Abstract
Myeloperoxidase (MPO) catalyzes a reaction between chloride and hydrogen peroxide to generate hypochlorous acid and other reactive compounds that have been linked to DNA damage. The MPO gene is expressed at high levels in normal myeloid precursors and in acute myeloid leukemias (AMLs) which are clonal derivatives of myeloid precursors that have lost the ability to differentiate into mature blood cells. Two MPO alleles differ at -463 G/A within a cluster of nuclear receptor binding sites in an Alu element. The -463 G creates a stronger SP1 binding site and retinoic acid (RA) response element (RARE) in the allele termed Sp. In this study, we investigate potential links between MPO genotype, MPO expression level, and myeloid leukemia. The SpSp MPO genotype is shown to correlate with increased MPO mRNA levels in primary myeloid leukemia cells. This higher-expressing SpSp genotype is further shown to be overrepresented in acute promyelocytic leukemia-M3 (APL-M3) and AML-M4, suggesting that higher levels of MPO are associated with an increased risk for this subset of leukemias.
THE MYELOPEROXIDASE (MPO) enzyme functions as an antimicrobial agent in neutrophils and monocytes by catalyzing the generation of hypochlorous acid and other reactive oxygen species.1,2 Expression of the MPO gene is restricted to the myeloid lineage and is highest in bone marrow precursors, peaking at the promyelocyte stage.3 MPO is also expressed in acute myeloid leukemia (AML) cells,4 which represent clonal expansions of myeloid precursors which have lost the ability to differentiate into mature blood cells. Two MPO alleles termed Sp and N (formerly AML and MPO) differ at one known position (-463 G/A) in an upstream Alu sequence within a cluster of hormone response elements (AluHREs).5,6 The AluHRE consists of four hexamer half-sites, related to the consensus nuclear receptor binding site AGGTCA,7 oriented as direct repeats spaced by two, four, and two basepairs (bps) (DR-2-4-2). The first DR-2 element contains the G/A base difference, which in the Sp allele creates the core binding site for the SP1 transcription factor (GGCGGG) and a perfect match for a 10-bp consensus SP1 binding site (TGAGGCGGGT v TGAGGCAGGT). This DR-2/SP1 element increases transcription of a reporter gene by 25-fold in cellular transfection assays, whereas the corresponding DR-2 element from the N allele is a less effective transactivator.5 The highest transactivation is observed in the presence of retinoic acid (RA) and cotransfected RA receptors (RAR and RXR). This RA response is apparently indirect, since RAR-RXR heterodimers synthesized in vitro do not bind this site in vitro. However, the 3′ DR-2 element does bind RAR-RXR and is a functional RA response element (RARE) in transfection assays in which these elements are linked to a reporter gene,5 whereas the central DR-4 element binds TR-RXR and is a functional thyroid hormone response element.
In the previous study, we noted that the SpSp homozygous genotype was overrepresented in 18 AML cases.5 The study had been prompted by a prior report that position -463 was a G residue in most cases of AML but was uniformly A in the normal population, suggesting that -463 G was a somatic mutation linked to leukemia.8 Our analysis of normal blood donors indicated that the -463 G/A base difference was instead an allelic polymorphism and that the G-containing Sp allele was predominant in the population. Nevertheless, the homozygous SpSp genotype was present in a higher proportion in AML cases than in normal donors.5 This overrepresentation of the allele with the stronger SP1 site and RARE initially suggested a possible link between MPO expression levels and AML.
In the normal myeloid lineage in the bone marrow, MPO is expressed in a narrow developmental window around the promyelocyte stage, and expression is sharply curtailed as these progenitors differentiate toward granulocyte or monocyte lineages.3 Most studies of MPO expression have used AML cell lines induced to differentiate in vitro, with accompanying loss of MPO gene expression.9-16 Consistent with those findings, peripheral blood neutrophils and monocytes lack detectable MPO mRNA,13 although the MPO protein continues to be stored at high levels in cytoplasmic lysosomes.17
The MPO gene encodes an 83-kD precursor polypeptide that is posttranslationally processed to yield 13.5-kD and 59-kD subunits.12,18 Two of each subunit are assembled with heme groups to produce the tetrameric enzyme. MPO catalyzes the reaction of chloride and hydrogen peroxide to yield hypochlorous acid (HOCl), a strong oxidant.1,2 In the presence of superoxide (O2−⋅), hypochlorous acid generates hydroxyl radicals,19 a highly reactive radical species. MPO also generates free chlorine (Cl2 ), which acts as a potent halogenating agent.20 MPO and its reactive by-products have been linked to DNA-strand breakage,21 generation of carcinogens,22,23 and inhibition of DNA repair.24 MPO has been linked to disease states mediated by later-stage myeloid cells, neutrophils, or monocytes/macrophages, including atherosclerosis,25 multiple sclerosis,26 and cystic fibrosis.27 These findings suggest that the antimicrobial functions of MPO are linked to inadvertent cytotoxic side effects, such as DNA damage, which could promote leukemogenesis.
AMLs are classified by the French-American-British (FAB) morphologic scheme as subtypes M0 through M6,28 approximating early to later stages of myeloid development, with M0 and M1 characterized as myeloblastic, M3 as promyelocytic, and M5 as monocytic leukemia. The highest level of MPO expression is seen in acute promyelocytic leukemia (APL-M3) and can be up to 20 times higher than in M1 or M5 subtypes.4 This indicates that myeloid-specific transcription factors essential for MPO transcription are present at the highest levels at the promyelocyte stage. Because the AluHRE with the SP1 and RARE is a non–cell type-specific enhancer, this element is unlikely to affect transcription in the absence of stage-specific factors required for MPO transcription. This predicts that the SpSp genotype, if linked to AML, might be more strongly linked to subtypes around the promyelocyte stage, such as APL-M3, that have the essential transcription factor background.
APL-M3 is the most homogeneous of the AML subtypes: almost all cases have undergone the reciprocal translocation t(15; 17) that interrupts the RARα gene, fusing it to the PML gene that encodes a putative zinc-finger transcription factor.29,30 The PML-RAR fusion protein retains the DNA-binding and ligand-binding domains of RARα and most of the PML protein, and is thought to be a dominant-negative oncoprotein that interferes with the expression of genes involved in normal myeloid differentiation.29 Disruption of the normal RA response appears to be key to the blockage of APL cell maturation: Treatment with all-trans-RA induces APL cells to differentiate into mature granulocytes,31 and accordingly, treatment of APL patients with RA results in complete remission in most cases.32 As further evidence that RARα function is important for normal myeloid differentiation, disruption of the RARα gene by another translocation, t(11; 17)(q23;q21), also leads to APL.33 One event induced by RA treatment of APL cells is rapid shutdown of MPO transcription.34 This raises the question of whether the Alu-encoded RARE, which differs in the Sp and N alleles, might be involved in RA-mediated regulation of MPO expression in leukemic versus differentiating cells.
In the current study, we analyzed MPO genotype in 46 AML cases classified by subtype to determine if the SpSp genotype is most strongly associated with subtypes that express significant levels of MPO. The findings indicate that the SpSp genotype is overrepresented in high MPO-expressing subtypes APL-M3 and -M4. We further show that primary AML cells of the SpSp genotype contain higher steady-state levels of MPO mRNA than SpN or NN genotypes. Overrepresentation of the higher-expressing MPO genotype with APL-M3 and -M4 implies that high MPO levels increase the incidence of these leukemias.
MATERIALS AND METHODS
Polymerase chain reaction for determination of MPO genotype of AML patients.Patients donated leukemia cells for research purposes with provision of informed consent and under the auspices of the Investigational Review Boards of the University of Pittsburg Medical Center and the University of Southern California Medical School. Cases were classified according to FAB criteria for morphology and histochemical staining.28 Most of the APL-M3 leukemias were also identified karyotypically by the t15; 17 translocation. Leukocytes were isolated from peripheral blood or bone marrow samples from AML patients by Ficoll-Hypaque (GIBCO-BRL, Gaithersburg, MD) density gradient centrifugation. DNA was isolated from the leukocytes as previously described.5 For determination of MPO genotype, polymerase chain reaction (PCR) was performed with 200 ng genomic DNA and 0.5 μg of each primer in a 50-μL reaction volume containing 50 mmol/L KCl, 10 mmol/L Tris hydrochloride, pH 8.3, 1.5 mmol/L MgCl2 , 200 μmol/L nucleotides, and 2.5 U Taq polymerase (Pharmacia, Piscataway, NJ). Nested primers (GIBCO-BRL) were synthesized to amplify a region extending from -829 to position +310 relative to the transcription start site. The first primer set was 5′ CTTGGTCCTGCGCCCACAGTCCCC 3′ and 5′ TCCCACCTTGGGAACTGTTACCTG 3′, and the second set was 5′ GCTGCCCATTGGGTGGCTGTTGGA 3′ and 5′ AGAGGGCTGGGGCGTGGCCAGAAT 3′. The cycling conditions were 94°C for 6 minutes followed by 30 cycles at 94°C for 1 minute, 55°C for 2 minutes, and 72°C for 2 minutes. Five microliters of the first reaction was used in a second PCR with the second primer set, internal to the first set. The resultant 1,148-bp PCR product (−829 to +319) was electrophoresed in agarose gels, purified, and sequenced directly (Sequenase kit; Amersham, Arlington Heights, IL). Each DNA sample was analyzed at least twice. For statistical analysis, the χ2 test was performed, and P values less than .05 were considered significant.
Reverse transcriptase-PCR of MPO mRNA sequences from primary AML cells.Frozen leukocytes isolated from AML blood samples were placed in culture in RPMI medium supplemented with 10% fetal calf serum, 2 mmol/L glutamine, and penicillin/streptomycin. After at least 2 days in culture, cells (approximately 105) were collected by centrifugation, and RNA was isolated by lysis in Trizol reagent (GIBCO-BRL). The RNA was used to make cDNA using reverse transcriptase (RT) and random hexamer primers (First strand synthesis kit; Pharmacia). PCR was performed with approximately 200 ng cDNA and 0.5 μg of each primer in a 50-μL reaction volume containing 50 mmol/L KCl, 10 mmol/L Tris hydrochloride, pH 8.3, 1.5 mmol/L MgCl2 , 200 mmol/L nucleotides, and 2.5 U Taq polymerase (Pharmacia). Nested primers (GIBCO) were synthesized to amplify a region extending from position +1 to +600 of the mRNA sequence in the first reaction and then from +30 to +583 in the second reaction. The first primer set was (1) 5′ AGCTGACAATATCAGGTGAGCTGTGG 3′ and (4) 5′ CAGTGACATTGAATGGCCT 3′. The second set was (2) 5′ TCCTTGGAAGCTGGATGACAGCAGCT 3′ and (3) 5′ TTCGCCACAGGGACCGCAGCTTCC 3′. Cycling conditions for the first set of primers were 94°C for 6 minutes followed by 10 cycles at 94°C for 1 minute, 55°C for 2 minutes, and 72°C for 2 minutes. Five microliters of the first PCR was used in a second PCR with the second, nested primer set for up to 25 cycles. In the second reaction, primer 3 was radiolabeled by polynucleotide kinase and γ 32P-ATP. The radiolabeled PCR product was electrophoresed in 5% polyacrylamide gels along with radiolabeled marker DNA fragments. The product of the second primer set was 553 bp if derived from mRNA and 1,067 bp if derived from genomic DNA. Relative cDNA concentrations for the various cDNA samples were determined using actin cDNA primers. To aid normalization between different PCR experiments, an internal cDNA standard prepared from NB4 cells was also included in all experiments. To ensure that the PCR was in the linear range of amplification, reactions were performed for 15, 20, and 25 cycles. Quantitation of relative amounts of radiolabeled PCR amplification products was performed with an Ambis radioisotope detector (Ambis, San Diego, CA).
Western blot analysis of MPO from AML cell extracts.AML cells in culture were collected and lysed in sodium dodecyl sulfate (SDS) containing buffer. Equal amounts of protein were electrophoresed on SDS-polyacrylamide gels, transferred to nylon membrane, reacted with rabbit polyclonal antibodies against human MPO (Dako, Carpinteria, CA), and visualized using the ECL system (Amersham). Quantitation was made by densitometric scans of the photographic films.
RESULTS
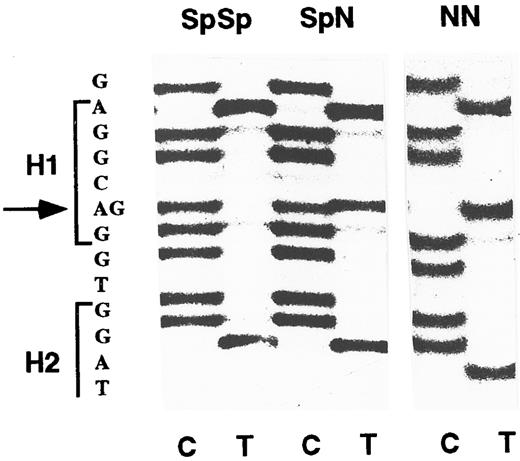
SpSp MPO genotype is overrepresented in APL-M3 and -M4 subtypes.To establish the ratio of Sp and N alleles in the normal population, we analyzed 111 DNA samples including 20 known Hispanic-American and 20 known Caucasian-American donors. The MPO upstream region was PCR-amplified, and the DNA sequence of the PCR product was directly determined. The -463 position appears as G in SpSp homozygotes, A in NN homozygotes, or both A and G at half-intensity in SpN heterozygotes (Fig 1). The results indicated that 61% of the general population are SpSp homozygotes, 33% are SpN heterozygotes, and 6% are NN homozygotes (Fig 2A), a distribution fitting the Hardy-Weinberg equilibrium. Hispanic and Caucasion ethnic-genetic subgroups were 60% to 63% SpSp, comparable to the overall population. Because of evidence of a higher incidence of APL in Hispanics,35 it was important to determine that the normal Hispanic population is not overrepresented by the SpSp genotype.
MPO genotypes determined by direct sequencing of PCR products. A DNA fragment including the -463 G/A base difference was amplified by PCR and directly sequenced using an internal primer. Positions of hexamers 1 and 2 (H1 and H2) are indicated in the sequence at left. The sequence of the opposite strand was determined using dideoxynucleotide C and T termination reactions as indicated at bottom. The SpSp homozygous genotype is identified by the G residue at -463 (arrow); the heterozygous SpN genotype has both G and A residues, and the homozygous NN genotype has only A.
MPO genotypes determined by direct sequencing of PCR products. A DNA fragment including the -463 G/A base difference was amplified by PCR and directly sequenced using an internal primer. Positions of hexamers 1 and 2 (H1 and H2) are indicated in the sequence at left. The sequence of the opposite strand was determined using dideoxynucleotide C and T termination reactions as indicated at bottom. The SpSp homozygous genotype is identified by the G residue at -463 (arrow); the heterozygous SpN genotype has both G and A residues, and the homozygous NN genotype has only A.
Schematic representation of MPO genotypes in the normal population and in AML subtypes. (A) Relative percentage of SpSp, SpN, and NN genotypes in 111 normal donors. (B) Relative number of SpSp, SpN heterozygotes, or NN homozygotes in the various AML subtypes. Cases are from Table 1. Subtypes M0 through M6 approximate early to late stages of myeloid differentiation. The highest proportions of the SpSp genotype are seen in APL-M3 and -M4 subtypes, which have high MPO expression.
Schematic representation of MPO genotypes in the normal population and in AML subtypes. (A) Relative percentage of SpSp, SpN, and NN genotypes in 111 normal donors. (B) Relative number of SpSp, SpN heterozygotes, or NN homozygotes in the various AML subtypes. Cases are from Table 1. Subtypes M0 through M6 approximate early to late stages of myeloid differentiation. The highest proportions of the SpSp genotype are seen in APL-M3 and -M4 subtypes, which have high MPO expression.
We previously found that the SpSp genotype was overrepresented in a sample of 18 AML cases, most of which had not been diagnosed as to subtype.5 To extend those findings, we analyzed 46 AML cases diagnosed as subtypes M0 to M6. These included 19 APL-M3, 11 APL-M4, 8 APL-M2 cases, and 8 cases of low MPO-expressing subtypes M0-1-5-6. Analysis of the MPO promoter sequence indicated that the SpSp genotype was overrepresented in high MPO-expressing subtypes APL-M3 and -M4; of 30 cases, 24 were SpSp, or 80% (P < .05; Table 1 and Fig 2B). APL-M3 cases were 79% SpSp (15 of 19 cases) and APL-M4 cases were 82% SpSp (9 of 11 cases). The eight cases categorized as low-expressing subtypes, M0, 1, 5, and 6, were 62% SpSp, equivalent to the normal population. The M2 subtype may be anomalous in that MPO expression is relatively high but the SpSp genotype was not overrepresented, and three of eight cases were the rare NN genotype. Examination of a greater number of M2 cases, especially the defined t8; 21 subset, will be necessary to draw conclusions. Another potentially interesting observation is that the female M3 to M4 cases appear to be more frequently of the SpSp genotype than males. As will be discussed, a female bias in SpSp association has been observed in multiple sclerosis (MS) cases.26
Overrepresentation of the SpSp Genotype in AML-M3 and -M4 Subtypes
| . | APL-M3 . | M4 . | M2 . | M0156 Low Expressors . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | Case No. . | Sex . | Karyotype . | Case No. . | Sex . | Karyotype . | Case No. . | Sex . | Karyotype . | Case No. . | Sex . | Karyotype . |
| SpSp | 801 | Male | N/D | 921 | Male | N/D | 1010 | Male | N/D | 3001 (M0) | Female | Normal |
| 803 | Male | t15; 17 | 933 | Male | Abnormal 7, 13, 14 | 1015 | Female | Normal | 3005 (M6) | Male | N/D | |
| 812 | Male | N/D | 935 | Female | N/D | 1020 | Female | N/D | 3010 (M5) | Male | Normal | |
| 823 | Male | t15; 17 | 940 | Male | Normal | 1027 | Male | inv 16 | 3020 (M1) | Male | Tetraploid | |
| 1637 | Male | t15; 17 | 947 | Male | Normal | 3030 (M1) | Female | N/D | ||||
| 1787 | Female | t15; 17 | 960 | Male | N/D | |||||||
| 1941 | Male | t15; 17 | 966 | Female | N/D | |||||||
| 2008 | Male | t15; 17 | 972 | Male | N/D | |||||||
| 2098 | Female | t15; 17 | 980 | Male | N/D | |||||||
| 2262 | Male | t15; 17 | ||||||||||
| 2312 | Female | t15; 17 | ||||||||||
| 2343 | Female | t15; 17 | ||||||||||
| 2354 | Male | t15; 17 | ||||||||||
| 2379 | Female | t15; 17 | ||||||||||
| 2429 | Male | t15; 17 | ||||||||||
| SpN | 2399 | Male | N/D | 927 | Male | Deletion 10.16 | 1030 | Female | N/D | 3034 (M1) | Male | Normal |
| 840 | Male | t15; 17 | 955 | Male | Trisomy 21 | 3038 (M5) | Male | Normal | ||||
| 3045 (M5) | Male | N/D | ||||||||||
| NN | 2394 | Male | t15; 17 | 1035 | Male | N/D | ||||||
| 1848 | Female | t15; 17 | 1038 | Male | t8; 21 | |||||||
| 1040 | Female | 46xx,i(14q) | ||||||||||
| . | APL-M3 . | M4 . | M2 . | M0156 Low Expressors . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | Case No. . | Sex . | Karyotype . | Case No. . | Sex . | Karyotype . | Case No. . | Sex . | Karyotype . | Case No. . | Sex . | Karyotype . |
| SpSp | 801 | Male | N/D | 921 | Male | N/D | 1010 | Male | N/D | 3001 (M0) | Female | Normal |
| 803 | Male | t15; 17 | 933 | Male | Abnormal 7, 13, 14 | 1015 | Female | Normal | 3005 (M6) | Male | N/D | |
| 812 | Male | N/D | 935 | Female | N/D | 1020 | Female | N/D | 3010 (M5) | Male | Normal | |
| 823 | Male | t15; 17 | 940 | Male | Normal | 1027 | Male | inv 16 | 3020 (M1) | Male | Tetraploid | |
| 1637 | Male | t15; 17 | 947 | Male | Normal | 3030 (M1) | Female | N/D | ||||
| 1787 | Female | t15; 17 | 960 | Male | N/D | |||||||
| 1941 | Male | t15; 17 | 966 | Female | N/D | |||||||
| 2008 | Male | t15; 17 | 972 | Male | N/D | |||||||
| 2098 | Female | t15; 17 | 980 | Male | N/D | |||||||
| 2262 | Male | t15; 17 | ||||||||||
| 2312 | Female | t15; 17 | ||||||||||
| 2343 | Female | t15; 17 | ||||||||||
| 2354 | Male | t15; 17 | ||||||||||
| 2379 | Female | t15; 17 | ||||||||||
| 2429 | Male | t15; 17 | ||||||||||
| SpN | 2399 | Male | N/D | 927 | Male | Deletion 10.16 | 1030 | Female | N/D | 3034 (M1) | Male | Normal |
| 840 | Male | t15; 17 | 955 | Male | Trisomy 21 | 3038 (M5) | Male | Normal | ||||
| 3045 (M5) | Male | N/D | ||||||||||
| NN | 2394 | Male | t15; 17 | 1035 | Male | N/D | ||||||
| 1848 | Female | t15; 17 | 1038 | Male | t8; 21 | |||||||
| 1040 | Female | 46xx,i(14q) | ||||||||||
| Total | Male | Female | Total No. | Male | Female | Total | Male | Female | Total | Male | Female | ||||
| SpSp (n) | 15 | 10 | 5 | 9 | 7 | 2 | 4 | 2 | 2 | 5 | 3 | 2 | |||
| SpN (n) | 2 | 2 | 0 | 2 | 2 | 0 | 1 | 0 | 1 | 3 | 3 | 0 | |||
| NN (n) | 2 | 1 | 1 | 0 | 0 | 0 | 3 | 2 | 1 | 0 | 0 | 0 | |||
| 19 | 11 | 8 | 8 | ||||||||||||
| %SpSp | 79 | 77 | 83 | 82 | 78 | 100 | 44 | 50 | 50 | 62 | 50 | 100 | |||
| Highest MPO | High MPO | High MPO | Low MPO | ||||||||||||
| Total | Male | Female | Total No. | Male | Female | Total | Male | Female | Total | Male | Female | ||||
| SpSp (n) | 15 | 10 | 5 | 9 | 7 | 2 | 4 | 2 | 2 | 5 | 3 | 2 | |||
| SpN (n) | 2 | 2 | 0 | 2 | 2 | 0 | 1 | 0 | 1 | 3 | 3 | 0 | |||
| NN (n) | 2 | 1 | 1 | 0 | 0 | 0 | 3 | 2 | 1 | 0 | 0 | 0 | |||
| 19 | 11 | 8 | 8 | ||||||||||||
| %SpSp | 79 | 77 | 83 | 82 | 78 | 100 | 44 | 50 | 50 | 62 | 50 | 100 | |||
| Highest MPO | High MPO | High MPO | Low MPO | ||||||||||||
APL-M3 cases with identification numbers between 1637 and 2429 were Hispanic.35 MPO genotypes are listed at left as SpSp homozygotes, SpN heterozygotes, or NN homozygotes. The percentage with the SpSp genotype is also shown. Most of the APL-M3 cases were karyotyped as t(15; 17) translocations. In subtypes other than APL, karyotypes listed as normal indicate that no karyotypic abnormality was detected. Approximate relative amounts of MPO gene expression indicated at bottom are from published studies.3,4 27
Abbreviation: ND, not determined.
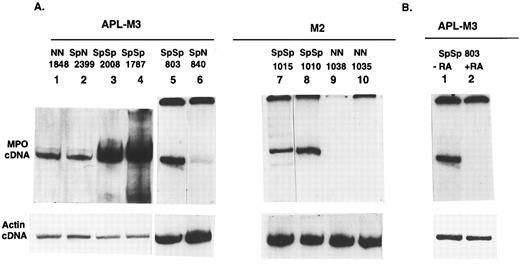
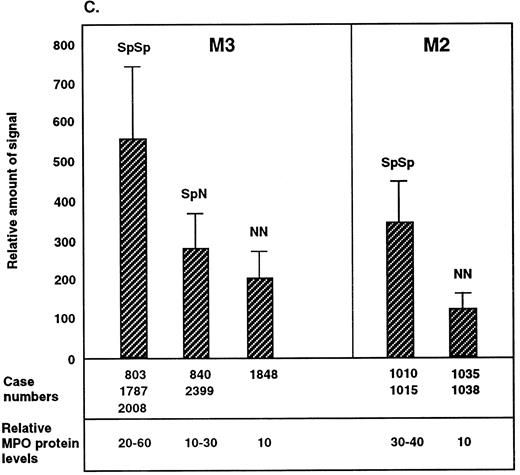
SpSp genotype is associated with higher levels of MPO mRNA expression.Overrepresentation of the SpSp genotype in APL-M3 and -M4 cases implies that this genotype increases the risk to develop these leukemias. One possible explanation is that the SpSp genotype is linked to higher levels of MPO gene expression, resulting in increased DNA damage, thereby increasing the risk of leukemia. There is reason to suspect that the Sp allele is linked to higher MPO expression: the G/A base difference in the AluHRE creates a stronger SP1 binding site and RARE in the Sp allele.5 To directly determine if the SpSp genotype is linked to higher MPO gene expression, we compared steady-state levels of MPO mRNA in primary AML cells of known MPO genotype. AML cells from donors listed in Table 1 were obtained as frozen aliquots and placed in culture medium to allow for cell recovery and RNA expression. Ten donor cell samples recovered, including six APL-M3 cases: three SpSp, two SpN, and one NN. Four M2 cases included two SpSp and two NN genotypes. M4 cells of SpSp and SpN genotypes did not survive in culture. RNA was isolated by Trizol reagent, and cDNA was prepared with RT (First strand kit; Pharmacia). The cDNAs were used in PCR with radiolabeled primers designed to amplify a region between position +30 and +580 of the MPO cDNA and spanning two introns in order to distinguish the cDNA-derived product (550 bp) from potential products of genomic DNA (1,067 bp). The different samples were normalized for overall cDNA concentrations using actin primers in separate PCRs. Comparisons of MPO cDNA levels for the different genotypes were made within the subtypes, not between M2 and M3 subtypes. The RT-PCR method allows a semiquantitative determination of steady-state levels of MPO mRNA in cells of different MPO genotype. Examples of PCR products are shown in Fig 3A, and the range of values from repeated experiments is indicated in Fig 3C. For each leukemic cell tested, the level of MPO mRNA varied with time in culture or cell density. Leukemic cells of the same genotype but from different donors also differed in MPO mRNA levels, presumably reflecting other genetic differences. Differences in MPO expression levels between M2 cell samples could also reflect different causative events, whereas APL-M3 cells all exhibited the t15; 17 translocation. Overall, the results indicate that AML cells of the SpSp genotype contain higher steady-state levels of MPO mRNA than SpN or NN cells. The difference in MPO mRNA levels between SpSp and SpN heterozygotes was approximately twofold, and the difference between SpSp and NN cells was approximately threefold.
Relative amount of MPO mRNA detected by RT-PCR in primary AML cells of the M3 or M2 subclasses. (A) Autoradiograph shows radiolabeled RT-PCR products from primary AML cells. Lanes 1 to 10, AML-M3 or -M4 cases from Table 1. The top panel shows 550-bp PCR products derived from random primed cDNA using MPO primers. The bottom panel shows products from a separate PCR using actin primers. The reactions in lanes 1 to 4 and 5 to 10 are from different experiments. (B) APL cells from patient no. 803 were cultured for 20 hours in the presence (+RA) or absence (−RA) of RA (10−6 mol/L). RT-PCR was performed with MPO primers. Lane 1 is the same as lane 5 in A. The actin control reactions were repeated to include both cDNAs. (C) Range of MPO transcript levels in the different AML donor cells in multiple experiments is shown with standard deviations indicated. Time in culture ranged from 48 hours to 2 weeks before RNA isolation. Different RT-PCR experiments were normalized to β-actin cDNA levels and also to the MPO signal from a cDNA standard prepared from the NB4 cell line (APL-derived). Relative amount of MPO protein in the different AML cell samples was determined by SDS gel electrophoresis of whole-cell extracts. Proteins were blotted onto nylon membrane, reacted with anti-MPO antibodies, and quantified. Values shown are an average of 3 separate experiments.
Relative amount of MPO mRNA detected by RT-PCR in primary AML cells of the M3 or M2 subclasses. (A) Autoradiograph shows radiolabeled RT-PCR products from primary AML cells. Lanes 1 to 10, AML-M3 or -M4 cases from Table 1. The top panel shows 550-bp PCR products derived from random primed cDNA using MPO primers. The bottom panel shows products from a separate PCR using actin primers. The reactions in lanes 1 to 4 and 5 to 10 are from different experiments. (B) APL cells from patient no. 803 were cultured for 20 hours in the presence (+RA) or absence (−RA) of RA (10−6 mol/L). RT-PCR was performed with MPO primers. Lane 1 is the same as lane 5 in A. The actin control reactions were repeated to include both cDNAs. (C) Range of MPO transcript levels in the different AML donor cells in multiple experiments is shown with standard deviations indicated. Time in culture ranged from 48 hours to 2 weeks before RNA isolation. Different RT-PCR experiments were normalized to β-actin cDNA levels and also to the MPO signal from a cDNA standard prepared from the NB4 cell line (APL-derived). Relative amount of MPO protein in the different AML cell samples was determined by SDS gel electrophoresis of whole-cell extracts. Proteins were blotted onto nylon membrane, reacted with anti-MPO antibodies, and quantified. Values shown are an average of 3 separate experiments.
RA treatment of primary APL cells resulted in rapid loss of MPO transcription (Fig 3B), consistent with similar findings for the APL-derived cell line NB434 and with the observation that RA induces differentiation of primary APL cells,31 since differentiation is accompanied by loss of MPO transcription.13 15
To determine if the higher levels of MPO mRNA correlate with higher protein levels, Western blot analysis was performed. Cellular extracts were electrophoresed in SDS-polyacrylamide gels, transferred to nylon membranes, and reacted with rabbit polyclonal antibodies directed against human MPO (Dako) to detect the 59-kD large subunit. These data indicate that cells of the SpSp genotype contain higher levels of MPO protein than SpN or NN cells.
DISCUSSION
These findings indicate that the SpSp genotype is overrepresented in APL-M3 and -M4 subtypes. The normal population is 61% SpSp, whereas these subtypes are 79% to 82% SpSp. The SpSp genotype is further shown to be associated with higher levels of MPO mRNA than the SpN or NN genotypes in primary AML cells. Overrepresentation of the higher-expressing SpSp MPO genotype in APL-M3 and -M4 implies that higher MPO levels increase the incidence of these leukemias. In APL-M3 and -M4, the ratio of SpSp to other genotypes was 4:1, 2.6-fold higher than the 1.5:1 ratio observed in the general population, suggesting that the SpSp genotype is a 2.6-fold risk factor. Because 61% of the normal population is SpSp, this genotype clearly does not constitute a high risk factor for leukemia. Nevertheless, these findings suggest that if APL occurs in one of 200,000 individuals overall, the incidence in SpSp individuals might be one in 100,000 and in SpN or NN individuals it might be one in 300,000. Causative events such as DNA mutations or translocations may occur about 2.6-fold more frequently in cells of the SpSp genotype, due to the higher levels of MPO-generated free radicals. MPO is present at high levels in promyelocytes, comprising 2% to 4% of the cellular protein, such that MPO-generated radicals could exist at levels sufficient to generate significant DNA cross-linking, mutation,22 or strand breakage,21 thereby increasing the likelihood of translocations such as the APL-associated t15; 17.
As a second possible mechanism by which MPO might increase the risk for leukemia, the oxidizing free radicals produced by the MPO pathway could interfere with the natural killer cell response to emerging cancer cells. Free radicals generated by monocytes have been shown to inhibit the ability of natural killer cells to lyse AML cells in culture.36 Thus, SpSp progenitor leukemic cells may evade the natural killer defense approximately 2.6 times more frequently than other genotypes. According to this model, the SpSp genotype would not increase the incidence rate of translocations or other causative events, but would increase the number of progenitor cells that succeed in evading immune surveillance.
These findings suggest that the SpSp genotype is most strongly associated with AML subtypes around the promyelocyte stage that express the myeloid stage-specific factors required for MPO gene transcription. Such factors or their cognate promoter elements have not been identified, although a number of upstream or intronic DNA elements have been found to bind cellular proteins or to act as promoter elements in transfection assays when linked to a reporter gene.37 There are also potential estrogen response elements (EREs) in intron 7 (which happens to contain an Alu element38 ) and intron 9.39 The upstream AluHRE including -463 G/A is one of the better-characterized promoter elements, with SP1, TR-RXR, and RAR-RXR binding sites.5 This site functions as a strong RARE in transfection assays, and RA strongly regulates MPO transcription in APL cells (Fig 3B). The most compelling evidence that this AluHRE functions as a MPO promoter element is the selection of the SpSp genotype in leukemic cells: This implies that the -463 G/A base difference significantly alters MPO transcription levels, thereby providing the basis for allelic selection. The -463 G results in a stronger RARE in the Sp allele, which may enhance MPO expression in APL leukemic cells and may also be involved in the downregulation of MPO in RA-treated APL cells.34
There is no evidence that the Sp and N alleles differ in the MPO coding region. A number of MPO cDNAs have been sequenced,10,16,40 and there is only one reported allelic polymorphism, a C to T substitution in exon 10 (R569W mutation) that results in hereditary MPO deficiency.41,42 This R569W allele is too rare to correspond to the N allele (22% of haplotypes). As regards potential differences in the MPO genomic sequence, the only complete sequence of the MPO gene including the -463 region is of the N allele.38 Other MPO gene sequences have been reported but cannot be identified as to allele in the absence of the upstream sequence.10 As regards the association of the MPO gene with APL, it should be noted that the MPO gene has been localized to 17q23.1,43 relatively distant in linkage terms from the APL-associated chromosomal breakpoint at 17q21 within the RARα gene.44
The SpSp genotype is not only associated with AML. Recent findings link this genotype to early-onset MS in females.26 Macrophages and microglia, which are descendants of the myeloid precursors giving rise to AML, play a key role in the demyelination of nerve axons in MS.45 Our findings indicate that brain macrophages/microglia at MS lesions contain MPO and that females with the SpSp genotype exhibit symptoms at an earlier age, suggesting that higher levels of MPO in microglia lead to accelerated demyelination. Females with early-onset MS were 86% SpSp, but the SpSp genotype was not overrepresented in males with MS. This lends credence to the preliminary observation that females with APL-M3 or -M4 are more highly associated with the SpSp genotype than males. The MPO gene may be upregulated through estrogen, possibly through the reported intronic ERE39 or through the upstream AluHRE, based on findings that some AluHREs can function as EREs.46
ACKNOWLEDGMENT
We thank Prescott L. Deininger for providing normal donor DNA for analysis.
Supported by a grant from the National Institutes of Health (RR09118-CA72995 to W.F.R.).
Address reprint requests to Wanda F. Reynolds, PhD, Sidney Kimmel Cancer Center, 3099 Science Park Rd, Suite 200, San Diego, CA 92121.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal