Abstract
Identification and characterization of mutations that disrupt normal hematopoiesis are essential for understanding the genetic pathways that control the development and regulation of the mammalian hematopoietic system. Previously, the fitness 1 gene was identified by five, independent mutations in N-ethyl-N-nitrosourea (ENU) saturation mutagenesis experiments within the albino (c) region of mouse chromosome 7 (MMU7). We report here that fit1 mutants are anemic, display numerous peripheral blood defects, and are deficient in early hematopoietic progenitor cell populations. The number of both erythroid and myeloid progenitors, as well as B cells, are reduced. These results implicate fit1 involvement in normal hematopoiesis and suggest that further characterization of the fit1 gene, and the five presumed point mutations of the gene, will lead to an improved understanding of normal hematopoiesis in the mouse.
MUCH OF OUR current knowledge of hematopoiesis has come from the molecular and phenotypic characterization of genetic mutations that disrupt the normal course of hematopoietic development in experimental systems. Indeed, several spontaneous and induced mutations resulting in anemia have been identified in the mouse,1-3 and characterization of these mutations has provided key insights into the complex pathways regulating hematopoiesis.
The induction of mutations in the mouse germline by chemical and radiation mutagenesis has provided a rich resource of new mutations affecting a wide range of developmental pathways, including hematopoiesis.4-7 Recently, five alleles of a gene, designated fitness 1 (fit1 ), were identified in an N-ethyl-N-nitrosourea (ENU) saturation mutagenesis screen of a defined region of mouse chromosome 7 (MMU 7).8 9 The fit1 mutants were originally detected in the screen as growth retarded and “less fit,” or viable, than other littermates; other than the observed growth retardation and reduced viability at the time of weaning, no other phenotypic consequence was obvious.
This study represents the initial phenotypic characterization of mice expressing the fit1 mutations. These analyses have shown that fit1 mice suffer from a severe hypochromic anemia, characterized by anisocytosis and poikilocytosis. Also, hematopoiesis is defective in fit1 mutants at an early progenitor stage, and cells representative of multiple hematopoietic lineages are significantly reduced. In addition to these hematopoietic defects, fit1 mutants are growth retarded in utero and remain dwarfed throughout their shortened lifespan. These results indicate that the fit1 gene plays a crucial role in fetal and adult hematopoiesis, and normal prenatal and postnatal growth and viability.
MATERIALS AND METHODS
Mice.All mice used in this study were bred at the Biology Division of the Oak Ridge National Laboratory. The ENU saturation mutagenesis experiments from which the fit1 mutations were recovered have been previously described.8 The fit1 locus is closely linked to the albino (c ) locus on MMU 7 (≤2 centimorgan [cM])8,9, and ENU-induced fit1 mutations are maintained in a c fit chromosome heterozygous with a chromosome carrying the chinchilla (cch ) allele of the albino (c ) locus (ie, cch +/c fit1 ). For the experiments described in this report, the fit1 mutations were maintained by mating carriers to the 47BS inbred strain, which is homozygous for the chinchilla allele (cch +/cch +). Alternatively, + c fit1 +/p cch + fr carriers were mated to the noninbred FR-p cchfr/p cchfr strain, which carries mutations at three linked loci: the pink-eyed dilution mutation (p ), the chinchilla allele at c, and the frizzy mutation (fr ) (the genetic distances between these loci are: p–14 cM–cch–10-20 cM–fr ). In these matings, only a rare double crossover event could produce cch/c mice that did not also carry the fit1 mutation.
To generate fit1 mutants for analysis, presumed fit1 carriers (from either the 47BS or FR line) were mated to cch +/c26DVT deletion heterozygotes. The c26DVT deletion mutation removes a 6- to 11-cM region surrounding the c locus, and by design of the mutagenesis stategy, must include the fit1 locus.8 Therefore, one quarter of the progeny from such a cross will express the albino mutation of the tyrosinase gene, as well as the fit1 mutation, opposite the c26DVT deletion (c fit1/c26DVT ). Deletion heterozygotes (cch +/c26DVT ) display a light-chinchilla coat color (and are indistinguishable from fit1 carriers [cch +/c fit1 and p cch + fr/+ c fit1 +]), in contrast to the darker, full-chinchilla coat color in cch/cch mice. Mice expressing the fit1 mutations opposite the c26DVT deletion were used due to availability of the cch +/c26DVT stock. cch+/c26DVT mice are phenotypically normal, which suggests the phenotype associated with c fit1/c26DVT mice results from the hemizygosity of fit1 mutations.
Growth analyses.Carriers (from the 47BS lines) of each of the five fit1 mutations (cch +/c fit1494SB, cch +/c fit1764SB, cch +/c fit13452SB, cch +/c fit14226SB, cch +/c fit14397SB ) were mated to mice heterozygous for the c26DVT deletion (cch +/c26DVT ). The number of offspring was recorded at birth for individual litters, and each pup was sexed and genotyped (based on eye pigmentation: cch +/cch +, full eye color; cch +/c fit1 or cch +/c26DVT, light eye color; c fit1/c26DVT, no eye color). Litter sizes were reduced at birth to four or five pups to enhance the survival of fit1 mutants. Also, reducing the litter sizes to a common number may minimize variability among litters due to different numbers of offspring.10 Additionally, the weights of male and female offspring were grouped, as all mice analyzed were weaning-aged or younger and did not differ significantly. The weight of each pup was recorded on the day of birth and then every other day thereafter up to at least 32 days of age or until the fit1 mutant(s) in each litter died. Data were collected from all phenotypic classes in each litter, however, no significant difference was seen between measurements of homozygous and heterozygous wild-type offspring (cch +/cch +; cch +/c fit1 and cch +/c26DVT ) and, therefore, only data for homozygous wild-type mice and fit1 mutants are shown.
The weights of 14.5-day (E14.5) fetuses were recorded only for fit14397SB mutants and their normal littermates. Males carrying the fit14397SB mutation (cch +/c fit14397SB ) were mated to cch +/c26DVT females, and females were checked daily for vaginal plug. Plugged females were killed at E14.5, fetuses were removed, genotyped (based on eye pigmentation [see above]), and weighed.
Histology.All tissues were removed at time of death (mice aged 3 to 5 weeks, unless otherwise noted) to a solution of 10% buffered formalin. Tissues were then embedded in paraffin, cut into 5- to 10-mm sections, mounted on glass slides, and stained with hematoxylin and eosin or with Prussian blue for iron.
Blood samples for scanning electron microscopy (SEM) were collected in a heparinized capillary tube and then fixed in 2% gluteraldehyde in 0.1 mol/L sodium cacodylate buffer, pH 7.3, for 1 hour. One drop of each suspension was placed on Thermanox tissue culture cover slips for 1 hour to allow the cells to settle and stick to the cover slips. The cells were then washed, dehydrated, air dried, and sputter coated with gold. The specimens were viewed in a JEOL JSM-840 scanning electron microscope.
Hematology.Blood samples were collected by removing blood from the intraorbital sinus of anesthetized fit1 mutants and wild-type littermates. The blood was then placed directly on coverslips and spread. Air-dried blood films were stained with Wright's stain and mounted on glass slides for examination. Hematologic data were determined by analysis on an Ortho ELT-15 laser-based Hematology Analyzer (Ortho Diagnostics, Raritan, NJ), modified to optimize the analysis of mouse blood.11 Reticulocyte counts and some hematocrits and white blood cell (WBC) counts were determined manually by standard protocols.12
In vitro progenitor cell assays.All in vitro progenitor cell assays were performed as previously described.13 14 Briefly, single-cell suspensions of spleen, bone marrow, and fetal liver tissues in α minimum essential media (αMEM; GIBCO-BRL, Gaithersburg, MD) were cultured in 0.8% Alpha Methylcellulose-Complete Ready-Mix, containing methylcellulose, fetal calf serum, bovine serum albumin, erythropoietin, and conditioned medium in alpha medium plus nucleosides (Stem Cell Technologies, Inc, Vancouver, British Columbia, Canada). For colony-forming unit-erythorid (CFU-E) analyses, 0.1-mL aliquots of a 4 × 105 cells/mL suspension were plated in 96-well culture plates and incubated at 37°C and 5% CO2 in air for 2 days. For bone marrow burst-forming unit-erythorid (BFU-E) quantitation, 1-mL aliquots of a 5 × 104 cells/mL suspension were plated in 6-well culture plates and cultured at 37°C and 5% CO2 in air for 7 to 10 days. The quantitation of splenic BFU-E was performed as described for bone marrow with the exception that 1-mL aliquots of a 5 × 105 cells/mL suspension were plated. Five wells were plated for each sample, and the values presented are based on the mean number of progenitors obtained from counting 2 to 4 wells of each sample.
Colony-forming unit-spleen (CFU-S) assay.The CFU-S assay was performed as described.15 Single-cell suspensions of bone marrow cells in calcium- and magnesium-free phosphate-buffered saline (PBS) were prepared from one femur and one tibia from each mouse, and groups of irradiated scid/scid mice (0.7 Gy/min, for a total of 2.5 Gy) received either 1 × 105 wild-type (cch +/cch +) cells (n = 6) in PBS per mouse, 2 × 105fit1 (c fit14397SB/c26DVT ) cells (n = 6) per mouse, or PBS only (n = 5) (this served as a radiation control group), and were kept in isolation for 14 days. The fit1 mice used in this study were from outbred stocks; therefore, immunocompromised scid/scid mice were used as bone marrow recipients to limit or prevent an immune response against the allogenic transferred bone marrow cells.16 After 14 days, all mice were killed, spleens were removed, fixed in Telly's fixative (100 parts 70% ethanol, 5 parts formaldehyde, 5 parts acetic acid), and CFU-S colonies were enumerated. Two colonies were found in the control group (PBS only) (0.4 CFU-S/spleen).
Flow cytometry.Cells from spleen and bone marrow were suspended in Ca++- and Mg++-free PBS, washed, and diluted to 107 cells/mL in PBS supplemented with 0.1% sodium azide and 0.1% bovine serum albumin. A total of 106 cells were incubated on ice with saturating quantities of monoclonal antibodies specific for T cells using fluoroisothiocyanate (FITC)-conjugated anti-Thy 1.2 antibody or for B cells using phycoerythrin (PE)-conjugated anti-B220 antibody (Pharmingen, San Diego, CA). Nonspecific binding to Fc receptor sites was blocked by using anti-CD16/CD32 antibody (Pharmingen). The cells were washed with supplemented PBS and analyzed using a FACStarPlus flow cytometer (Becton Dickinson, Mississauga, Ontario, Canada). List mode data were analyzed using Multi2D software (Phoenix Flow Systems). Light scatter and propidium iodide uptake were used to exclude red cells and dead cells from the analysis. The data are expressed as a percent of live cells positive for either antibody.
RESULTS
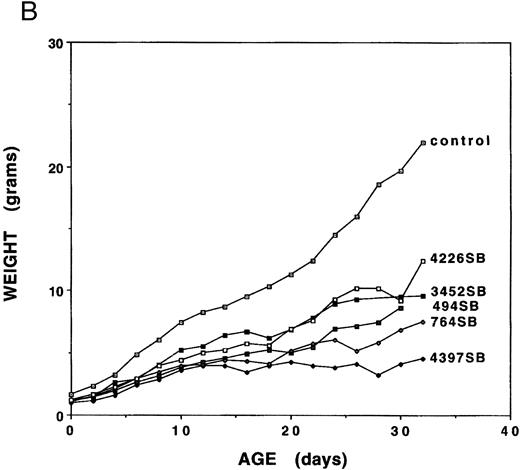
Growth analyses.Mice expressing any of the five independent fit1 mutations (designated fit1494SB, fit1764SB, fit13452SB, fit14226SB, and fit14397SB ) were smaller in size as compared with their wild-type littermates (Fig 1A), and usually died between birth and weaning age. To determine the characteristics of the fit1 growth retardation, the body weight was measured for fit1 mutants (c fit1m/c26DVT; where m represents any one of the fit1 alleles) and wild-type littermates (cch +/cch +, cch +/c fit1m, and cch +/c26DVT ). Weights of fit1 and wild-type mice show that all fit1 mutants are growth retarded at birth and remain smaller throughout their shortened lifespan (Fig 1B). Mice expressing the fit1 mutant alleles displayed a range of growth retardation, with mutants expressing the fit14397SB allele more severely affected than mice expressing any of the other four alleles. fit14397SB mutants were, on average, 61.1% the size of normal littermates at birth and 20.4% of the normal size at weaning (26 to 30 days of age). In contrast, mutants expressing the fit14226SB allele appear to be the least growth retarded of the five fit1 mutants. fit14226SB mutants were, on average, 75.0% of control weight at birth and 56.3% of control weight at weaning. The remaining three alleles were intermediate to these extremes, with fit1494SB mutants more severely affected than either fit1764SB or fit13452SB mutants. Other growth parameters (body length and tail length) measured for each mouse reflected the same patterns of severity among each of the alleles (data not shown).
Growth retardation of fit1 mutants. (A) Representative fit1 mutants, each expressing one of the five fit1 alleles, shown with one wild-type littermate (far right). The genotypes of the mice, beginning at the left, are: c fit14397SB/c26DVT; c fit1494SB/c26DVT; c fit1764SB/c26DVT; c fit13452SB/c26DVT; c fit14226SB/c26DVT; and cch +/c26DVT. All mice shown are 26 days old. The wild-type littermate is from the mating that generated the fit1494SB mutant shown. (B) Growth curves of fit1 mutants and wild-type littermates (cch +/cch +). The graph shows the mean weight of each group of mice (mutants or wild-type) plotted versus the day of weight measurement (the date of birth was designated day 0).
Growth retardation of fit1 mutants. (A) Representative fit1 mutants, each expressing one of the five fit1 alleles, shown with one wild-type littermate (far right). The genotypes of the mice, beginning at the left, are: c fit14397SB/c26DVT; c fit1494SB/c26DVT; c fit1764SB/c26DVT; c fit13452SB/c26DVT; c fit14226SB/c26DVT; and cch +/c26DVT. All mice shown are 26 days old. The wild-type littermate is from the mating that generated the fit1494SB mutant shown. (B) Growth curves of fit1 mutants and wild-type littermates (cch +/cch +). The graph shows the mean weight of each group of mice (mutants or wild-type) plotted versus the day of weight measurement (the date of birth was designated day 0).
Because fit1 mutants were growth retarded at birth, the fit1 gene may also be necessary for normal prenatal growth. To address this possibility, the weights of fit14397SB mutants were compared with those of their wild-type littermates at 14.5 days of gestation (E14.5). The fit14397SB mutant fetuses were approximately 70.2% the size of normal littermates (fit14397SB/c26DVT = 0.17 ± 0.04 g [n = 19]; littermate controls = 0.24 ± 0.06 g [n = 81], P < .05), suggesting that the fit1 gene product is also required for normal prenatal growth.
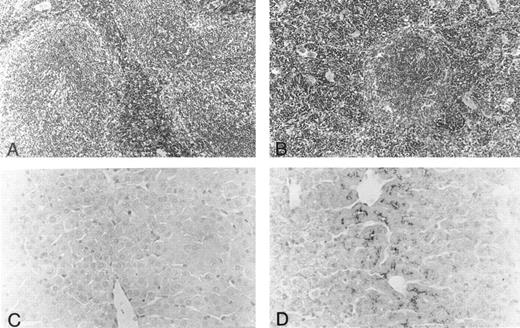
Histology.Body organs of fit1 mice appeared normally developed; however, fit1 mutant livers were detectably pale, and splenomegaly was present in most fit1 hemizygotes. Indeed, spleens from fit14397SB mutants composed an average of 1.87% ± 0.78 (n = 5) of the total body weight, whereas the mean weight of the normal littermate (cch +/cch +) spleens was 0.42% ± 0.09 (n = 5) of mean body weight (means are significantly different, P < .05). The spleens of fit14397SB mice had significant extramedullary hematopoiesis (EMH), characterized by a greater number of erythroblasts and poorly defined borders between the red pulp and white pulp, as opposed to the characteristic pattern seen in the spleens of wild-type mice (cch +/cch +) (Figs 2A and B). No EMH was seen in fit1 mutant liver (data not shown). Additionally, the fit1 mutant livers stained strongly for iron, compared with tissues from their wild-type littermates (Figs 2C and D). To a lesser degree, iron deposits were also observed in fit1 mutant spleens, and no iron deposits were seen in fit1 mutant bone marrow (data not shown).
Histology of spleen and liver from fit1 mutant mice. (A and B) Photomicrographs of hematoxylin and eosin-stained sections of (A) wild-type littermate (cch +/cch +) and (B) fit14397SB spleen. Erythroid precursors (darker staining cells) appear to be present in larger numbers in the fit1 mutant spleen, and in the mutant spleen, the white pulp areas appear smaller and less well defined. (C and D) Photomicrographs of Prussian blue stained sections from wild-type littermate liver (C) and fit14397SB mutant liver (D). Increased iron stores can be seen in the fit1 mutant liver as dark staining within several cells.
Histology of spleen and liver from fit1 mutant mice. (A and B) Photomicrographs of hematoxylin and eosin-stained sections of (A) wild-type littermate (cch +/cch +) and (B) fit14397SB spleen. Erythroid precursors (darker staining cells) appear to be present in larger numbers in the fit1 mutant spleen, and in the mutant spleen, the white pulp areas appear smaller and less well defined. (C and D) Photomicrographs of Prussian blue stained sections from wild-type littermate liver (C) and fit14397SB mutant liver (D). Increased iron stores can be seen in the fit1 mutant liver as dark staining within several cells.
In contrast to the fit1 mutant spleens, there was no increase in hematopoietic activity in fit1 mutant bone marrow (data not shown). Additionally, thymic sections from fit14397SB mutants showed no gross structural abnormalitites, however, thymic mass varied greatly among individual mutants (data not shown). Also, no lesions or other evidence of cell destruction were detected in fit1 mutant skin or other organs.
Hematology.Peripheral blood smears from fit14397SB mutants and wild-type littermates were also examined. The fit14397SB peripheral blood displayed hypochromic red blood cells (RBCs) with marked anisocytosis and poikilocytosis (Fig 3). Blood from mice expressing the other fit1 mutations showed similar characteristics, but to varying degrees (data not shown). Additionally, as shown in Table 1, the number of reticulocytes was significantly increased in fit14397SB mutants.
Morphology of RBCs from fit1 mutant mice. Peripheral blood smear from a cch +/cch + wild-type littermate (A) and from a cfit14397SB/c26DVT mutant (B). RBCs from fit1 mutants were typically hypochromic and varied greatly in size. Scanning electron micrographs of wild-type (cch +/cch +) blood (C) and mutant (c fit14397SB/c fit14397SB ) blood (D) more clearly show the size and shape variations of the fit1 mutant RBCs. The SEM bar represents 10 mm.
Morphology of RBCs from fit1 mutant mice. Peripheral blood smear from a cch +/cch + wild-type littermate (A) and from a cfit14397SB/c26DVT mutant (B). RBCs from fit1 mutants were typically hypochromic and varied greatly in size. Scanning electron micrographs of wild-type (cch +/cch +) blood (C) and mutant (c fit14397SB/c fit14397SB ) blood (D) more clearly show the size and shape variations of the fit1 mutant RBCs. The SEM bar represents 10 mm.
Hematologic Values From fit1 Mutants and Wild-Type Littermates
| Group . | n . | Retic . | WBC . | RBC . | HGB . | HCT . | MCV . | MCH . | MCHC . |
|---|---|---|---|---|---|---|---|---|---|
| . | . | (%) . | (×106/mL) . | (×109/mL) . | (g/dL) . | (%) . | (FL) . | (pg) . | (g/dL) . |
| Control | 24 | 4.4 ± 1.1* | 6.5 ± 1.3 | 9.2 ± 0.9 | 13.9 ± 1.4 | 44.7 ± 3.7 | 47.5 ± 3.1 | 15.2 ± 0.8 | 32.0 ± 1.8 |
| 494SB | 1 | — | 6.2 | 4.1 | 4.4 | 19.8 ± 4.9† | 32.0 | 10.7 | 33.3 |
| 764SB | 14 | — | 3.8 ± 1.8 | 6.3 ± 1.2 | 7.3 ± 1.5 | 24.7 ± 5.4 | 33.2 ± 2.5 | 11.1 ± 0.7 | 33.5 ± 1.9 |
| 3452SB | 5 | — | 6.6 ± 1.4 | 9.4 ± 1.6 | 13.1 ± 1.8 | 34.5 ± 5.4 | 44.0 ± 3.7 | 14.1 ± 1.1 | 32.1 ± 0.4 |
| 4226SB | 11 | — | 5.7 ± 1.5 | 7.3 ± 1.0 | 8.1 ± 1.3 | 26.7 ± 3.6 | 35.3 ± 1.5 | 11.2 ± 0.6 | 31.5 ± 0.9 |
| 4397SB | 10 | 15.8 ± 4.2* | 3.5 ± 1.5 | 5.6 ± 1.3 | 5.6 ± 1.0 | 16.7 ± 5.3 | 28.2 ± 2.5 | 10.3 ± 0.7 | 36.9 ± 3.8 |
| Group . | n . | Retic . | WBC . | RBC . | HGB . | HCT . | MCV . | MCH . | MCHC . |
|---|---|---|---|---|---|---|---|---|---|
| . | . | (%) . | (×106/mL) . | (×109/mL) . | (g/dL) . | (%) . | (FL) . | (pg) . | (g/dL) . |
| Control | 24 | 4.4 ± 1.1* | 6.5 ± 1.3 | 9.2 ± 0.9 | 13.9 ± 1.4 | 44.7 ± 3.7 | 47.5 ± 3.1 | 15.2 ± 0.8 | 32.0 ± 1.8 |
| 494SB | 1 | — | 6.2 | 4.1 | 4.4 | 19.8 ± 4.9† | 32.0 | 10.7 | 33.3 |
| 764SB | 14 | — | 3.8 ± 1.8 | 6.3 ± 1.2 | 7.3 ± 1.5 | 24.7 ± 5.4 | 33.2 ± 2.5 | 11.1 ± 0.7 | 33.5 ± 1.9 |
| 3452SB | 5 | — | 6.6 ± 1.4 | 9.4 ± 1.6 | 13.1 ± 1.8 | 34.5 ± 5.4 | 44.0 ± 3.7 | 14.1 ± 1.1 | 32.1 ± 0.4 |
| 4226SB | 11 | — | 5.7 ± 1.5 | 7.3 ± 1.0 | 8.1 ± 1.3 | 26.7 ± 3.6 | 35.3 ± 1.5 | 11.2 ± 0.6 | 31.5 ± 0.9 |
| 4397SB | 10 | 15.8 ± 4.2* | 3.5 ± 1.5 | 5.6 ± 1.3 | 5.6 ± 1.0 | 16.7 ± 5.3 | 28.2 ± 2.5 | 10.3 ± 0.7 | 36.9 ± 3.8 |
Control mice [cch +/cch+, cch+/c26DVT, and cch+/c fit1m (where m designates the individual fit1 alleles)] and mutants (c fit1m/c26DVT, shown in the table by the allele designations) were analyzed at 3 to 5 weeks of age. Data are mean ± standard deviation.
Abbreviations: Retic, reticulocytes; WBC, white blood cells; RBC, red blood cells; HGB, hemoglobin; HCT, hematocrit; MCV, mean cell volume; MCH, mean cell hemoglobin; MCHC, mean cell hemoglobin concentration.
n = 3.
n = 8.
Further analyses of peripheral blood from fit1 mutants showed numerous abnormalities in blood cell parameters (Table 1). As shown in Table 1, all the fit1 mutants were moderately to severely anemic and had much lower hemoglobin (Hgb) levels than their control littermates. It is interesting to note that the mean hematocrit (Hct) and Hgb values for each of the fit1 mutant groups displayed a range of severity, which correlated in rank order with the degree of growth retardation observed for each allele (Fig 2B). The mean cell volume (MCV) and the mean cell hemoglobin (MCH) were also significantly reduced in fit1 mutants. The severity of the reduction of these values reflected the same pattern as the Hct and Hgb values, with fit14397SB being the most affected and fit13452SB the least (Table 1).
Hematopoietic progenitors.Because of the marked alterations in the mature red blood cell populations in these mice, we next investigated whether mutations in fit1 affected the hematopoietic progenitor populations. To determine the numbers of erythroid, as well as other myeloid, progenitors in hematopoietic tissues from fit1 mutants, bone marrow and spleen cells from weaning-age fit14397SB mutants and wild-type littermates were assayed. As shown in Table 2, the numbers of both CFU-E and the earlier progenitor, BFU-E, were reduced in fit14397SB weaning age hematopoietic tissues. The total numbers of CFU-E in the bone marrow and spleens of fit14397SB mutants were less than 20% that observed in wild-type littermate tissues. Similarly, the number of BFU-E in adult bone marrow and spleen was less than 10% that of controls. Additionally, a myeloid progenitor of granulocytes and macrophages, the CFU-granulocyte macrophage (GM), was also reduced in fit14397SB mutants, although not as severely as the erythroid progenitors (17% and 44% that of controls in the bone marrow and spleen, respectively). The hematopoietic defect in fit14397SB mutants is also apparent at 7 and 19 days of age (data not shown). Additionally, hematopoietic progenitors were decreased in tissues from mice expressing other fit1 mutations (494SB, 764SB, and 4226SB), as well (data not shown). These data suggest mutations in the fit1 gene affect both erythroid and myeloid progenitor cell populations.
Total No. of Hematopoietic Progenitors in fit1 Mice and Wild-Type Littermates
| Tissue . | . | Total Cells . | CFU-E . | BFU-E . | CFU-GM . |
|---|---|---|---|---|---|
| . | . | (×106) . | (×104) . | (×103) . | (×103) . |
| Spleen | + (7) | 180.4 ± 19.2 | 7.6 ± 1.8 | 2.9 ± 1.2 | 21.0 ± 1.5 |
| m (8) | 247.9* ± 81.3 | 1.0 ± 0.4 | 0.1 ± 0.1 | 9.4 ± 2.3 | |
| Bone marrow† | + (7) | 43.7 ± 15.6 | 26.0 ± 1.5 | 12.0 ± 1.6 | 110.0 ± 8.9 |
| m (8) | 17.2 ± 5.6 | 1.9 ± 0.5 | 0.8 ± 0.4 | 19.0 ± 4.0 | |
| E14.5 liver | + (6) | 28.3 ± 11.8 | 27.0 ± 1.3 | 6.1 ± 1.0 | 46.0 ± 7.5 |
| m (5) | 14.2 ± 11.6 | 5.3 ± 2.3 | 0.4 ± 0.2 | 12.0 ± 3.0 |
| Tissue . | . | Total Cells . | CFU-E . | BFU-E . | CFU-GM . |
|---|---|---|---|---|---|
| . | . | (×106) . | (×104) . | (×103) . | (×103) . |
| Spleen | + (7) | 180.4 ± 19.2 | 7.6 ± 1.8 | 2.9 ± 1.2 | 21.0 ± 1.5 |
| m (8) | 247.9* ± 81.3 | 1.0 ± 0.4 | 0.1 ± 0.1 | 9.4 ± 2.3 | |
| Bone marrow† | + (7) | 43.7 ± 15.6 | 26.0 ± 1.5 | 12.0 ± 1.6 | 110.0 ± 8.9 |
| m (8) | 17.2 ± 5.6 | 1.9 ± 0.5 | 0.8 ± 0.4 | 19.0 ± 4.0 | |
| E14.5 liver | + (6) | 28.3 ± 11.8 | 27.0 ± 1.3 | 6.1 ± 1.0 | 46.0 ± 7.5 |
| m (5) | 14.2 ± 11.6 | 5.3 ± 2.3 | 0.4 ± 0.2 | 12.0 ± 3.0 |
The mean total number of cells per organ listed and hematopoietic progenitors listed are shown for wild-type littermates (+) and for c fit14397B/c26DVT mutants (m). The number of mice in each group is shown in parentheses.
The mean of the mutant spleen cellularity is not significantly different from the mean cellularity of control spleens.
The values for bone marrow reflect the total number of cells and progenitors derived from one tibia and one femur from each animal.
The defect in hematopoiesis was also evident during fetal development. As shown in Table 2, the numbers of both erythroid and myeloid progenitors were markedly reduced in fit14397SB mutant fetal livers at E14.5. The numbers of CFU-E in fit14397SB E14.5 liver were approximately 20% that of normal littermates, and BFU-E were reduced to an even greater extent (to approximately 7% of control). The numbers of CFU-GM were also reduced, although, as in weaning-age bone marrow and spleen, not as severely as the erythroid progenitors (Table 2). These data suggest that the fit1 gene is necessary for both fetal and adult hematopoiesis, at least as early as E14.5 of gestation.
Because CFU-E, BFU-E, and CFU-GM are all affected by the fit1 mutations, it is possible that an earlier, common progenitor is also influenced by mutations in fit1. To test this hypothesis, 14-day CFU-S assays were performed with bone marrow cells from fit14397SB mutants injected into irradiated scid/scid mice (see Materials and Methods). The numbers of CFU-S in fit14397SB bone marrow (2.4 ± 0.65 per 105 cells) were significantly reduced compared with the number in wild-type littermate bone marrow (6.8 ± 1.9 per 105 cells) (P < .05). Histological sections from the spleen colonies showed cells of mixed lineages in both control- and fit14397SB-derived colonies. However, in addition to being fewer in number, the colonies that developed from fit14397SB mutant bone marrow were much smaller than those from wild-type cells (data not shown).
Flow cytometry analyses.The data presented above show that fit1 mutations have a severe effect on both erythropoiesis and myelopoiesis. Because the number of circulating WBCs was significantly reduced in fit1 mice, the fit1 gene product may also be necessary for normal lymphoid development. To establish a preliminary measure of lymphoid cell populations in fit14397SB mutants, fit14397SB hematopoietic tissues were analyzed by flow cytometry with monoclonal antibodies specific to B (CD45R/B220) and T (Thy1.2) cells (Table 3; data not shown). The numbers of B220+ cells in fit14397SB/c26DVT bone marrow were approximately 50% of that in their wild-type littermates (cch +/cch +) bone marrow. Similarly, the numbers of B220+ cells in fit14397SB/c26DVT spleens were reduced by 70% compared with that found in wild-type spleens. The numbers of Thy1.2-positive cells in fit14397SB spleen tissue were also reduced; however, the differences from control were not statistically significant (data not shown).
Flow Cytometric Analysis of B-Cell and T-Cell Populations in Spleen and Bone Marrow of fit 14397SB Mutants
| . | . | B220 . | Thy-1 . |
|---|---|---|---|
| Bone marrow | c (12) | 39.7 ± 8.4 | ND |
| m (12) | 21.2 ± 13.43-150 | ND | |
| Spleen | c (10) | 49.5 ± 15.2 | 14.1 ± 3.5 |
| m (11) | 15.3 ± 9.33-150 | 11.3 ± 4.1 |
| . | . | B220 . | Thy-1 . |
|---|---|---|---|
| Bone marrow | c (12) | 39.7 ± 8.4 | ND |
| m (12) | 21.2 ± 13.43-150 | ND | |
| Spleen | c (10) | 49.5 ± 15.2 | 14.1 ± 3.5 |
| m (11) | 15.3 ± 9.33-150 | 11.3 ± 4.1 |
Single cell suspensions from 26- to 32-day-old fit 14397SB mutants (c fit 14397SB/c26DVT) and wild-type littermates (cch +/cch + ) were analyzed with the listed antibodies as described in Materials and Methods. The values represent the mean percentage of antibody-positive cells ± SD, and the number of mice analyzed in each group is shown in parentheses.
Abbreviation: ND, not determined.
Significance of the difference from control mean: P < .05.
DISCUSSION
The studies reported here represent the initial characterization of mice expressing any of five independent mutations in the fit1 gene. Hematopoiesis was markedly defective in fit1 mutants at least as early as the CFU-S stage, and cells representative of multiple hematopoietic lineages (erythroid, myleoid, and B cell) were significantly reduced in numbers. Moreover, the circulating RBCs of fit1 mutants were hypochromic and displayed varying degrees of anisocytosis and poikilocytosis. In addition to these hematopoietic defects, fit1 mutants were growth retarded in utero and remain dwarfed throughout their shortened lifespan. We conclude that the fit1 gene plays a crucial role in hematopoiesis, fetal and adult growth, and viability.
The morphology of circulating RBC, marked reticulocytosis, and splenic hyperplasia in fit1 mutants indicates that the fit1 hematopoietic system is under significant proliferative stress. Additionally, the increased iron reserves observed in fit1 mutant livers and spleens suggests increased RBC turnover. However, despite the apparent increased numbers of erythroblasts in fit1 mutant spleens and the increased numbers of reticulocytes in fit1 mutant blood, these mice remain extremely anemic. Interestingly, no increase in cellularity was observed in fit1 bone marrow, and the numbers of erythroid progenitors were significantly reduced in both fit1 mutant bone marrow and spleen. Indeed, both CFU-E and the earlier erythroid-progenitor, BFU-E, are significantly reduced to a similar extent in the bone marrow, spleen, and fetal liver of fit1 mutants. The numbers of CFU-S and CFU-GM are also significantly reduced in all fit1 mutant hematopoietic tissues. The results presented here also suggest that fit1 mutations affect B-, but not T-cell development. WBC counts were reduced in fit1 mutant blood, and B220+ cells were reduced approximately 50% in fit14397SB mutants. Together, these results suggest that the fit1 gene either has pleiotropic effects on several hematopoietic lineages or perhaps affects an early, pluripotent progenitor cell population.
Several other mutations in the rodents produce a phenotype similar to that of fit1 mutants. The anemias resulting from mutations in the flexed-tail (f ), microcytic anemia (mk ), and sex-linked anemia (sla ) loci in the mouse are all thought to result from defects in iron metabolism and all produce a hypochromic, microcytic anemia with accompanying reticulocytosis.2 It is also interesting that f, mk, and sla mutants are all growth retarded in utero.2 Additionally, experimentally induced iron deficiency anemias (IDA) produce a phenotype similar to that of fit1 mutants (growth retardation and anemia)17,18 However, although iron is an indespensable component of a large number of biochemical reactions, the vast majority of iron is required for hemoglobin synthesis.19 Because hemoglobin synthesis predominates in the later stages of erythropoiesis, it is unclear how iron deficiency may affect early erythroid progenitors and other hematopoietic lineages. In experimentally induced IDA in the rat, the numbers of CFU-E in bone marrow and spleen increased, whereas the numbers of BFU-E and CFU-GM remained unchanged.17 However, the Belgrade rat anemia presumably results from an iron deficiency, and CFU-Es and BFU-Es are reduced in these animals.20 Additional studies have shown that granulopoiesis, megakaryocytopoiesis, and T cells are affected by the Belgrade rat mutation.21-24 Thus, in Belgrade rat anemia, the wide range of hematopoietic defects may reflect the importance of iron for cellular proliferation,20 or alternatively, to an acquired disorder of hematopoietic stem cells due to prolonged hypoxia.24 In an experimentally induced IDA in mice,18 both B- and T-cell populations were significantly reduced. Indeed, similar to the results found in the fit1 mice, the number of B220+ cells in the spleens of iron-deficient mice were reduced approximately 50%; however, in contrast to the phenotype seen in fit1 mutants, splenic T cells were reduced to an even greater extent.18 Although abundant iron stores are present in fit1 mutant livers, it is possible that some aspect of utilization of these iron stores is defective in fit1 mutants. Indeed, several cytokines (eg, tumor necrosis factor [TNF ]α, interleukin-1 [IL-1], interferon [IFN]γ) induce anemia and suppress erythroid progenitor populations by altering iron utilization.25 Whether fit1 is involved, directly or indirectly, awaits further analyses.
The fit1 phenotype shares some of the phenotypic characteristics associated with thalassemias in mice.26-30 Mice with βo-thalassemia exhibit a hypochromic, microcytic anemia, with reticulocytosis and splenomegaly.30 Additionally, these mice have abundant tissue stores of iron resulting from increased erythropoietic activity. However, in contrast to fit1 mutants, these mice have increased bone marrow cellularity, and presumably, early hematopoietic progenitors are not affected. Although the fit1 locus maps to a location distinct from the Hbb locus on MMU 7, it is possible that the fit1 gene could play some role in hemoglobinization.
The fit1 gene has been mapped to a specific subregion of the c deletion complex in MMU 79; no other known mouse anemia, or candidate genes, currently map in this region. However, recent physical-mapping experiments have narrowed the interval containing at least a portion of the fit1 gene to a region, defined by deletion breakpoints, of approximately 340 kb that is spanned by two yeast artificial chromosomes31; therefore, molecular identification of the fit1 gene should be forthcoming.
The fit1 mutations were recovered through saturation mutagenesis experiments using the spermatogonial stem cell point mutagen, ENU.4 It is likely, therefore, that the fit1 mutations represent independent, single amino acid changes that result in defects in growth, viability, and hematopoiesis. Series of allelic mutations that induce subtle changes in gene function, thereby resulting in a range of phenotypic variations, are extremely useful tools for deciphering the roles that specific genes play in development.32 33 The range of phenotypic severity observed among the five fit1 alleles, suggest that each of the fit1 mutations has altered the functional capacity and/or the expression of the fit1 gene product to varying degrees. Consequently, molecular characterization of the fit1 gene and the five mutant alleles, should lead to an increased understanding of the role of this gene in hematopoiesis and to the elucidation of important structural motifs in the fit1 protein.
ACKNOWLEDGMENT
We thank B. Fike, K. Harpel, D. Holmyard, K. Houser, C. Long, and S. Vesely for excellent technical assistance. In addition, we are grateful to J. Bodine, J. Bollekens, L. Fine, M.C. Howard, R. Paulson, A.O. Taylor, and E. Wilkinson for helpful discussions and critical review of the work.
Supported by the Office of Health and Environment Research, US Department of Energy, under contract number DE-AC05-84OR21400 with Martin Marietta Energy Systems, Inc, by the National Center for Human Genome Research (HG 00370), the Medical Research Council of Canada, and Bristol Myers-Squibb.
Address reprint requests to Dabney K. Johnson, PhD, Biology Division, Oak Ridge National Laboratory, PO Box 2009, Oak Ridge, TN 37831-8077.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal