Abstract
Infection of monocytes with human immunodeficiency virus type 1Ba-L (HIV-1Ba-L ) is significantly inhibited by treatment with the serine protease inhibitor, secretory leukocyte protease inhibitor (SLPI). SLPI does not appear to act on virus directly, but rather the inhibitory activity is most likely due to interaction with the host cell. The current study was initiated to investigate how SLPI interacts with monocytes to inhibit infection. SLPI was found to bind to monocytes with high affinity to a single class of receptor sites (∼7,000 receptors per monocyte, KD = 3.6 nmol/L). The putative SLPI receptor was identified as a surface protein with a molecular weight of 55 ± 5 kD. A well-characterized function of SLPI is inhibition of neutrophil elastase and cathepsin G. However, two SLPI mutants (or muteins) that contain single amino acid substitutions and exhibit greatly reduced protease inhibitory activity still bound to monocytes and retained anti–HIV-1 activity. SLPI consists of two domains, of which the C-terminal domain contains the protease inhibiting region. However, when tested independently, neither domain had potent anti–HIV-1 activity. SLPI binding neither prevented virus binding to monocytes nor attenuated the infectivity of any virus progeny that escaped inhibition by SLPI. A polymerase chain reaction (PCR)-based assay for newly generated viral DNA demonstrated that SLPI blocks at or before viral DNA synthesis. Therefore, it most likely inhibits a step of viral infection that occurs after virus binding but before reverse transcription. Taken together, the unique antiviral activity of SLPI, which may be independent of its previously characterized antiprotease activity, appears to reside in disruption of the viral infection process soon after virus binding.
THE VIRTUAL ABSENCE of oral transmission of human immunodeficiency virus type 1 (HIV-1)1-3 and reports of antiviral activity in human saliva4-10 led to identification of secretory leukocyte protease inhibitor (SLPI) as a potent antiviral factor in saliva.11-13 Recent confirmation of the antiviral role of saliva-derived SLPI14 suggests that this molecule may in fact contribute to the limited evidence for any salivary transmission of HIV-1. SLPI is a low–molecular-weight salivary protein also found in certain other mucous secretions (eg, cervical and bronchial secretions).15 SLPI inhibits HIV-1 infection of human monocytes at physiologic concentrations (1 to 10 μg/mL), and its depletion from saliva results in a significant loss of antiviral activity.11 SLPI does not appear to interact with virus directly, in that it does not bind to viral proteins gp120, gp160, aspartyl protease, or reverse transcriptase (RT).11 Based on these findings, the antiviral activity of SLPI is most likely due to interaction with host cells. However, the inhibitory activity exhibited by SLPI does not involve the HIV-1 cellular receptor CD4, as the protease inhibitor failed to downregulate CD4 expression and did not bind to a recombinant form of the cell-surface protein CD4.11 Many reports suggested that accessory molecules for HIV-1 infectivity may be proteases or have proteolytic activity,16-20 although new evidence suggests this may not be a requisite step.21-24 Nonetheless, SLPI is a potent serine protease inhibitor with biologic activity against a broad range of proteases such as neutrophil elastase and cathepsin G,25 and as such, the antiviral activity exhibited by SLPI could be due to inhibition of a protease-mediated event that is required for virus entry/infectivity of susceptible cells.
This study was initiated to examine SLPI interaction with monocytes as target cells and to investigate the mechanism of SLPI anti–HIV-1 activity toward monocytes. HIV-1 infection of monocytes/macrophages plays a major role in the pathology of AIDS.26,27 Infection of these cells occurs at an early stage after viral transmission, and virus isolated from individuals very early after HIV transmission is monocytotropic in nature.26 Moreover, monocytes that are nondividing are thought to serve as virus reservoirs during the course of infection.26 In this investigation, it was observed that SLPI interacts with monocytes through binding to the monocyte cell surface. Binding of SLPI to monocytes was found to be specific, mediated through high-affinity interactions, and apparently necessary for the antiviral activity of SLPI. The antiviral effect mediated by SLPI was found to occur at an early step in the infection process (eg, virus entry or uncoating), since SLPI did not inhibit virus binding but could inhibit viral DNA synthesis. However, the antiviral activity surprisingly did not appear to depend on the potent antiprotease activity of SLPI. Thus, SLPI may inhibit another critical molecule(s) required during HIV-1 internalization or may function in a novel way to block HIV-1 entry.
MATERIALS AND METHODS
Monocyte isolation and culture.Peripheral blood mononuclear cells (PBMC) obtained by leukapheresis (Department of Transfusion Medicine, National Institutes of Health [NIH]) were separated from granulocytes and red blood cells by fractionation in Ficoll-Hypaque (Pharmacia-LKB, Stockholm, Sweden). PBMC were then separated by countercurrent centrifugal elutriation as previously described.28 By this method, 85% to 95% pure monocytes are obtained.28 Monocytes were diluted to 2 to 4 × 106/mL in Dulbecco's modified Eagle's medium ([DMEM] BioWhittaker, Walkersville, MD) with 2 mmol/L L-glutamine and 50 μg/mL gentamicin, and 2 mL/well was plated in two-well glass chamber slides (Nunc Inc, Naperville, IL). After adherence (overnight at 37°C and 5% CO2 ), 10% human AB− serum (Department of Transfusion Medicine, NIH) was added to the culture medium. Cells were routinely cultured 10 to 14 days before being infected with HIV-1.
Reagents assayed for anti–HIV-1 activity.α1-Protease inhibitor (PI) was obtained from Sigma (St Louis, MO). Either native or recombinant SLPI was used in assays of anti–HIV-1 activity. Native SLPI purified from unstimulated whole saliva (obtained by expectoration) by affinity chromatography using polyclonal goat anti-SLPI antibody (IgG) was found to be greater than 90% to 95% pure by silver stain on a 12% polyacrylamide sodium dodecyl sulfate (SDS) gel under reducing conditions. Recombinant SLPI29 was greater than 98% pure by SDS gel analysis. Recombinant SLPI is equivalent to native SLPI by the following criteria: (1) neither molecule is glycosylated; (2) both molecules have the same amino-terminal sequence (H2N-Ser-Gly-Lys-Ser-Phe-...) due to processing enzymes in Escherichia coli that remove the encoded amino-terminal methionine residue from the recombinant protein; and (3) they possess equivalent biologic activity.25,29 SLPI mutants Phe72 and Gly72 were prepared by mutating residue 72 of recombinant SLPI as described previously.29
To isolate individual domains of SLPI, the intact protein was treated with formic acid and the individual domains were isolated using gel filtration column chromatography.29 SLPI is susceptible to oxidation, which may or may not lead to degradation of the molecule,30-32 and was found to be unstable during repeated freezing/thawing (such as occurs during storage in frost-free freezers) or prolonged storage at 4°C, resulting in a loss of antiviral but not necessarily of antiproteolytic activity. Therefore, SLPI was frozen in 50- to 200-μL aliquots at 1 mg/mL and thawed immediately before use.
HIV-1 infection of monocytes.HIV-1Ba-L33 (Advanced Biotechnologies Inc, Columbia, MD) propagated in primary human monocyte/macrophages was diluted to 1 × 104 TCID50 /mL in DMEM and combined in a 1:1 ratio (vol/vol) with DMEM or purified inhibitory proteins for 30 minutes at 37°C and 5% CO2 . The virus with or without inhibitor was added to adherent monocytes in two-well glass chamber slides at 400 μL/well. After 60 minutes at 37°C in 5% CO2 , unbound virus was removed by washing the cells three times with 2 mL phosphate-buffered saline (PBS) and refeeding with 2 mL DMEM containing 10% human AB− serum, 50 μg/mL gentamicin, and 2 mmol/L L-glutamine (DMEM complete medium). Every 3 to 4 days, 0.5 mL medium was removed for virus assay and replaced with fresh complete medium. Cells were cultured for 3 to 3.5 weeks after infection. When the infection time course was complete, slides were stained with Diff-Quik (Baxter, Miami, FL) to monitor whether equivalent numbers of monocytes remained in each well. Cell culture conditions and infection typically did not affect monocyte viability. However, data are reported only from experiments in which equivalent cell numbers were documented.
Monitoring of viral infection.The RT assay was performed as previously described.34 p24 antigen was assayed using the p24 core profile enzyme-linked immunosorbent assay (ELISA) kit from Dupont (Wilmington, DE).11
Measurement of SLPI binding to monocytes.SLPI (300 μg) was radioiodinated using Iodogen (Pierce, Rockford, IL) and Na125I (0.5 mCi; ICN Biomedicals, Irvine, CA) in PBS, pH 7.4, according to the manufacturer's directions. Specific activity of the radioiodinated SLPI was 4.1 mCi/mg. Purity and integrity of the 125I-labeled SLPI were determined by SDS-PAGE and Western blot analysis using polyclonal goat antibodies to human SLPI. Migration of radioiodinated SLPI (by Coomassie blue staining and autoradiography) and detection with anti-SLPI antibody were equivalent to those of unlabeled SLPI (data not shown). Binding of radioiodinated SLPI to human monocytes was assayed by addition of 125I-labeled SLPI to 12 × 106 freshly isolated monocytes in 1 mL assay buffer (DMEM, 5 mmol/L HEPES, and 1 mg/mL bovine serum albumin (BSA) at either 4°C or 37°C. After a designated incubation time, cells were pelleted and washed twice with cold PBS, and cell-associated radioactivity was measured in the gamma counter. In DMEM alone, a second class of binding sites on monocytes exhibiting 10-fold less affinity could be detected. However, since these were not detected in the presence of BSA, the character of that class of binding sites was not examined further. For competition analysis, cells were incubated with 0.5 nmol/L 125I-labeled SLPI with increasing amounts of SLPI or other competitor in assay buffer overnight at 4°C with shaking, as in the method of Darbonne et al.35 Cells were then pelleted and washed twice with cold PBS, and cell-associated radioactivity was measured in the gamma counter. In all binding assays, a 200-fold excess of SLPI was included to determine nonspecific binding, which did not exceed 15% to 20% of total binding. Antibodies (IgG) used in competition assays included rabbit anti–human neutrophil elastase (Calbiochem, La Jolla, CA), sheep anti–human cathepsin G (Calbiochem), and mouse monoclonal anti–human CD14 (Becton Dickinson Monoclonal Centre Inc, Mountain View, CA). Antibodies used in competition assays were purified from serum by isolation on protein A–acrylamide beads (Pierce) before use in assays.
In additional studies, SLPI binding to monocytes was measured using a SLPI ELISA kit (R & D Systems Inc, Minneapolis, MN). Adherent monocytes (day 10 of culture) were infected with HIV-1Ba-L in the presence of SLPI (10 μg/mL), or alternatively, SLPI alone was added to the adherent cells. After exposure of the cells to SLPI plus virus for 1 hour, the free virus and SLPI were washed away with PBS (three times) and 2 mL complete DMEM was added to the cells in the two-well chamber slides. The cells were incubated at 37°C. At designated times, the supernatant was removed and the cells were solubilized in 150 μL 0.5% Triton X-100. Both the cell lysates and supernatants were assayed for the presence of SLPI.
Identification of the SLPI binding molecule on monocytes.Monocytes (30 to 60 × 106) in ice-cold PBS, which were greater than 95% viable using trypan blue exclusion, were placed in 20-mL glass scintillation vials coated with 100 μg Iodogen and then radioiodinated with 0.5 mCi Na125I for 20 minutes at room temperature. Cells were pelleted, washed twice with PBS, and lysed using 0.5% NP40 in 100 mmol/L Tris HCl, pH 7.4, 150 mmol/L NaCl, 5 mmol/L EDTA, and 1 mmol/L 1-o-phenanthroline on ice for 2 hours. SLPI (12 μg/mL) or buffer alone was added to the lysates, and the lysates were combined with goat anti-SLPI antibody coupled to agarose beads equilibrated in the lysing buffer. After rocking for 2 hours at 4°C, lysates were removed, beads were washed extensively with PBS containing 0.15% NP40, and SLPI binding molecules were eluted using 50 mmol/L glycine, pH 2.5, and 0.15% NP40.
Assay of virus binding to monocyte cell surface.HIV-1Ba-L (104 TCID50 /mL) was added to 4 × 106 monocytes in the presence or absence of SLPI for 30 minutes at 37°C in 1 mL DMEM. Alternatively, cells were preincubated in the presence of SLPI for 30 minutes before addition of virus and SLPI was not washed away before virus addition. Monocytes were pelleted and washed twice in PBS and resuspended in 0.5% Triton X-100. Virus was quantified by p24 ELISA.36
PCR-based assay for newly synthesized viral DNA.To determine whether SLPI targeted an early step in the virus life cycle, a PCR-based assay37 for newly synthesized viral DNA was adapted.38 DNase-treated HIV-1Ba-L was preincubated with SLPI for 15 minutes at 37°C and used to infect human monocytes that had been in culture for 11 days. After a 2.5-hour incubation at 37°C, the cells were washed twice with PBS, treated with trypsin-EDTA (0.5% trypsin and 0.53 mmol/L EDTA) for 5 minutes at room temperature to remove noninternalized virus particles, washed twice with PBS, and then incubated in culture medium (complete DMEM) for 18 hours. This period represents a single round of virus infection.37 Cells were then washed twice with PBS and harvested in lysis buffer (100 mmol/L KCl, 10 mmol/L Tris HCl, pH 8.0, 2.5 mmol/L MgCl2 , 0.5% Tween 20, and 0.5% NP40, 4 × 106 cells/150 μL). Cellular lysates (5 μL each) were subjected to nested-primer PCR amplification (50 μL total volume). The first 30-cycle round of amplification used primers corresponding to the env gene (nt 8838 to 8358, HIV-1HXB2 sequence) and the U3 region of the 3′ long terminal repeat (nt 9533 to 9558). Each cycle consisted of denaturation for 1 minute at 94°C, annealing for 1 minute at 55°C, and elongation for 1 minute at 72°C. PCR reactions (5 μL each) from the first amplification were then subjected to a second 30-cycle amplification round using primers (5′ primer, nt 8754 to 8782; 3′ primer, nt 9436 to 9457) located within the nef gene. PCR products (∼730 bp) from the second amplification were visualized by ethidium bromide staining after agarose gel electrophoresis. Equivalent amounts of cellular DNA per sample were analyzed as indicated by PCR amplification using β-globin–specific primers (data not shown). In additional control experiments, SLPI (0.1 to 100 pg/mL) did not interfere with PCR amplification of target HIV-1 DNA under similar conditions (data not shown).
Statistical analysis.Statistical analysis was performed using Abacus Concepts StatView software (Abacus Concepts Inc, Berkeley, CA) and one-way ANOVA and Fisher's protected least-significant difference test for comparison of data pairs. Data shown are representative of two or three independent experiments.
RESULTS
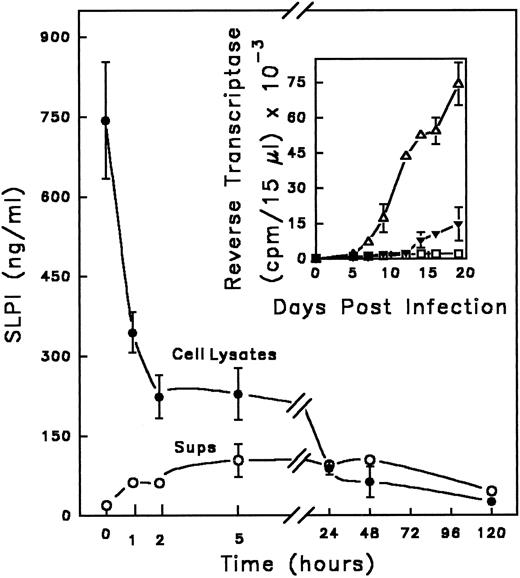
SLPI binds to monocytes in the presence of HIV-1.SLPI inhibits HIV-1 infection of monocytes, but does not appear to interact directly with isolated viral proteins,11 suggesting that the target of SLPI resides on or in the host cell. This hypothesis is consistent with the previously demonstrated retention of SLPI anti–HIV-1 activity on monocytes pretreated with the inhibitor, washed, and then exposed to HIV-1.11 To further explore this possibility, the interaction of SLPI with monocytes during infection with HIV-1 was examined. SLPI (0.6 to 0.9 μg/mL) bound to monocytes during the 1-hour infection period (Fig 1). Similar amounts of SLPI bound to monocytes in the absence of virus (data not shown), indicating that the presence of virus did not increase or alter SLPI binding. Cell-associated SLPI was rapidly degraded, tightly complexed, or processed in such a way as to be unrecognizable by ELISA in both infected and noninfected monocytes, such that only minimal amounts of SLPI could be detected after 24 hours, and did not reappear through day 5 of incubation. However, when present during infection, SLPI binding to monocytes effected a dramatic decrease in HIV-1 infectivity, as shown in an analogous experiment in Fig 1 (inset). Progeny virus production by infected monocytes in the absence of SLPI became measurable by day 5 postinfection, well after SLPI in the treated cultures had been reduced to negligible levels. These data imply that SLPI antiviral activity may occur during an early phase of viral infection.
Kinetics of SLPI interaction with monocytes in the presence of virus. Adherent monocytes were incubated with SLPI in the presence of HIV-1Ba-L for 1 hour at 37°C. (Equivalent results were found using either 5 or 10 μg/mL SLPI; data shown were obtained with 10 μg/mL SLPI.) Cells were washed 3 times with PBS and cultured in DMEM complete medium. At designated times, supernatants were removed, cells were solubilized, and both supernatant (○) and cell lysates (•) were assayed for the presence of SLPI using a SLPI ELISA (background of assay < 100 pg/mL). The time course starts immediately after the 1-hour incubation and is indicated by the zero time point. In parallel (inset), monocytes were infected in the absence (▵) or presence of SLPI 5 μg/mL (▴) or left uninfected (▪ □) and the culture supernatants were monitored for virus by RT activity for 20 days. Values are the mean ± SD; n = 3.
Kinetics of SLPI interaction with monocytes in the presence of virus. Adherent monocytes were incubated with SLPI in the presence of HIV-1Ba-L for 1 hour at 37°C. (Equivalent results were found using either 5 or 10 μg/mL SLPI; data shown were obtained with 10 μg/mL SLPI.) Cells were washed 3 times with PBS and cultured in DMEM complete medium. At designated times, supernatants were removed, cells were solubilized, and both supernatant (○) and cell lysates (•) were assayed for the presence of SLPI using a SLPI ELISA (background of assay < 100 pg/mL). The time course starts immediately after the 1-hour incubation and is indicated by the zero time point. In parallel (inset), monocytes were infected in the absence (▵) or presence of SLPI 5 μg/mL (▴) or left uninfected (▪ □) and the culture supernatants were monitored for virus by RT activity for 20 days. Values are the mean ± SD; n = 3.
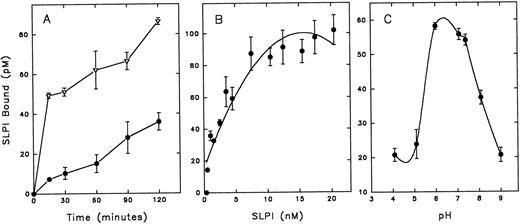
SLPI binds to monocytes with high affinity and specificity.Because the site of SLPI action appears to reside with the monocyte, radioiodinated SLPI was used to further characterize SLPI interaction with monocytes. SLPI bound in a temperature- and time-dependent manner to monocytes (Fig 2A). There was an initial rapid equilibration of SLPI with monocytes at 37°C followed by a slow increase over 2 hours of incubation. The binding rate was greatly reduced at 4°C, and binding was essentially eliminated at 0° to 2°C (data not shown). Since SLPI binding to monocytes increased with time, the remainder of the binding experiments were performed using overnight incubations at 4°C. SLPI binding to monocytes at 4°C overnight was found to reach equilibrium and to be reversible (data not shown).
Assay of SLPI binding to monocytes. Radioiodinated SLPI was added to monocytes (12 × 106) in binding buffer (DMEM, 5 mmol/L HEPES, and 0.1% BSA). Cells were then pelleted and washed twice with PBS, and cell pellets were assayed for 125I emission. Nonspecific binding of 125I-SLPI (binding in the presence of a 200-fold excess of unlabeled SLPI) was subtracted from the indicated results. (A) 2 nmol/L 125I-SLPI was added to monocytes at 37°C (▵) or 4°C (•) and incubated for the designated times before pelleting cells. (B) Increasing amounts of 125I-SLPI were added to monocytes and incubated overnight at 4°C. (C) 3 nmol/L 125I-SLPI was added to monocytes for 1 hour at 4°C in binding buffer adjusted to designated pH values. Values are the mean ± SD; n = 3.
Assay of SLPI binding to monocytes. Radioiodinated SLPI was added to monocytes (12 × 106) in binding buffer (DMEM, 5 mmol/L HEPES, and 0.1% BSA). Cells were then pelleted and washed twice with PBS, and cell pellets were assayed for 125I emission. Nonspecific binding of 125I-SLPI (binding in the presence of a 200-fold excess of unlabeled SLPI) was subtracted from the indicated results. (A) 2 nmol/L 125I-SLPI was added to monocytes at 37°C (▵) or 4°C (•) and incubated for the designated times before pelleting cells. (B) Increasing amounts of 125I-SLPI were added to monocytes and incubated overnight at 4°C. (C) 3 nmol/L 125I-SLPI was added to monocytes for 1 hour at 4°C in binding buffer adjusted to designated pH values. Values are the mean ± SD; n = 3.
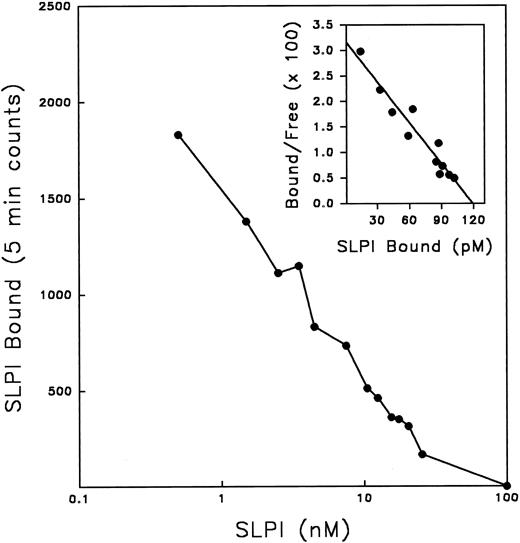
SLPI binding to monocytes was dose-dependent and saturable (Fig 2B). Additionally, SLPI binding to monocytes was pH-dependent, with peak binding in the slightly acidic to neutral range (pH 6 to 6.5; Fig 2C), perhaps reflecting favorable binding at the acidic pH of biologic fluids in which SLPI is found, eg, saliva and bronchial fluids. Radioiodinated SLPI binding to monocytes was competable by unlabeled SLPI (Fig 3). Scatchard analysis of these data indicated high-affinity binding (apparent KD = 3.6 ± 0.05 nmol/L, mean ± SE; n = 9) with a single class of high-affinity binding sites (∼7,000 sites per cell) (Fig 3, inset).
Competition of radioiodinated SLPI binding with unlabeled SLPI. 0.5 nmol/L radioiodinated SLPI plus increasing amounts of unlabeled SLPI were added to monocytes in binding buffer overnight at 4°C. Cells were washed twice with PBS, and cell pellets were assayed for 125I emission. Values shown are the mean of a data set in triplicate. Inset, data replotted by Scatchard method.
Competition of radioiodinated SLPI binding with unlabeled SLPI. 0.5 nmol/L radioiodinated SLPI plus increasing amounts of unlabeled SLPI were added to monocytes in binding buffer overnight at 4°C. Cells were washed twice with PBS, and cell pellets were assayed for 125I emission. Values shown are the mean of a data set in triplicate. Inset, data replotted by Scatchard method.
In initial attempts to determine the identity of the monocyte SLPI receptor, surface radioiodinated monocytes were lysed using 0.5% NP40 and potential SLPI binding proteins were coprecipitated using polyclonal goat anti-SLPI antibodies and either 12 μg/mL SLPI or medium alone. In the presence but not in the absence of SLPI, a major radioiodinated band of approximately 55 ± 5 kD was observed to bind to the SLPI/anti-SLPI antibody complex (Fig 4, right lane). The high–molecular-weight radioiodinated material at the top of the lane was found to be aggregated material, since it did not consistently appear in similar gels and its disappearance did not correspond to the appearance of additional lower–molecular-weight bands. Since the 55-kD band could not be chemically cross-linked to SLPI (data not shown), the identity of this molecule as the SLPI receptor remains tentative.
Coprecipitation of SLPI binding proteins from monocytes using anti-SLPI antibodies. Intact monocytes were surfaced-labeled with Na125I. Radioiodinated cells were lysed in 0.5% NP40, and lysate was added to immobilized anti-SLPI antibodies in the presence or absence of SLPI. Proteins binding to the antibodies were eluted using 50 mmol/L glycine, pH 2.5, and 0.15% NP40. Eluted proteins were assayed by PAGE on a 12% gel under nonreducing conditions. The gel was dried and an autoradiogram was made. A major radioiodinated band coprecipitating with SLPI migrated at 55 ± kD (arrow), whereas the same protein did not bind to the antibodies in the absence of SLPI.
Coprecipitation of SLPI binding proteins from monocytes using anti-SLPI antibodies. Intact monocytes were surfaced-labeled with Na125I. Radioiodinated cells were lysed in 0.5% NP40, and lysate was added to immobilized anti-SLPI antibodies in the presence or absence of SLPI. Proteins binding to the antibodies were eluted using 50 mmol/L glycine, pH 2.5, and 0.15% NP40. Eluted proteins were assayed by PAGE on a 12% gel under nonreducing conditions. The gel was dried and an autoradiogram was made. A major radioiodinated band coprecipitating with SLPI migrated at 55 ± kD (arrow), whereas the same protein did not bind to the antibodies in the absence of SLPI.
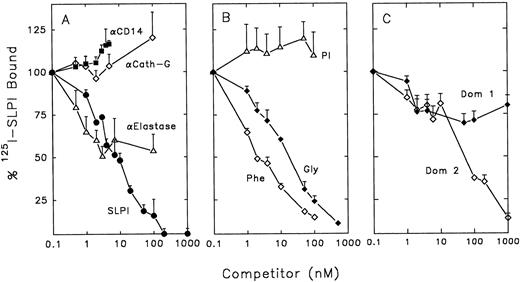
SLPI binding and anti–HIV-1 activity is not dependent on its protease-inhibiting activity.To characterize SLPI binding to monocytes and its relationship to SLPI anti–HIV-1 activity, competition assays were performed using radioiodinated SLPI. Two of the major target enzymes for SLPI inhibition in vivo are neutrophil elastase and cathepsin G.25 Since both of these enzymes may be associated with monocytes,39-41 it was possible that SLPI could bind to either or both of these enzymes on the surface of monocytes. SLPI binding to monocytes was not inhibited by polyclonal antibodies to human cathepsin G (Fig 5A). However, polyclonal antibodies to human neutrophil elastase could inhibit about 50% of SLPI binding to monocytes. Antibodies to CD14 (a constitutively expressed monocyte cell-surface marker), used as a control, did not show any inhibition of SLPI binding at the concentrations used.
Competition of radioiodinated SLPI binding to monocytes. 0.5 nmol/L radioiodinated SLPI was added to 12 × 106 monocytes in binding buffer with increasing amounts of competitors overnight at 4°C. Cells were washed twice with PBS and cell pellets were assayed for 125I emission. Data shown are the mean of triplicate values ± SD, and are representative of experiments performed ≥3 times. (A) Competitors included monoclonal anti-CD14 (▪), anti–human cathepsin G (⋄), anti–human neutrophil elastase (▵), or unlabeled SLPI (•). (B) Competitors included PI (▵), mutein Gly72 (♦), or mutein Phe72 (⋄). (C) Competotors included domain 1 of SLPI (Dom 1, ♦) and domain 2 of SLPI (Dom 2, ⋄).
Competition of radioiodinated SLPI binding to monocytes. 0.5 nmol/L radioiodinated SLPI was added to 12 × 106 monocytes in binding buffer with increasing amounts of competitors overnight at 4°C. Cells were washed twice with PBS and cell pellets were assayed for 125I emission. Data shown are the mean of triplicate values ± SD, and are representative of experiments performed ≥3 times. (A) Competitors included monoclonal anti-CD14 (▪), anti–human cathepsin G (⋄), anti–human neutrophil elastase (▵), or unlabeled SLPI (•). (B) Competitors included PI (▵), mutein Gly72 (♦), or mutein Phe72 (⋄). (C) Competotors included domain 1 of SLPI (Dom 1, ♦) and domain 2 of SLPI (Dom 2, ⋄).
To further investigate whether SLPI binding to elastase (or an elastase-like molecule) on the surface of monocytes was important for anti–HIV-1 activity, mutants of the SLPI molecule were investigated. These muteins contain single amino acid substitutions in the second domain of SLPI at residue 72, the active site of elastase, trypsin, and chymotrypsin inhibition.29 In the mutein Phe72, leucine at position 72 of the wild-type molecule is replaced with phenylalanine. Similarly, in the mutein Gly72, leucine is replaced with glycine. Replacement of leucine with phenylalanine increases the Ki of SLPI for elastase by almost 500-fold (from 0.15 nmol/L to 70 nmol/L.29 However, Phe72 was able to completely inhibit wild-type SLPI binding on monocytes (Fig 5B). Mutein Gly72 also competed with wild-type SLPI for binding to monocytes (Fig 5B). Replacement of leucine with glycine increases the Ki for elastase about 60-fold. However, it reduces even further the chymotrypsin and trypsin inhibitor activities of SLPI; the Ki for chymotrypsin is increased 1,500-fold (from 0.4 to 700 nmol/L) and the Ki for trypsin is increased 300-fold (from 3.0 to 1,000 nmol/L).29 Importantly, the muteins Phe72 and Gly72 retained anti–HIV-1 activity (Fig 6A and B). Interestingly, the apparent high-affinity binding of Phe72 to monocytes, which is greater than that of wild-type SLPI, is reflected in a corresponding modest increase in anti–HIV-1 activity over wild-type SLPI (McNeely TB, unpublished data, 1995).
Anti–HIV-1 activity of SLPI muteins. Adherent monocytes were infected 1 hour with HIV-1Ba-L in the presence of SLPI muteins, PI, or SLPI single domains. Free virus and inhibitor were washed away, and the infection was monitored by RT or p24 assay. Cells were infected in the absence of virus (Con), with virus only (DMEM), or in the presence of inhibitor. Values shown are at day 21 postinfection. *Significantly different from DMEM. (A) Cells were infected in the presence of PI 5 μg/mL or SLPI mutein Gly72 5 μg/mL. (B) Cells were infected in the presence of SLPI mutein Phe72 5 μg/mL. (C) Cells were infected in the presence of SLPI single domain 1 at concentrations of 10 or 20 μg/mL. (D) Cells were infected in the presence of SLPI single domain 2 at concentrations of 10, 20, or 50 μg/mL.
Anti–HIV-1 activity of SLPI muteins. Adherent monocytes were infected 1 hour with HIV-1Ba-L in the presence of SLPI muteins, PI, or SLPI single domains. Free virus and inhibitor were washed away, and the infection was monitored by RT or p24 assay. Cells were infected in the absence of virus (Con), with virus only (DMEM), or in the presence of inhibitor. Values shown are at day 21 postinfection. *Significantly different from DMEM. (A) Cells were infected in the presence of PI 5 μg/mL or SLPI mutein Gly72 5 μg/mL. (B) Cells were infected in the presence of SLPI mutein Phe72 5 μg/mL. (C) Cells were infected in the presence of SLPI single domain 1 at concentrations of 10 or 20 μg/mL. (D) Cells were infected in the presence of SLPI single domain 2 at concentrations of 10, 20, or 50 μg/mL.
In contrast, another potent elastase inhibitor, α1-protease inhibitor (PI), did not compete with SLPI binding to monocytes and did not inhibit HIV-1 (Figs 5B and 6A). Based on these data, the elastase-inhibiting activity of SLPI may not be essential for anti–HIV-1 activity.
Neither of the two domains of SLPI alone has anti–HIV-1 activity.X-ray crystallographic analysis of SLPI complexed with α-chymotrypsin has revealed that SLPI consists of two domains of similar mass and organization.42 To determine if a single domain of SLPI possessed antiviral activity, the wild-type SLPI was cleaved into its two homologous domains using formic acid.29 The N-terminal domain (domain 1) of SLPI only weakly inhibited SLPI binding to monocytes, while the C-terminal domain (domain 2, which retains elastase-inhibiting activity) could inhibit approximately 65% of SLPI binding when present at 200-fold molar excess over SLPI (Fig 5C). However, neither domain independently had potent anti–HIV-1 activity at concentrations tested (Fig 6C to D). Therefore, domain 2 binding to monocytes alone is not sufficient for antiviral activity, further indicating that SLPI antiviral activity does not depend solely on its antielastase activity.
SLPI binding to monocytes during infection interferes with an early step of viral infection.Taken together, the data thus far suggest that high-affinity SLPI binding to monocytes initiates antiviral activity and that SLPI may act early in the virus life cycle to inhibit virus infectivity. Additional experiments were initiated to examine how SLPI binding to monocytes inhibits virus infectivity. SLPI does not bind to the HIV-1 receptor CD4,11 nor does it appear to block HIV-1 binding to CD4, since no inhibition of virus binding to monocytes was observed regardless of whether SLPI was preincubated with cells before virus addition or added concurrently with virus (Fig 7A). Based on these data, SLPI did not appear to interfere with the initial virus-binding step of cellular infection.
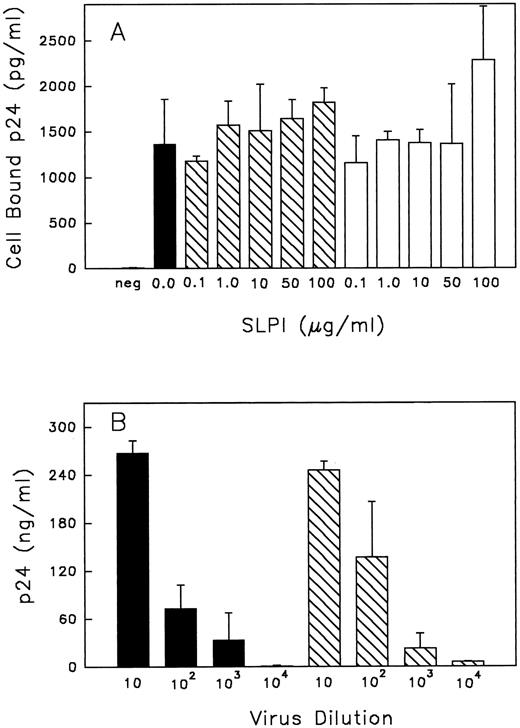
(A) Binding of HIV-1 to monocytes with or without SLPI. Monocytes were either pretreated with SLPI (▧) or DMEM (▪) for 30 minutes at 37°C and then virus was added, or alternatively, virus + SLPI was added to cells concurrently (□) for 30 minutes at 37°C. Cells were washed twice with PBS and resuspended in 0.5% Triton X-100, and cell-bound virus was assayed by p24 ELISA. The negative control (neg) was cells with no virus or SLPI added. Values are the mean ± SD; n = 4. (B) Virus titer produced from cells treated with SLPI. Monocytes infected with HIV-1Ba-L 1 hour in the absence or presence of SLPI 5 μg/mL were cultured for 18 days. Virus from the culture medium on day 18 was collected and titrated in fresh monocytes. For titration, virus was initially equalized based on RT values. Viral titers are from SLPI-treated cells (▧) or untreated cells (▪). Final dilution of virus is indicated under each bar. p24 values are the mean ± SD (n = 3) from day 21 postinfection.
(A) Binding of HIV-1 to monocytes with or without SLPI. Monocytes were either pretreated with SLPI (▧) or DMEM (▪) for 30 minutes at 37°C and then virus was added, or alternatively, virus + SLPI was added to cells concurrently (□) for 30 minutes at 37°C. Cells were washed twice with PBS and resuspended in 0.5% Triton X-100, and cell-bound virus was assayed by p24 ELISA. The negative control (neg) was cells with no virus or SLPI added. Values are the mean ± SD; n = 4. (B) Virus titer produced from cells treated with SLPI. Monocytes infected with HIV-1Ba-L 1 hour in the absence or presence of SLPI 5 μg/mL were cultured for 18 days. Virus from the culture medium on day 18 was collected and titrated in fresh monocytes. For titration, virus was initially equalized based on RT values. Viral titers are from SLPI-treated cells (▧) or untreated cells (▪). Final dilution of virus is indicated under each bar. p24 values are the mean ± SD (n = 3) from day 21 postinfection.
We also considered that SLPI binding to monocytes could result in inhibition of a cellular protease required for maturation of viral proteins, leading to viral progeny with attenuated infectivity. The furin-like enzyme involved in processing gp 160 to the mature viral envelope proteins gp120 and gp41 is an example of such an enzyme.43 To address this possibility, the virus produced by SLPI-treated monocytes, albeit at low levels, was collected and titrated against an equivalent amount of virus produced under similar conditions but in the absence of SLPI treatment. Starting titers were equalized based on RT activity. SLPI treatment during infection did not result in production of aberrant or noninfectious virus particles. That is, virus collected after SLPI treatment was as infectious as untreated virus (Fig 7B). Although SLPI treatment greatly reduced virus production in general, this inhibitor does not appear to alter the infectivity of progeny virus, suggesting that the requisite transcriptional, translational, and proteolytic events necessary for production of progeny virions were not affected by the presence of SLPI, and that SLPI inhibition of HIV-1 most likely occurs at an early step of infection.
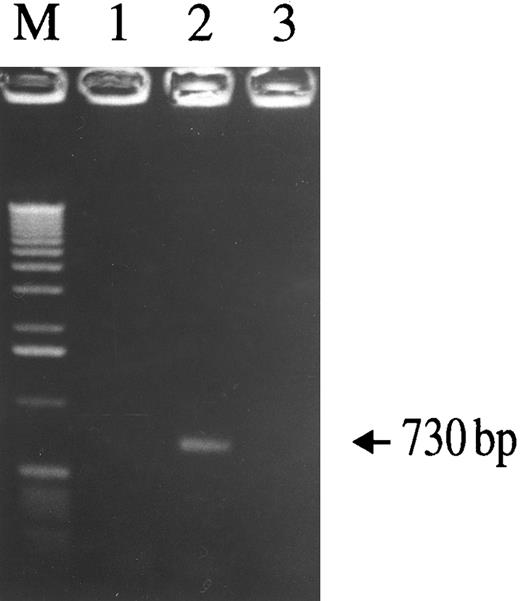
To determine whether SLPI inhibits virus infectivity after virus binding but before reverse transcription, cells were infected and the formation of nascent viral DNA was assessed using a nested-PCR–based assay. The presence of SLPI (5 μg/mL) during the initial virus inoculation period significantly inhibited viral DNA synthesis. As demonstrated by the HIV-1–specific 730-bp PCR product (Fig 8), DNA was detected in cells infected with HIV-1Ba-L alone (lane 2), but was not detected in either uninfected cells (lane 1) or cells infected in the presence of SLPI (lane 3). Therefore, SLPI inhibits HIV-1 infectivity after virus binding and before reverse transcription proceeds. Since it was previously shown that SLPI does not inhibit viral RT,11 SLPI inhibitory activity must precede reverse transcription. Therefore, SLPI antiviral activity can be localized to a step immediately after virus binding, probably during viral entry and/or uncoating.
Assay of nascent viral DNA synthesis in monocytes with or without SLPI treatment. Adherent monocytes were infected with HIV-1Ba-L for 2.5 hours, washed extensively, trypsinized to remove loosely bound virus particles, and then incubated for 18 hours. Cellular lysates were prepared and assayed for the presence of viral DNA using a nested-PCR–based assay. A 730-bp HIV-1–specific PCR product was detected in virus-infected cells (lane 2), but not in uninfected cells (lane 1). In the presence of 5 μg/mL SLPI, no virus-specific PCR product was generated (lane 3), indicating SLPI-mediated inhibition of viral DNA formation. Equivalent amounts of cellular DNA per sample were analyzed as indicated by PCR amplification using β-globin–specific primers (data not shown).
Assay of nascent viral DNA synthesis in monocytes with or without SLPI treatment. Adherent monocytes were infected with HIV-1Ba-L for 2.5 hours, washed extensively, trypsinized to remove loosely bound virus particles, and then incubated for 18 hours. Cellular lysates were prepared and assayed for the presence of viral DNA using a nested-PCR–based assay. A 730-bp HIV-1–specific PCR product was detected in virus-infected cells (lane 2), but not in uninfected cells (lane 1). In the presence of 5 μg/mL SLPI, no virus-specific PCR product was generated (lane 3), indicating SLPI-mediated inhibition of viral DNA formation. Equivalent amounts of cellular DNA per sample were analyzed as indicated by PCR amplification using β-globin–specific primers (data not shown).
DISCUSSION
Saliva contains a unique anti–HIV-1 activity that can be attributed in part to the presence of SLPI.11-14 SLPI plays an important role in the antiviral activity of saliva in vitro11 and possibly in vivo. Therefore, elucidation of the mechanism of its activity is central in our understanding of one of the body's endogenous defense mechanisms against this deadly virus. Although the physiologic role of SLPI is believed to center on its inhibition of neutrophil elastase and cathepsin G at sites of inflammation,25 SLPI may also exhibit broader functions as a host defense factor, consistent with its location in areas of the body routinely exposed to potential pathogens (eg, upper respiratory tract, cervix, and oral cavity). SLPI is found in equivalent amounts in the saliva and salivary glands of both normal and HIV-1–infected individuals,12,13 thus pointing to its potential importance in thwarting the spread of HIV-1 via the oral route. We have previously demonstrated the anti–HIV-1 activity of SLPI toward both the monocytotropic isolate (Ba-L) and lymphocytotropic isolates (IIIB and NL4-3), in purified primary monocytes (Ba-L) or T cells (IIIB), and in mixed peripheral blood mononuclear cells (Ba-L, IIIB, and NL4-3).12 14 The anti–HIV-1 mechanism of SLPI toward monocytes was examined in detail in this report.
Data from several sets of experiments indicate that SLPI antiviral activity resides in its ability to bind to monocytes and disrupt an early point of the virus life cycle. Using radioiodinated SLPI, specific and high-affinity binding of SLPI to monocytes was demonstrated. SLPI did not block virus binding to monocytes, nor was the infectivity of the low residual amount of virus produced by monocytes in the presence of SLPI significantly attenuated. Importantly, SLPI clearly inhibited nascent viral DNA formation. It therefore appears that SLPI anti–HIV-1 activity involves inhibition of the infection process just after virus binding to CD4 (eg, membrane fusion between viral and cellular membranes, or viral uncoating). Surprisingly, the antiviral activity of SLPI appeared not to depend on its known antiprotease activity.
Evidence generated through the use of recombinant SLPI muteins suggested that SLPI anti–HIV-1 activity may be distinct from its previously characterized antiprotease action. Muteins that have greatly reduced antiprotease activity competed for SLPI binding to monocytes and retained anti–HIV-1 activity. Domain 2 of SLPI, which possesses elastase inhibitory activity29 and partially inhibited SLPI binding to monocytes, did not independently retain anti–HIV-1 activity. These data are compatible with new evidence ruling out host cell proteases (eg, tryptase, elastase, cathepsin G, and dipeptidyl peptidase IV)16-20,40,41 as important factors for HIV-1 viral infectivity.22-24 Recent studies have identified several monocyte chemokines (RANTES, MIP-1α, and MIP-1β) as inhibitors for HIV-1,21 and chemokine receptors as coreceptors, together with CD4, for HIV-1.22-24 Although SLPI alone (10 μg/mL) is not chemotactic and does not block chemotaxis of monocytes in the presence of either RANTES, MIP-1α, or MCP1 (10 to 20 ng/mL) (McNeely TB, unpublished observations, 1996), additional studies in progress focus on the potential interaction of SLPI with these or other chemokine receptors. Taken together, intact SLPI binding with high affinity to its receptor(s) on monocytes appears to be necessary for anti–HIV-1 activity. The identification of the monocyte receptor(s) for SLPI is of much interest. This molecule(s) could be involved in early steps of viral entry into monocytes, such that complete blockage of that function by SLPI may lead to loss of infectivity. We have identified a putative SLPI receptor on monocytes with molecular mass of 55 ± 5 kD. This molecule is most likely a surface protein, based on the labeling method used. The identity of this protein remains unknown and is the object of ongoing investigation. The small molecular size, cationic charge, and two domains of SLPI may give it a unique ability to locate and bind to molecules important for early events of viral infectivity.
Having established that the point of viral infection at which SLPI antiviral activity occurs is early in the infection process, the mechanism of this endogenous inhibitor can be further dissected. Since SLPI appears to function primarily at a step preceding viral DNA synthesis, elucidation of its mechanism holds potential promise in the development of interventions to reduce viral spread during infection. Additionally, since SLPI antiviral activity is apparently effected through interaction with host cell molecules and not viral molecules, SLPI anti–HIV-1 activity may not lead to viral resistance in vivo. Its potential presence in other secretions of the body is therefore of great importance.
ACKNOWLEDGMENT
The authors gratefully acknowledge the many helpful discussions with David Dripps during preparation of the manuscript.
Supported by a Collaborative Research and Development Agreement between the National Institute of Dental Research (NIDR) and Amgen (Synergen) Inc, and in part by a Neurobehavioral Science Research Training Program grant (T32MH15177) and NIDR Grant No. R01DE12162 (D.C.S.).
Address reprint requests to Tessie B. McNeely, PhD, WP26B-1144, Box 4, Merck Inc, West Point, PA 19486.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal