 ovary cancer, 2 testis cancers, 1 retinoblastoma) and the blood of a healthy donor. All of these control samples were negative.
Analysis of IgH rearrangement was then performed by automated scanning in independently extracted DNA samples of 7 BM and 9 PBPC from NHL patients (when sufficient cells were available) and of 10 control products (the 8 patients with solid tumors, 1 PBPC and 1 BM from healthy donors). The results shown in Fig 6 indicate the presence of a clear monoclonal peak in 1 of 9 BM (patient 25) and 2 of 9 PBPC (patients 35 and 43) and a predominant peak in 5 of 9 PBPC of patients with NHL. All products were previously found to be positive using standard staining with ethidium bromide (Table 2). In contrast, the 6 of 7 BM and 2 of 9 PBPC (nos. 33 and 45) from patients with NHL and the control cells, which were all found negative with the standard technique, exhibited no predominant peak. Therefore, the results of automated scanning paralled those obtained by observation of photographs after ethidium bromide staining, suggesting the actual presence of cells with an abnormal IgH pattern in the samples from NHL patients.
IgH electrophoretograms produced by gene scanning software of PCR-amplified DNA extracts from unprocessed BM and PBPC products and from various control samples. Lines 1 to 8, samples 21, 22, and 24 to 28 from BM of NHL patients. Lines 3 and 4 corresponded to BM samples of patient 24 collected at a 3-month interval. Line 9, control BM of a healthy donor (HD). Lines 10 to 18, samples 33 to 35, 37, 39, 41 to 43, and 45 from PBPC of NHL patients. Lines 19 to 26, control samples from various solid tumors. Line 27, control blood from a healthy donor (HD). Abnormal peaks are indicated by an arrow (➭). Please note that in the ethidium bromide staining technique, samples 21, 22, 24, 26, 27, 28, 33, 45 and the controls were found negative, while samples 25, 34, 35, 37, 39, 41, 42, and 43 were found positive.
IgH electrophoretograms produced by gene scanning software of PCR-amplified DNA extracts from unprocessed BM and PBPC products and from various control samples. Lines 1 to 8, samples 21, 22, and 24 to 28 from BM of NHL patients. Lines 3 and 4 corresponded to BM samples of patient 24 collected at a 3-month interval. Line 9, control BM of a healthy donor (HD). Lines 10 to 18, samples 33 to 35, 37, 39, 41 to 43, and 45 from PBPC of NHL patients. Lines 19 to 26, control samples from various solid tumors. Line 27, control blood from a healthy donor (HD). Abnormal peaks are indicated by an arrow (➭). Please note that in the ethidium bromide staining technique, samples 21, 22, 24, 26, 27, 28, 33, 45 and the controls were found negative, while samples 25, 34, 35, 37, 39, 41, 42, and 43 were found positive.
Detection of MRD After CD34 Selection
After selection of CD34+ cells, BCL2/JH+ cells were still detected in 3 of 10 BM (patients nos. 14, 28, and 32) and in 2 of 2 initially positive PBPC (patients 45 and 47) (Table 2 and Figs 4 and 5). It is noteworthy that the BM of patient 24, which was harvested twice after a 3-month interval, was found both times to be BCL2/JH+ and twice negative after CD34 selection.
Abnormal IgH+ cells persisted in 1 of 1 BM (patient 25) and in 11 of 14 initially positive PBPC (1 with both BCL2/JH+ and abnormal IgH+ cells) (Table 2). It is also of interest that none of the samples that were MRD-negative at harvesting became positive after CD34+ cell selection, indicating that the processing did not result in an increased concentration of tumor cells.
In summary, CD34+ cell selection led to a reduction of BCL2/JH+ cells to below the level of detection in 7 of 12 (58%) of the initially positive BM and PBPC harvests. This reduction was not linked to the final CD34+ cell purity (Table 2). For instance, the BM of patient no. 19, which became BCL2/JH− after CD34+ cell selection, had a 28% CD34+ cell purity, while the BM of patient no. 32, which remained BCL2/JH+, had a 69% CD34+ cell purity. In contrast, a reduction to below the level of detection of abnormal IgH+ cells was not effective in 12 of 15 harvests (one BM, 11 PBPC).
DISCUSSION
PBPC have become the prevalent source of stem cells for hematologic rescue in patients given high-dose chemotherapy for the treatment of solid tumors and hematologic maligancies. The major advantage of PBPC transplants over BM is a faster hematologic recovery. Similarly, faster kinetics have been observed with CD34+ progenitors selected from PBPC as opposed to BM.1 4
In the present report, we compared the CD34+ cell selection achieved with the Ceprate SC device using BM and PBPC as CD34+ cell sources in patients with NHL. The data indicate that the final CD34+ cell purity was higher in PBPC selected fractions than in BM (70% v 50%), as well as the CD34+ cell recovery (42% v 24%). The heterogeneous results obtained in CD34+ cell purity with BM (range, 13% to 81%) were in part due to the low initial CD34+ cell concentration, but were also probably due to the regularly high hematocrit of the BM BC. We think that a high hematocrit combined with a low initial CD34+ cell content, likely impeded in some instances a good capture of CD34+ cells by the column. Indeed, in independent experiments, where we have performed a preliminary step of Ficoll-Hypaque separation on BM BC, the CD34+ cell purity of the selected fractions reached levels obtained with PBPC (M. Lopez, unpublished results, June 1995).
The content of the more immature cell populations (CD34+/CD33−, CD34+/HLA-DR−, CD34+/CD38−) in the selected fractions was low in both BM and PBPC samples. However, the median number of CD34+/CD117+ and CD34+/CD38− cells was significantly higher in BM than in PBPC selected fractions. It is noteworthy that in the selected CD34+ cell fractions, medians of 14% and 1.6% of CD34+/CD19+ cells remained present in BM and PBPC, respectively. However, there was no correlation between the proportion of these residual CD34+/CD19+ cells and the presence of cells with BCL2/JH marker in the selected fractions.
Surprisingly, the recoveries of early and late CFU-GM progenitors were higher with BM than with PBPC, in contrast with the CD34+ cell recoveries. This could mean that in selected PBPC, a proportion of CD34+ cells were not clonogenic and therefore not involved in the CFU-GM recoveries.
Concerning the tumor cell reduction in the BM by the CD34+ cell selection procedure, our results with the BCL2/JH detection were in agreement with our preliminary results: from 10 initially positive BM, only 3 remained positive after Ceprate SC processing. As expected from previous reports,7 8 the BCL2/JH+ cell contamination was lower in PBPC collections than in BM: at harvesting, 2 of 16 PBPC only contained BCL2/JH+ (1 with both BCL2/JH+ and IgH+ cells). Taking BM and PBPC products together, the BCL2/JH+ marker was present in 12 before Ceprate SC processing, and still found in 5 after processing. The BCL2/JH+ cell reduction was not correlated with the final CD34+ cell purity of the selected fractions.
Concerning the detection of cells bearing an abnormal IgH pattern, contrasting results were observed between BM and PBPC before CD34+ cell selection: only 1 of 25 BM, but 14 of 16 PBPC contained abnormal IgH+ cells. Because BM was collected in patients in steady state and PBPC were collected after a course of chemotherapy followed by G-CSF administration, we first hypothesized that we were dealing with false positivity emerging from a G-CSF disturbance of the hematopoietic equilibrium.19 Therefore, we tested samples from eight patients with solid tumors who received a course of chemotherapy followed by G-CSF and none were positive, suggesting that the presence of these abnormal cells is restricted to NHL samples. However, because lymphoma tissue specimens from patients at diagnosis were not available, the malignant character of the observed IgH rearrangements could not be confirmed by sequencing and probing with allele-specific nucleotides. In addition, we ignored whether these cells were already present before PBPC mobilization. Hence, a correlation between the detection of cells with an abnormal IgH pattern and the presence of tumor cells remains to be established. However, because a risk of mobilizing tumor cells into the circulation by chemotherapy and growth factor treatment was already shown for solid tumors and myelomas,20-22 the persistance of these abnormal cells in the majority of the CD34+ cells selected from PBPC raises concern. Indeed, it has been recently shown23 that after autologous BM transplantation (ABMT), eradication of PCR detectable IgH rearrangement is associated with decreased relapse in patients with NHL.
Whether lymphoma cells bear the CD34 antigen is still a matter of discussion. Berenson et al6 initially reported that lymphoma cells do not express CD34. But recently, Macintyre et al24 sorted CD34+/CD19+ and CD34+/CD19− cells from marrow samples of patients with follicular lymphoma at diagnosis or at progressive disease, with tumor contamination by histopathology and demonstrated by PCR that 3 of 3 of the CD34+/CD19+ samples were positive for BCL2/JH rearrangement, while 4 of 5 of the CD34+/CD19− samples were negative. Although their number of samples studied was low, this questionned the potentiality of CD34+ cell selection alone to deplete adequately lymphoma cells from BM or PBPC. However, in our experience, there was no correlation between the presence of tumor cells as detected by BCL2/JH rearrangement and the number of CD34+/CD19+ cells in the BM and PBPC products of NHL patients (the majority of them being in complete or partial remission at the time of harvesting, without BM involvement by histopathology).
In conclusion, our data indicate that CD34+ cell selection using an avidin-biotin immunoadsorption technique for hematopoietic stem cell products in NHL patients resulted in better CD34+ cell purity and recovery with PBPC. However, the significance of the high proportion of cells with an abnormal IgH pattern in PBPC and their persistance after CD34+ cell selection call for additional investigations to understand whether the detection of IgH rearrangement in the context of PB mobilization in NHL means either tumor contamination or, if so, tumor cell mobilization.
ACKNOWLEDGMENT
We thank Prof B. Granchamp and Dr H. Cavé for performing IgH rearrangement analysis using fluorescent automated laser DNA sequencing, D. Bardinet, C. Martinache, H. Nguyen, J. Rocsin, S Bouchet, M.C. Burland, C. Baillou, V. Jobard, and F. Siotto for their expert technical assistance.
Supported in part by Grant No. 930408 from INSERM to F.M.L.
Address reprint requests to M. Lopez, PhD, Inserm U76, Laboratoire d'Hématologie, CHU St Antoine, 27 rue de Chaligny, 75012, Paris, France.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal