Abstract
The H genes, encoding an α1,2fucosyltransferase, which defines blood groups with the H structure, of four Bombay and 13 para-Bombay Japanese individuals were analyzed for mutations. Four Bombay individuals were homologous for the same null H allele, which is inactivated by a single nonsense mutation at position 695 from G to A (G695A), resulting in termination of H gene translation. The allele inactivated by the G695A was designated h1. The other 13 para-Bombay individuals possessed a trace amount of H antigens on erythrocytes regardless of their secretor status. Sequence analysis of their H genes showed four additional inactivated H gene alleles, h2, h3, h4, and h5. The h2 allele possesed a single base deletion at position 990 G (990-del). The h3 and h4 alleles possessed a single missense mutation, T721C, which changes Tyr 241 to His, and G442T, which changes Asp148 to Tyr, respectively. The h5 allele possessed two missense mutations, T460C (Tyr154 to His) and G1042A (Glu348 to Lys). The h2, h3, h4, and h5 enzymes directed by these alleles were not fully inactivated by the deletion and the missense mutations expressing some residual enzyme activity resulting in synthesis of H antigen on erythrocytes. Thirteen para-Bombay individuals whose erythrocytes retained a trace amount of H antigen were determined to be heterozygous or homozygous for at least one of h2, h3, h4, or h5 alleles. This clarified that the levels (null to trace amount) of H antigen expression on erythrocytes of Bombay and para-Bombay individuals are determined solely by H enzyme activity. These mutations found in the Japanese H alleles differ from a nonsense mutation found in the Indonesian population. To determine the roles of the H, Se, and Le genes in the expression of H antigen in secretions and Lewis blood group antigen on erythrocytes, the Lewis and secretor genes were also examined in these Bombay and para-Bombay individuals. The Lewis blood group phenotype, Le(α- b+), was determined by the combinatorial activity of two fucosyltransferases, the Lewis enzyme and the secretor enzyme, and the secretor status was solely determined by the secretor enzyme activity, not by H enzyme activity. Bombay individuals were confirmed to be homozygous for the inactivated H and Se genes. As expected from the very low frequency of Bombay and para-Bombay individuals in the population, ie, approximately one in two or 300,000, the H gene mutations were found to be very variable, unlike the cases of the point mutations in the other glycosyltransferase genes; the ABO genes, the Lewis gene, and the secretor gene.
RECENT PROGRESS IN molecular genetic analysis of the human blood group systems has clarified that blood group antigen polymorphism is determined mostly by point mutations and sometimes by deletion and recombination of the responsible genes.1-11 Carbohydrate structure polymorphisms of blood group antigens, such as the ABO and Lewis antigens, are determined by changes in substrate specificity or inactivation of the glycosyltransferases responsible for synthesis of the carbohydrate antigenic epitopes.1,2,5-11 Point mutations within the glycosyltransferase genes have been found to determine the enzyme substrate specificity and activity. In the ABO system, four missense mutations, which are conserved in all ethnic groups were found to determine the A or B alleles. Of these mutations, an amino acid substitution at position 268 of the glycosyltransferase is critical to determine donor specificity for either the A or B antigen.1,2 Our recent finding of three missense mutations in the Lewis gene (Le gene) in Lewis antigen-negative (Le[-]) individuals showed that the loss of expression of Lewis antigen on erythrocytes in Le(-) individuals is caused by inactivation of the Le enzyme by a single amino acid substitution in the catalytic domain of the enzyme.7 Molecular genetic analysis of the mutant Le genes verified that the Le gene is further responsible for determining type-1 chain Lewis antigen expression, including sialyl Lewis a antigen expression as a tumor marker, in intestinal tissues such as colon tissue.12
The human H gene encoding the H enzyme, an α1,2fucosyltransferase (α1,2FT) transferring fucose to galactose with α1,2 linkage, was first cloned by the expression cloning method.13,14 The human secretor gene (Se gene), which determines secretion of ABH antigens into secretions such as saliva, was classically considered to regulate the H gene expression in such secretory tissues.15 However, to date, two different kinds of human α1,2FTs have been isolated, and biochemical analysis of these purified α1,2FTs has strongly suggested that the Se gene is a structural gene encoding an α1,2FT distinct from the H enzyme.16-18 Recent success in cloning of the Se gene clearly verified that the Se gene is a structural gene encoding an α1,2FT whose substrate specificity is very similar to that of the H enzyme.10,19,20 Inactivation of the Se enzyme was also found to be caused by point mutations in the Se gene.10,11,21 22
There is a very rare mutation of the H gene, and persons with this mutation, referred to as Bombay or para-Bombay individuals, lack the H enzyme and the H antigens on erythrocytes.23,24 Bombay individuals are defined as those who lack ABH antigens either on erythrocytes or in secretions such as saliva and milk. Para-Bombay individuals possess similar phenotypes to Bombay individuals. There are two different definitions of para-Bombay individuals. One defines them as the individuals who lack ABH antigens on erythrocytes, but possess them in secretions. Another definition includes, in addition to the above individuals, people who possess very few ABH antigens on erythrocytes compared with ordinary O-type individuals regardless of the presence or the absence of ABH antigens in secretions. The H genes of one Bombay individual and one para-Bombay individual in an Indonesian sample were analyzed,23 and point mutations were found that terminate translation and result in complete inactivation of the H enzyme. Although the H gene was proven to be responsible for the H antigen expression on erythrocytes by this study,23 it is not known what determines various levels of H antigen expression on the erythrocytes of para-Bombay individuals who possess a trace amount of ABH antigens, how precisely the H and Se enzymes are involved in the antigen expression on erythrocytes and in saliva, and whether any correlation exists among H, Se, and Le genes.
To explore these problems and to identify H gene mutation(s) in the Japanese population, we analyzed the H, Se, and Le genes of four Bombay individuals and 13 para-Bombay individuals, who were classified by the second definition. In addition, we had data on the Se and Le genes from more than 200 Japanese individuals.7 11 We found one mutant allele with G695A (Trp232 to termination) in the H genes of the Bombay individuals, and four mutated alleles with G990-del, T721C (Tyr241 to His), G442T (Asp148 to Tyr) and T460C (Tyr154 to His) plus G1042A (Glu348 to Lys), respectively, in the H genes of the para-Bombay individuals. These results are described together with results indicating linkage between the H and Se genotypes, and we discuss the phylogenic aspects of H gene mutations.
MATERIALS AND METHODS
Peripheral blood samples and saliva samples.Peripheral blood samples that were donated by volunteers to Red Cross Blood Centers in Japan for use in blood transfusion were routinely subjected to ABO blood typing by hemagglutination tests. The donors whose erythrocytes showed the rare phenotypes having none or few ABH antigens were suspected to be Bombay or para-Bombay individuals. A total of 5 mL of peripheral blood was bled with heparin from the suspected Bombay and para-Bombay individuals who also donated saliva samples. ABH antigens were determined on erythrocytes and in saliva.
Detection of ABH antigens on erythrocytes.Two hemagglutination methods were employed to detect ABH antigens on erythrocytes. In the ordinary method, one drop of antibody was placed on a glass slide and mixed with washed red blood cells (RBCs). After a 1-hour incubation at room temperature, hemagglutination was carefully observed. A more sensitive method, the adsorption-elution test, was employed for detection of trace amounts of ABH antigens. For detection of H antigen, 100 μL of washed and packed RBCs was incubated with 100 μL of 10% Ulex lectin at 37°C for 7 minutes with continuous agitation. After centrifugation, the supernatant was discarded and the RBCs were washed five times with phosphate-buffered saline (PBS) (pH 7.2). The lectin bound to RBCs was eluted by 30 minutes incubation at 50°C. The amount of the lectin contained in the eluate was measured by the routine hemagglutination of O-type RBCs. The same adsorption-elution test was used to detect the trace amount of A and B antigens on the RBCs of Ah, Bh, and ABh individuals using anti-A or B antibody instead of Ulex lectin. The first incubation was done at 4°C and the elution of the antibody bound to RBCs was done at 50°C for 30 minutes. Antibody in the eluate was assayed by the hemagglutination of each type RBC.
Detection of ABH antigens in saliva.The presence or absence of ABH antigens in saliva was determined by hemagglutination inhibition using standard serological techniques.25
Primers and the first polymerase chain reaction (PCR) amplification.PCR primers and PCR conditions used in this study are listed in Table 1. To amplify the full-length H, Se, and Le genes, the 5′- and 3′-flanking sequences of each gene were used for sense mk1, tk1, and sn1 primer sequences, and antisense mk2, tk2, and sn2 primer sequences, respectively. Each primer (1 μmol/L) was added to 1 μg of genomic DNA in a total volume of 50 μL containing 200 μmol/L of each deoxynucleoside triphosphate (dNTP), 10 mmol/L Tris-HCl (pH 8.8), 50 mmol/L KCl, 2.5 mmol/L MgCl2, and 0.1 mg/mL gelatin. Thirty cycles in the conditions described in Table 1 were run, and the PCR product from each set of primers was used as template in the genotyping of each gene by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) described later. For sequencing the full-length H genes, the products by mk1 and mk2 primers were subcloned into pBluescript SK(-) (pBS). The mk1 and mk2 primers were flanked by EcoR I and Hind III sequences, respectively, for convenient subcloning into pBS vector.
Primer Pairs and PCR Conditions for Screening of Mutation in H and Se Genes
| . | Name . | Primers (5′-3′) . | Inactivated Allele Detected . | Annealing Temperature . | Fragment Sizes . | Enzyme . |
|---|---|---|---|---|---|---|
| H gene | mk1 | CTCGAATTCCTCAGCCTCAGAGCATTTG EcoRI | (h4) | 60°C | 1,170 bp | — (Eam1105 I) |
| mk2 | CTCAAGCTTCTACTTCAGAAAGTCTCCCTG HindIII | |||||
| mk3 | CTATCTGCAGGTTATGCCTCAGCTCT | h1 | 62°C | 78 bp | Xba I | |
| mk4 | AACCAGTCCATGGCCTGCCGGAGG | |||||
| mk5 | GCAACCACACCATTATGACCATTG | h2 | 62°C | 116 bp | Xho I | |
| mk6 | GATCTTCAGGAACTCAGAGTCTCG | |||||
| mk7 | AAGGGTGTGGTGGGCGACAGGGCC | h3 | 62°C | 101 bp | Apa I | |
| mk8 | TCCATGCCGTTGCTGGTGACCACGA | |||||
| mk9 | TTCACGACTGGATGCGGAGGTG | h5 | 62°C | 126 bp | ApaLI | |
| mk10 | GCAGGGTGAACTCTCTGCGGATC | |||||
| mk11 | AACTTCACCCTGCCAGACTCTGAG | h5 | 62°C | 125 bp | BstXI | |
| mk12 | GGCTCTCAAGGCTTAGCCAATGTC | |||||
| Se gene | tk1 | CTCGAATTCGGGCCTCCATCTCCCAGCTAAC EcoRI | 65°C | 1,170 bp | ||
| tk2 | CTCAAGCTTGCTTCTCATGCCCGGGCACTC HindIII | |||||
| tk5 | CAGGATCCCCTGGCAGAACTACCACATTAA | sej | 65°C | 98 bp | Ase I | |
| tk6 | AGCAGGGGTAGCCGGTGAAGCGGACGTACT | |||||
| tk7 | AACGACTGGATGGAGGAGGAATACCGCAGC | Se2, sej | 65°C | 80 bp | Alu I | |
| tk8 | AAGGTCCAGGAGCAGGGGTAGCCGGTGAAG | |||||
| Le gene | sn1 | CTCGAATTCTAAGCAGGAGATTGTCATCACTGACC EcoRI | 60°C | 1,615 bp | ||
| sn2 | CTCAAGCTTCGTGCCGTGATGATCTCTCTGCAC HindIII | |||||
| sn3 | CCATGGCGCCGCTGTCTGGCCGCCC | le1, le2 | 62°C | 93 bp | Msp I | |
| sn4 | AGTGGCATCGTCTCGGGACACACG | |||||
| sn6 | CGCTCCTTCAGCTGGGCACTGGA | le2 | 60°C | 109 bp | HindIII | |
| sn7 | CGGCCTCTCAGGTGAACCAAGAAGCT | |||||
| sn8 | ACTTGGAGCCACCCCCTAACTGCCA | le1 | 70°C | 206 bp | Pvu II | |
| sn9 | TGAGTCCGGCTTCCAGTTGGACACC |
| . | Name . | Primers (5′-3′) . | Inactivated Allele Detected . | Annealing Temperature . | Fragment Sizes . | Enzyme . |
|---|---|---|---|---|---|---|
| H gene | mk1 | CTCGAATTCCTCAGCCTCAGAGCATTTG EcoRI | (h4) | 60°C | 1,170 bp | — (Eam1105 I) |
| mk2 | CTCAAGCTTCTACTTCAGAAAGTCTCCCTG HindIII | |||||
| mk3 | CTATCTGCAGGTTATGCCTCAGCTCT | h1 | 62°C | 78 bp | Xba I | |
| mk4 | AACCAGTCCATGGCCTGCCGGAGG | |||||
| mk5 | GCAACCACACCATTATGACCATTG | h2 | 62°C | 116 bp | Xho I | |
| mk6 | GATCTTCAGGAACTCAGAGTCTCG | |||||
| mk7 | AAGGGTGTGGTGGGCGACAGGGCC | h3 | 62°C | 101 bp | Apa I | |
| mk8 | TCCATGCCGTTGCTGGTGACCACGA | |||||
| mk9 | TTCACGACTGGATGCGGAGGTG | h5 | 62°C | 126 bp | ApaLI | |
| mk10 | GCAGGGTGAACTCTCTGCGGATC | |||||
| mk11 | AACTTCACCCTGCCAGACTCTGAG | h5 | 62°C | 125 bp | BstXI | |
| mk12 | GGCTCTCAAGGCTTAGCCAATGTC | |||||
| Se gene | tk1 | CTCGAATTCGGGCCTCCATCTCCCAGCTAAC EcoRI | 65°C | 1,170 bp | ||
| tk2 | CTCAAGCTTGCTTCTCATGCCCGGGCACTC HindIII | |||||
| tk5 | CAGGATCCCCTGGCAGAACTACCACATTAA | sej | 65°C | 98 bp | Ase I | |
| tk6 | AGCAGGGGTAGCCGGTGAAGCGGACGTACT | |||||
| tk7 | AACGACTGGATGGAGGAGGAATACCGCAGC | Se2, sej | 65°C | 80 bp | Alu I | |
| tk8 | AAGGTCCAGGAGCAGGGGTAGCCGGTGAAG | |||||
| Le gene | sn1 | CTCGAATTCTAAGCAGGAGATTGTCATCACTGACC EcoRI | 60°C | 1,615 bp | ||
| sn2 | CTCAAGCTTCGTGCCGTGATGATCTCTCTGCAC HindIII | |||||
| sn3 | CCATGGCGCCGCTGTCTGGCCGCCC | le1, le2 | 62°C | 93 bp | Msp I | |
| sn4 | AGTGGCATCGTCTCGGGACACACG | |||||
| sn6 | CGCTCCTTCAGCTGGGCACTGGA | le2 | 60°C | 109 bp | HindIII | |
| sn7 | CGGCCTCTCAGGTGAACCAAGAAGCT | |||||
| sn8 | ACTTGGAGCCACCCCCTAACTGCCA | le1 | 70°C | 206 bp | Pvu II | |
| sn9 | TGAGTCCGGCTTCCAGTTGGACACC |
Sequencing of H genes and determination of H genotypes by PCR-RFLP method.To distinguish genuine point mutations from artificial misincorporation by Taq DNA polymerase, at least eight H gene subclones in the pBS vector from each individual were fully sequenced by the dideoxy chain termination method. The point mutation shared in multiple clones among eight clones was considered to be a genuine mutation, and the mutations found in a single clone were disregarded. Seven point mutations found during sequencing of the 40 clones from five individuals in total were regarded as false mutations. They were confirmed afterward by the PCR-RFLP method.
After finding a point mutation in the H gene, we designed primers for detection of the mutation by the PCR-RFLP method. The principle of the PCR-RFLP method has been previously described in detail.7 11 For example, we first found the G695A nonsense mutation in a Bombay individual by sequencing as described in the Results section. The allele with the G695A mutation was named the h1 allele because this mutation was identified for the first time in this study. To detect the h1 allele by PCR-RFLP, we designed primers, mk3 and mk4, to amplify the 78-bp PCR product using the first full-length PCR product as a template. The sense mk3 primer was designed as a primer mismatched to the original H gene sequence by changing G to T at position 693 to create an Xba I site, TCTAGA, in the PCR product from the mutant h1 allele. The product from the h1 allele was separated into two fragments, 54 and 24 bp, by Xba I digestion. Thus, all 17 individuals, four Bombay and 13 para-Bombay, were tested for the h1 allele by PCR-RFLP. After determination of h1 allele distribution in the 17 individuals, individuals without the h1 allele were selected for full sequencing of the H gene to find different point mutations. The G990-del was found in an individual and the allele with the G990-del was named h2. Again the PCR-RFLP method for detection of this mutation was established using the mk5 and mk6 primers shown in Table 1. In the same manner, inactivated alleles by point mutations were successively found, and the five inactivated alleles in total, h1 to h5, were identified in the 17 individuals. All primers, the PCR conditions for determination of each allele, and the enzymes used for digestion are listed in Table 1.
Electrophoresis of the digested second PCR products for detection of the point mutations was performed on a 4% agarose gel (NuSieve GTG agarose, FMC Corp BioProducts, Rockland, ME), and DNA was visualized by ethidium bromide staining.
Determination of Se and Le genotypes.The PCR-RFLP method for the determination of Japanese Se and Le genotypes was described in detail in our previous reports.7 11 The principle of the PCR-RFLP method is the same as described in the previous section. In brief, PCR-RFLPs for the detection of the C357T and A385T mutations in the Se gene were performed using the tk5 and tk6 primer set and the tk7 and tk8 primer set, respectively. The PCR-RFLPs for detection of the T59G, G508A and T1067A mutations in the Le gene were performed using the primer sets, sn3 and sn4, sn5 and sn6, and sn7 and sn8, respectively. These primers, conditions of PCR, and enzymes used for digestion of PCR products are shown in Table 1.
H gene expression in COS-1 cells.Modification of the pCDM8 vector to introduce an EcoR I site was done to facilitate subcloning. Inserts with real mutations were excised by EcoRI and Xho I digestion from pBS vector and subcloned into the EcoRI and Sal I sites of the expression vector pCDM8. COS-1 cells were transfected with 15 μg of each vector with insert in combination with 1 μg of the β-actin promoter driven luciferase expression vector as an indicator of transfection efficiency.
Assays of α1,2FT and luciferase activities.Luciferase activity was measured as previously described.7 The detailed method for measuring α1,2FT activity was described in a previous report.11 In brief, phenyl-β-D-galactose (Phe-Gal) was used as an acceptor substrate, and assays were performed in a total volume of 100 μL containing 50 mmol/L Tris-HCl (pH 7.2), 0.3% Triton X-100, 10 mmol/L NaN3, 10 mmol/L MnCl2, 5 mmol/L adenosine triphosphate (ATP), 25 μmol/L guanosine diphosphate (GDP)-fucose, 0.2 μmol/L 3H-GDP–fucose, 30 mmol/L Phe-Gal, and 10 μg protein of soluble fraction of homogenate of the transfected COS-1 cells. After incubation at 37°C for 1 hour, the reaction mixture supernatant was diluted with 5 mL of water and applied to Sep-Pak plus C18 (Waters, MA) for binding of Phe-Gal. 3H-fucose–incorporated Phe-Gal was eluted from the Sep-Pak plus C18 with 5 mL of methanol, and the radioactivity in the eluate was counted in a liquid scintillation counter. The actual radioactivity was divided by luciferase activity directed by cotransfected luciferase cDNA to normalize α1,2FT activity of the transfected COS-1 cells. The luciferase activity was relatively constant from transfection to transfection with errors within plus or minus 20%.
RESULTS
RBC- and saliva-phenotypes of Bombay and para-Bombay individuals.The RBC and saliva phenotypes of two control ordinary O-type individuals (YN and KN), four Bombay individuals (KK, KaT, JM, and GO) and 13 para-Bombay individuals (AI, MO, AM, HN, YH, MY, HF, KT, HiN, NK, HA, SK, and YT) were determined to confirm the alleged phenotypes, and results are summarized in Table 2. ABH antigens on erythrocytes of all Bombay and para-Bombay individuals were not detected by the ordinary method of hemagglutination. Using a more sensitive method, the adsorption-elution test described in Materials and Methods, 13 para-Bombay individuals, including two Omh (B)-type (MO and AI), two Ah-type (YT and AM), two Bh-type (NK and HiN), one ABh-type (HF), and six Omh-type (HN, YH, MY, KT, HA, and SK) individuals, were determined to have a trace amount of ABH antigens on erythrocytes, whereas three Oh-type (KK, KaT, and JM) and one Oh (A)-type individual (GO) completely lacked erythrocyte expression of the ABH antigens. Among the 13 para-Bombay individuals, AI, MO, HA, SK, and YT possessed fewer H antigens than the others. Their secretor status was also determined by detection of ABH antigens in saliva, and six persons, KK, KaT, JM, GO, HA, and SK, were determined to be nonsecretors. Thus, three Oh-type and one Oh (A)-type individuals, KK, KaT, JM, and GO, were confirmed to be Bombay individuals who lack ABH antigens either on erythrocytes or in saliva. The other 13 were classified as para-Bombay individuals because they had different phenotypes from the Bombay individuals. They were determined to be para-Bombay individuals, as they had markedly decreased levels of ABH antigens on erythrocytes, regardless of their secretor status.
Genotypes and Phenotypes of Bombay and Para-Bombay Individuals
| Initials . | Birthplace . | . | Antigens on Erythrocytes . | Antigens in Saliva . | Adsorption-Elution Test . | Anti-H or -HI Antibody . | Lewis Phenotype . | Saliva . | Alleles . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | A or B . | H . | A or B . | H . | H . | in Serum . | . | . | H . | Se . | Le . |
| YN | O | − | + | − | + | 3+ | — | ||||||
| KN | O | − | + | − | + | 3+ | — | ||||||
| KK | Hamamatsu | Oh | − | − | − | − | 0 | Anti-H | Le(a+b−) | Nonsecretor | h1/h1 | sej/sej | Le/le2 |
| KaT | Aichi | Oh | − | − | − | − | 0 | Anti-H | Le(a+b−) | Nonsecretor | h1/h1 | sej/sej | Le/Le |
| JM | Niigata | Oh | − | − | − | − | 0 | Anti-H | Le(a+b−) | Nonsecretor | h1/h1 | sej/sej | Le/Le |
| GO | Gifu | Oh(A) | − | − | − | − | 0 | Anti-H | Le(a+b−) | Nonsecretor | h1/h1 | sej/sej | Le/le1 |
| AI | Osaka | Omh(B) | − | − | + | NT | +w | Anti-HI | Le(a−b+) | Secretor | h2/h2 | Se1/Se1 | Le/le1 |
| MO | Kita-Osaka | Omh(B) | − | − | + | NT | +w | Anti-HI | Le(a−b−) | Secretor | h2/h2 | Se1/Se1 | le1/le2 |
| AM | Miyazaki | Ah | + | +w | NT | NT | NT | Anti-H | Le(a−b+) | Secretor | h3/h3 | Se2/Se2 | Le/le1 |
| HN | Kanagawa | Omh | − | − | − | + | 2+ | Anti-HI | Le(a−b−) | Secretor | h3/h3 | Se2/Se2 | le1/le1 |
| YH | Fukui | Omh | − | − | − | + | 2+ | Anti-HI | Le(a−b+) | Secretor | h3/h3 | Se2/Se2 | Le/Le |
| MY | Fukui | Omh | − | − | − | + | 2+ | Anti-HI | Le(a−b+) | Secretor | h3/h3 | Se2/Se2 | Le/Le |
| HF | Gunma | ABh | + | +w | + | + | NT | Anti-H | Le(a−b+) | Secretor | h3/h3 | Se2/Se2 | Le/le1 |
| KT | Gifu | Omh | − | − | − | + | 2+ | Anti-HI | Le(a−b+) | Secretor | h1/h3 | Se2/sej | Le/le2 |
| HiN | Gunma | Bh | ± | − | + | − | 1+ | Anti-H | Le(a−b+) | Secretor | h1/h3 | Se2/Se2 | Le/le1 |
| NK | Gunma | Bh | ± | − | + | NT | 1+ | Anti-H | Le(a−b+) | Secretor | h3/h5 | Se1/Se2 | Le/Le |
| HA | Tokushima | Omh | − | − | − | − | +w | Anti-HI | Le(a+b−) | Nonsecretor | h4/h4 | sej/sej | Le/le2 |
| SK | Tokushima | Omh | − | − | − | − | +w | Anti-HI | Le(a+b−) | Nonsecretor | h4/h4 | sej/sej | Le/le2 |
| YT | Okayama | Ah | − | − | + | + | +w | Anti-H | Le(a−b+) | Secretor | h5/h5 | Se1/Se1 | Le/Le |
| Initials . | Birthplace . | . | Antigens on Erythrocytes . | Antigens in Saliva . | Adsorption-Elution Test . | Anti-H or -HI Antibody . | Lewis Phenotype . | Saliva . | Alleles . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | A or B . | H . | A or B . | H . | H . | in Serum . | . | . | H . | Se . | Le . |
| YN | O | − | + | − | + | 3+ | — | ||||||
| KN | O | − | + | − | + | 3+ | — | ||||||
| KK | Hamamatsu | Oh | − | − | − | − | 0 | Anti-H | Le(a+b−) | Nonsecretor | h1/h1 | sej/sej | Le/le2 |
| KaT | Aichi | Oh | − | − | − | − | 0 | Anti-H | Le(a+b−) | Nonsecretor | h1/h1 | sej/sej | Le/Le |
| JM | Niigata | Oh | − | − | − | − | 0 | Anti-H | Le(a+b−) | Nonsecretor | h1/h1 | sej/sej | Le/Le |
| GO | Gifu | Oh(A) | − | − | − | − | 0 | Anti-H | Le(a+b−) | Nonsecretor | h1/h1 | sej/sej | Le/le1 |
| AI | Osaka | Omh(B) | − | − | + | NT | +w | Anti-HI | Le(a−b+) | Secretor | h2/h2 | Se1/Se1 | Le/le1 |
| MO | Kita-Osaka | Omh(B) | − | − | + | NT | +w | Anti-HI | Le(a−b−) | Secretor | h2/h2 | Se1/Se1 | le1/le2 |
| AM | Miyazaki | Ah | + | +w | NT | NT | NT | Anti-H | Le(a−b+) | Secretor | h3/h3 | Se2/Se2 | Le/le1 |
| HN | Kanagawa | Omh | − | − | − | + | 2+ | Anti-HI | Le(a−b−) | Secretor | h3/h3 | Se2/Se2 | le1/le1 |
| YH | Fukui | Omh | − | − | − | + | 2+ | Anti-HI | Le(a−b+) | Secretor | h3/h3 | Se2/Se2 | Le/Le |
| MY | Fukui | Omh | − | − | − | + | 2+ | Anti-HI | Le(a−b+) | Secretor | h3/h3 | Se2/Se2 | Le/Le |
| HF | Gunma | ABh | + | +w | + | + | NT | Anti-H | Le(a−b+) | Secretor | h3/h3 | Se2/Se2 | Le/le1 |
| KT | Gifu | Omh | − | − | − | + | 2+ | Anti-HI | Le(a−b+) | Secretor | h1/h3 | Se2/sej | Le/le2 |
| HiN | Gunma | Bh | ± | − | + | − | 1+ | Anti-H | Le(a−b+) | Secretor | h1/h3 | Se2/Se2 | Le/le1 |
| NK | Gunma | Bh | ± | − | + | NT | 1+ | Anti-H | Le(a−b+) | Secretor | h3/h5 | Se1/Se2 | Le/Le |
| HA | Tokushima | Omh | − | − | − | − | +w | Anti-HI | Le(a+b−) | Nonsecretor | h4/h4 | sej/sej | Le/le2 |
| SK | Tokushima | Omh | − | − | − | − | +w | Anti-HI | Le(a+b−) | Nonsecretor | h4/h4 | sej/sej | Le/le2 |
| YT | Okayama | Ah | − | − | + | + | +w | Anti-H | Le(a−b+) | Secretor | h5/h5 | Se1/Se1 | Le/Le |
Abbreviations: NT, not tested; +w, weakly positive.
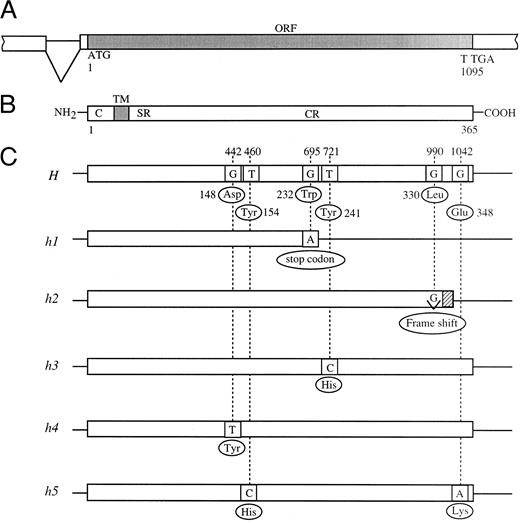
Point mutations in H genes of Bombay and para-Bombay individuals found by sequencing.The full-length sequences of the H genes of one Bombay, KK in Table 2, and four para-Bombay individuals, MO, HN, HA, and NK, in Table 2, were determined to identify point mutations. Identification of point mutations in the H genes of these individuals was performed sequentially as described in Materials and Methods by combining full-length sequencing of the H gene for identification of mutations with PCR-RFLP assay for the detection of mutations. The sequence of the H gene of KK was first determined. The PCR products of the full-length H gene from KK amplified by mk1 and mk2 primers were cloned into a pBS vector and then eight clones were fully sequenced. The eight clones shared the same point mutation, G695A, which creates a termination codon, changing TGG (Trp232) to a termination codon TAG, and terminates the H gene translation. The allele having this mutation was named h1. As all eight clones of KK shared the G695A nonsense mutation, he was expected to be a homozygote for the h1 allele. PCR-RFLP assay was performed to detect the G695A mutation on the 17 Bombay and para-Bombay individuals. From the individuals who were determined not to have h1 allele by this assay, the H gene of MO was fully sequenced and was found to have a single base deletion, G990-del, which we named the h2 allele. The G990-del, located at the C-terminal, changes the reading frame and the original 35 amino acids are replaced by an additional five amino acids.
In this manner, we found three more mutant alleles in the H genes of para-Bombay individuals. The H genes of HN and HA were found to have a single missense mutation each for the catalytic region of the enzyme, T721C (Tyr241 to His) and G442T (Asp148 to Tyr), which we named the h3 and h4 alleles, respectively. The H gene of NK was found to have two missense point mutations, T460C (Tyr154 to His) and G1042A (Glu348 to Lys), in one allele which we named h5. The five novel mutant alleles, h1 to h5, identified in this study are schematically presented in Fig 1.
Schematic diagrams of mutant H alleles found in Bombay and para-Bombay individuals. (A) Partial genomic structure encompassing the open reading frame (ORF) of the H gene. (B) Schematic presentation of the H enzyme. C, TM, SR, and CR indicate cytoplasmic tail, transmembrane domain, stem region, and catalytic region, respectively. (C) The locations of the point mutations found in mutant H alleles. The nucleotide sequence of the active H allele in Japanese population was determined to be the same as that reported by Larsen et al.14
Schematic diagrams of mutant H alleles found in Bombay and para-Bombay individuals. (A) Partial genomic structure encompassing the open reading frame (ORF) of the H gene. (B) Schematic presentation of the H enzyme. C, TM, SR, and CR indicate cytoplasmic tail, transmembrane domain, stem region, and catalytic region, respectively. (C) The locations of the point mutations found in mutant H alleles. The nucleotide sequence of the active H allele in Japanese population was determined to be the same as that reported by Larsen et al.14
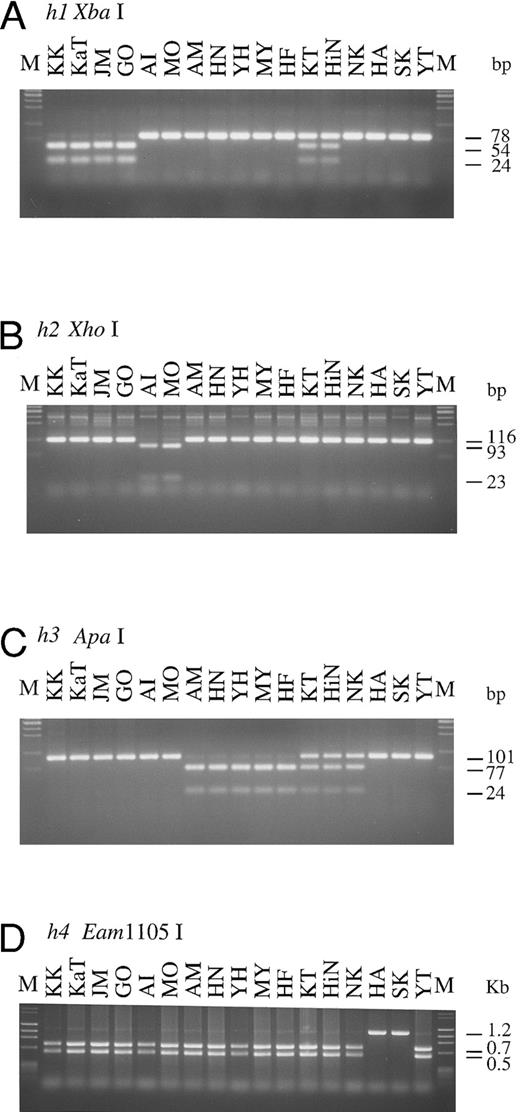
H, Se, and Le genotypes of Bombay and para-Bombay individuals determined by PCR-RFLP.The results of genotyping by PCR-RFLP performed on four Bombay and 13 para-Bombay individuals are presented in Fig 2. For detection of G695A in the h1 allele, PCR-RFLP analysis was performed using the mk3 and mk4 primer set for PCR and Xba I for digestion of the PCR product. The 78-bp PCR product of the h1 allele was separated into two fragments of 54 and 24 bp by Xba I digestion. The PCR-RFLP products shown in Fig 2A indicate that KK, KaT, JM, and GO were homozygous for the h1 allele, and KT and HiN were heterozygous. The other 11 individuals do not have the h1 allele, as their PCR product did not cut with Xba I.
PCR-RFLPs for detection of each of mutant H and Se alleles. Profiles of PCR-RFLPs for detection of h1 (A), h2 (B), h3 (C), h4 (D), and h5 (E and F) alleles in H gene, and profiles of PCR-RFLPs for genotyping of Se gene (G and H) are presented. Initials of each individual are at the top. M, marker.
PCR-RFLPs for detection of each of mutant H and Se alleles. Profiles of PCR-RFLPs for detection of h1 (A), h2 (B), h3 (C), h4 (D), and h5 (E and F) alleles in H gene, and profiles of PCR-RFLPs for genotyping of Se gene (G and H) are presented. Initials of each individual are at the top. M, marker.
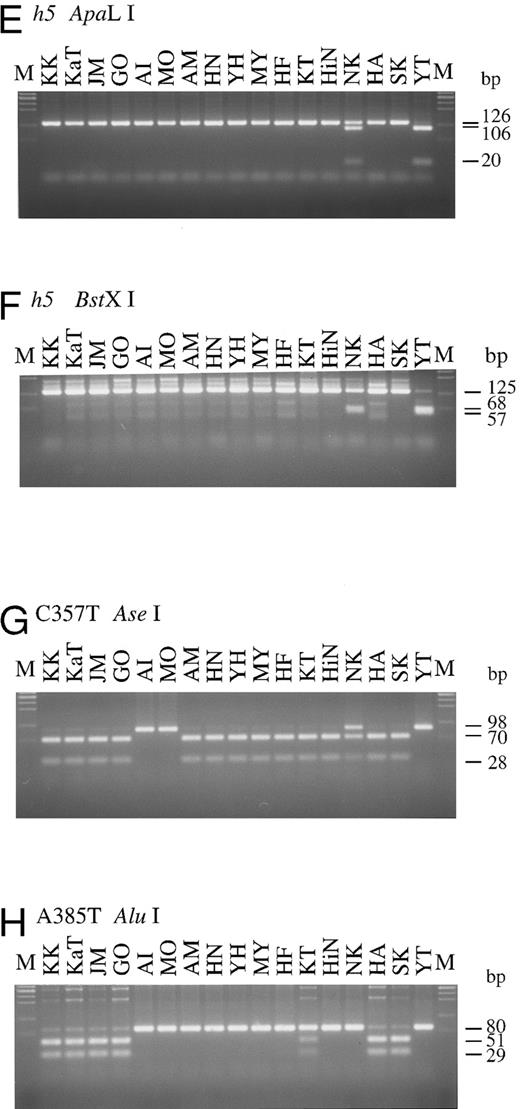
For detection of the G990-del in the h2 allele, the mk6 primer, mismatched at position 992 to the original H gene sequence, was designed to create an Xho I site in the h2 allele 116 bp PCR product. As seen in Fig 2B, both alleles of AI and MO were cleaved into two fragments, 93 and 23 bp, by Xho I digestion, indicating that these individuals were homozygotes with the h2/h2 genotype, whereas the other 15 individuals did not have the h2 allele. Detection of the h3 allele was performed with the mk7 and mk8 primer set, and the result is shown in Fig 2C. Five para-Bombay individuals, AM, HN, YH, MY, and HF, were determined to have an h3/h3 genotype because both their alleles were cleaved into two fragments, 77 and 24 bp, by Apa I digestion. The other three para-Bombay individuals, KT, HiN, and NK, were heterozygotes having a single h3 allele because Apa I digestion of their PCR products resulted in three fragments of 101, 77, and 24 bp. KK, KaT, JM, GO, AI, MO, HA, SK, and YT did not possess the h3 allele.
The original H gene sequence possesses an Eam1105 I site (GACNNNNNGTC) at position 442. The G442T missense mutation in the h4 allele removes this Eam1105 I site. The full-length H genes of Bombay and para-Bombay individuals amplified with the mk1 and mk2 primer set were subjected to Eam1105 I digestion for detection of the G442T mutation in the h4 allele. As seen in Fig 2D, the 1.2-kb full-length H genes of all 15 individuals, except for HA and SK, were cleaved into two fragments, 0.7 and 0.5 kb, while the H gene of HA and SK showed an undigested 1.2 kb band. This indicated that HA and SK are homozygotes for the G442T mutation and that the others were negative for the h4 allele. Determination of the two h5 allele mutations, T460C and G1042A, by PCR-RFLP was performed using the mk9 and 10 primer set and the mk11 and 12 primer set, for detection of each mutation, respectively. PCR with mk9 and mk10 creates an ApaLI site on the h5 allele PCR product, and the product is separated into 106 and 20 bp fragments by ApaLI digestion. The G1042A mutation creates a BstXI site (CCANNNNNNTGG) in the H gene sequence, so the 125-bp PCR product of the mutated gene with the mk11 and mk12 primer set, which amplify the fragment encompassing the position 1042, was cleaved by BstXI. As seen in Fig 2E and F, YT was homozygous for the h5 allele, possessing both the mutations in both alleles and NK was heterozygous for the h5 allele. The H genotypes of all 17 individuals examined are summarized in Table 2.
To explore the role of H and Se genes on the expression of H antigens on erythrocytes and in saliva and to examine the relationship betwen H, Se, and Le genes, the Se and Le genes of the four Bombay and 13 para-Bombay individuals was determined. In our previous studies on more than 200 Japanese individuals randomly sampled,7,11 Japanese Se genes were classified into two active alleles, Se1 with no mutation and Se2 with the C357T silent mutation, and one inactivated allele, sej having the C357T and A385T missense mutations. Le genes were classified into one active allele, Le with no mutation, and two inactivated alleles, le1 with the T59G and G508A mutations and le2 with the T59G and T1067A mutations. The Se genes of the 17 Bombay and para-Bombay individuals were examined by PCR-RFLP as described previously.7 11 As shown in Fig 2G, 13 of the 17 individuals were homozygous for the C357T mutation, as both Se alleles amplified by PCR were cleaved into a 70- and a 28-bp fragment by Ase I digestion. The other individual (NK) was determined to be heterozygous for this mutation. AI, MO, and YT were determined to be homozygotes with the Se1/Se1 genotype because they did not have the C357T mutation. Results shown in Fig 2H indicate that KK, KaT, JM, GO, HA, and SK were A385T homozygotes, as both alleles of their amplified Se genes were cut by Alu I. From the results in Fig 2G and H, both Se gene alleles of KK, KaT, JM, GO, HA, and SK had the C357T and A385T mutations. Their Se genotypes were assigned to be sej/sej. Figure 2H also shows that KT was heterozygous for the A385T Se mutation.
The Se genotypes of the 17 Bombay and para-Bombay individuals identified in this experiment are summarized and presented in Table 2 together with Le genotypes, which were also determined by PCR-RFLP.
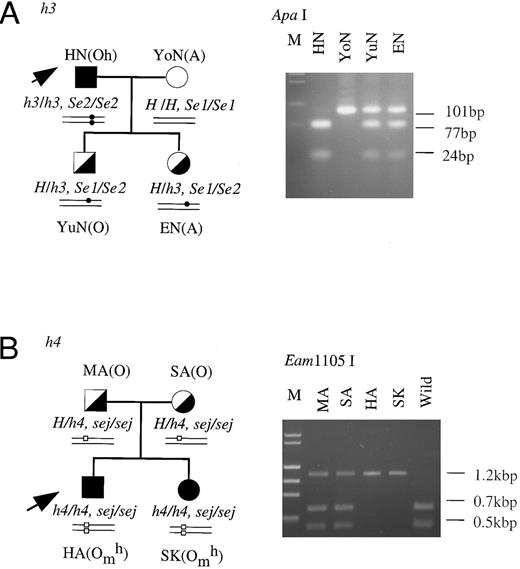
Family studies of the mutant h genes.Family studies were performed to confirm that the inactivated h alleles showed Mendelian inheritance. The H genes of members of two families, HN's family and HA's family, were studied by PCR-RFLP-method. As seen in Fig 3A, HN was an h3/h3 homozygote and his wife was a homozygote with an active H gene. Both of HN's children were confirmed to be heterozygotes for the h3 allele (H/h3). Their Se genotypes were also analyzed. HN was an Se2/Se2 homozygote, his wife was an Se1/Se1 homozygote, and their children were Se1/Se2 heterozygotes. In HA's (h4/h4) family shown in Fig 3B, both of HA's parents were identified to be heterozygotes for the h4 allele (H/h4) and homozygotes for the sej allele. HA and his sister, SK, were homozygous for both the h4 and sej alleles. These results indicated that two haplotypes of the inactivated H gene, h3, probably linked with Se2 and h4, probably linked with sej, were inherited according to the Mendelian rules.
Family studies on the h3 and h4 alleles. Erythrocyte phenotypes, and H and Se genotypes were determined in the members of the families of HN (A) and HA (B). The H genotypes of the members of the two families were identified by PCR-RFLP tests for detection of T721C in the H genes of HN's family members and for detection of G442T in H genes of HA's family members. The Se genotypes of the members were determined as described in a previous report.11 Square, male; circle, female; filled symbol, homozygote of the mutant H allele; half-filled symbol, heterozygote of wild-type H and mutant H alleles; open symbol, homozygote of wild-type H allele. Small closed circles and open squares in schematic alleles represent T721C in h3 allele and G442T in the h4 allele, respectively.
Family studies on the h3 and h4 alleles. Erythrocyte phenotypes, and H and Se genotypes were determined in the members of the families of HN (A) and HA (B). The H genotypes of the members of the two families were identified by PCR-RFLP tests for detection of T721C in the H genes of HN's family members and for detection of G442T in H genes of HA's family members. The Se genotypes of the members were determined as described in a previous report.11 Square, male; circle, female; filled symbol, homozygote of the mutant H allele; half-filled symbol, heterozygote of wild-type H and mutant H alleles; open symbol, homozygote of wild-type H allele. Small closed circles and open squares in schematic alleles represent T721C in h3 allele and G442T in the h4 allele, respectively.
Haplotypes of H and Se genes found in Bombay and para-Bombay individuals.It is well-established by a physical mapping analysis that the H and Se genes are closely located (within 40 kb) on chromosome 19q13.3.26 27 As summarized in Table 2, all four Bombay individuals, KK, KaT, JM, and GO, shared H and Se genotypes, both of which were inactive (h1/h1 and sej/sej), while their Le genotypes differed. Five para-Bombay individuals, AM, HN, YH, MY, and HF, shared the H and Se genotypes, h3/h3 and Se2/Se2, and one of the para-Bombay individuals, KT, was heterozygous for both the H and Se genes (h1/h3 and Se2/sej). These results indicate that in relation to the Se gene, the h1 and h3 alleles form h1-sej and h3-Se2 haplotypes, respectively. The G695A point mutation in the h1 allele and T721C in the h3 allele must have occurred on the chromosome with the sej allele or the Se2 allele, respectively. The Se genotype of the remaining one h3 allele heterozygote, HiN (h1/h3), was determined to be Se2/Se2. To explain HiN's H and Se genotypes, we assume that a recombinatorial event between the H gene and the Se gene must have previously given rise to the chromosome possessing the h1 and Se2 alleles.
In the present study, we could find only two h2/h2 (AI and MO), two h4/h4 (HA and SK), one h5/h5 (YT) homozygotes, and one h3/h5 heterozygote (NK). Although numbers of individuals with h2, h4, or h5 gene were limited, determination of Se genotypes of these individuals seemed to indicate that h2 allele forms h2-Se1 haplotype, h4 allele forms h4-sej haplotype, and h5 allele forms h5-Se1 haplotype.
From the results in Table 2, no haplotype linkage was observed between the H gene and the Le gene or between the Se gene and the Le gene.
α1,2FT activity directed by each of the mutant h alleles.We first measured the luciferase activity directed by the cotransfected luciferase cDNA for normalization of transfection efficiency and the amount of H gene transcripts by Northern blot analysis in the COS cell transfection assays (data not shown). It was confirmed that the mRNA expression of the H genes was almost at the same level between the wild type gene and the five mutant genes. Translation must occur at the same level between the wild type and the mutant enzymes because it is very hard to consider that only a few amino acid substitutions in h3, h4, and h5 enzymes and a frameshift in h2 enzyme affects the translational efficiency.
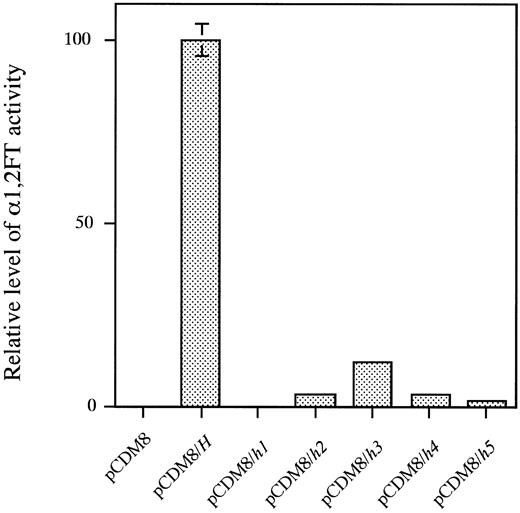
As shown in Fig 4, lysates of the COS-1 cells transfected with the wild type H allele showed full α1,2FT activity. The COS-1 cell lysates transfected with the mutant allele, h1, did not exert any activity at all, whereas those transfected with the h2, h3, h4, or h5 allele showed quite low, but positive activity. The h2, h3, h4, and h5 enzymes showed 3.3%, 11.8%, 3.5%, and 1.6% of the activity of the wild type H-transfected cell lysates, respectively. The single base deletion, G990-del in the h2 allele, and the missense mutations, T721C (Tyr241 to His) in the h3 allele, G442T (Asp442 to Tyr) in the h4 allele and T460C (Tyr154 to His) and G1042A (Glu348 to Lys) in the h5 allele did not cause complete inactivation of the encoded enzymes, and exerted residual activity.
α(1,2)FT activity directed by each of the wild-type H and five mutant H alleles. From left to right, pCDM8, activity of lysate of COS-1 cells mock-transfected with pCDM8 vector; pCDM8/H, activity of H-transfected COS-1 cell lysate; pCDM8/h1, h2, h3, h4 or h5, activity of h1, h2, h3, h4, or h5-transfected COS-1 cell lysate, respectively. The activity in each lysate was normalized by transfection efficiency, which was determined by expression of cotransfected luciferase expression vector. Data represent means and standard deviation (SD) of three independent assays.
α(1,2)FT activity directed by each of the wild-type H and five mutant H alleles. From left to right, pCDM8, activity of lysate of COS-1 cells mock-transfected with pCDM8 vector; pCDM8/H, activity of H-transfected COS-1 cell lysate; pCDM8/h1, h2, h3, h4 or h5, activity of h1, h2, h3, h4, or h5-transfected COS-1 cell lysate, respectively. The activity in each lysate was normalized by transfection efficiency, which was determined by expression of cotransfected luciferase expression vector. Data represent means and standard deviation (SD) of three independent assays.
H antigen levels on erythrocytes are determined solely by H enzyme activity, while expression of Lewis b (Leb) antigen on erythrocytes is determined by Le and Se enzyme activities and secretor status is determined solely by Se enzyme activity.It is now clear that the H gene and the Se gene encode different α1,2FTs. The results summarized in Table 2 indicate clearly that the H enzyme determines the expression of H antigens on erythrocytes, while the Se enzyme determines the secretor status and, in cooperation with the Le enzyme, the expression of Leb antigen on erythrocytes.
The residual H enzyme activity encoded by the h2, h3, h4, or the h5 allele was in good agreement with the level of ABH antigen expression on erythrocytes shown in Table 2. The 13 para-Bombay individuals, eight of whom having the Omh phenotype, two of whom having the Ah phenotype, two of whom having the Bh phenotype and one of whom having the ABh phenotype, possessed a trace amount of H antigen on their erythrocytes, which could be detected by the adsorption-elution test as shown in Table 2. All of the 13 para-Bombay individuals possessed at least one of the weakened H alleles, h2, h3, h4, or h5. AI, MO, HA, SK, and YT having fewer RBC ABH antigens than the other eight individuals as determined by the adsorption-elution test, were homozygotes with the h2, h4, or h5 allele, respectively. This agrees with the finding that the H enzyme activity directed by h2, h4, and h5 was weaker than that directed by h3. All of the individuals who completely lack H antigens on erythrocytes, KK, KaT, JM, and GO (Table 2) were determined to be completely inactivated h1 allele homozygotes (Table 2 and Fig 4).
All 11 secretors in Table 2 who secreted H antigen into saliva possessed at least one of the active Se genes, Se1 or Se2, and all six nonsecretors possessed homozygous alleles of the inactivated Se gene, sej (Table 2). Nine individuals, AI, AM, YH, MY, HF, KT, HiN, NK, and YT, having the Le(a- b+) phenotype were confirmed to possess both of the active Le and Se genes, homozygously or heterozygously. Six individuals, KK, KaT, JM, GO, HA, and SK, having the Le(a+ b-) phenotype, possessed at least one of the active Le genes, but their Se genes were inactive with the homozygous sej/sej genotype. Two Le(a- b-) individuals, MO and HN, were confirmed to lack the active Le gene, regardless of the Se genotype.
DISCUSSION
Both the H and Se enzymes are α1,2FTs with subtle differences in substrate specificity. It was clearly shown in this study that the H enzyme activity determines the amount of ABH antigens on erythrocytes, while the Se enzyme activity determines the secretor status and, in cooperation with Le enzyme, Leb expression on erythrocytes. Both the H and Se genes of Bombay individuals were confirmed to be inactivated by point mutations. The absence of ABH antigens on erythrocytes of four Bombay individuals was explained by complete inactivation of their H genes due to the nonsense mutation. A variety of phenotypes of the 13 para-Bombay individuals in this study were clearly explained by independent expression of H and Se enzymes. The weak expression of ABH antigen on erythrocytes of para-Bombay individuals was explained by residual h2, h3, h4, and h5 enzyme activity and the presence of ABH antigens in the saliva of some para-Bombay individuals was explained by the Se enzyme activity.
It is of interest to know where the active sites of the enzyme lie within the known primary structure. One study28 analyzed the donor substrate binding site of α1,3fucosyltransferases using sulfhydryl-group reagents, but the enzyme active sites of many fucosyltransferases have not been analyzed. The enzymes retaining residual activity like the h3, h4, and h5 enzymes might have a folded structure, and the positions of mutations might exist at active sites of the enzyme, such as the acceptor- or the donor-binding site. Together with missense mutations in h3, h4, and h5 alleles, further study of missense mutations differing from those in h3, h4, and h5 alleles in the other para-Bombay individuals would clarify the active sites of the H enzyme.
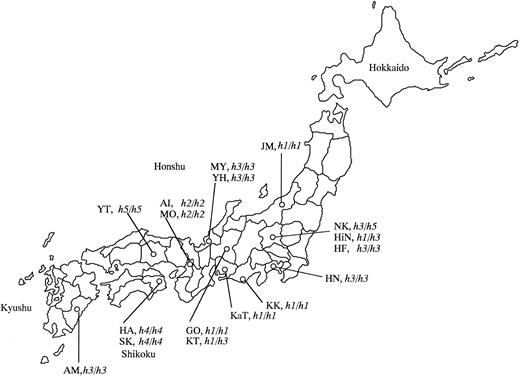
Map of Japan indicating the birthplaces of the 17 Bombay and para-Bombay individuals.
Map of Japan indicating the birthplaces of the 17 Bombay and para-Bombay individuals.
The H and Se genes form a gene cluster on 19q13.3 and are located only 35 kb apart.26,27 The Le gene also maps to 19p13.3 within the gene cluster of the α1,3fucosyltransferase family,29 30 but is distant from the H and Se genes. In this study, determination of the H, Se, and Le genotypes of the four Bombay and 13 para-Bombay individuals showed interesting results that each of the mutant h alleles was closely associated with specific haplotype of Se gene, ie, h1-sej, h2-Se1, h3-Se2, h4-sej, and h5-Se1 haplotypes, in the Japanese population, although the number of individuals with h2, h4, or h5 gene was limited.
The sej allele inactivated by the single A385T missense mutation is known to be widely distributed in the Japanese, the Chinese in Taiwan, and Polynesians,11,21,22 although its distribution in other ethnic groups has not been analyzed. In our previous study,11 all 35 Japanese nonsecretors examined were homozygotes with the inactive allele, sej/sej. The Se enzyme-inactivating G428A mutation found in Caucasians10 has never been found in Japanese samples.11 From the observation of ethnic group specific point mutations, which inactivate Se gene, we assumed that Se gene inactivation must have occurred after these ethnic groups diverged.11 As for the mutations in the H gene, we conclude that mutations that inactivate the H gene occurred after Se gene inactivation, as all of the Bombay and nonsecretor para-Bombay individuals examined in this study shared the same inactivated Se allele, sej, but possessed a variety of inactivated H alleles. Le gene inactivation might be similar to that of Se gene inactivation, as the le1 allele inactivated by the G508A missense mutation, which is rather widely distributed in the Japanese population,7,11 is uncommon among Caucasians.9
In the present study, we collected the 17 Bombay or para-Bombay individuals through routine determination of ABO blood types on a large number of randomly sampled volunteers who donated their blood to Red Cross Blood Centers. Thus, the incidence of Bombay and para-Bombay individuals was determined to be very rare and a doctor in the Red Cross Blood Center conjectured that it would be approximately one in two or 300,000 in the whole Japanese population. In contrast to the wide distribution of the single mutant alleles of Se and Le genes, it was found that the point mutations inactivating the H gene are very variable and are specific to the individuals and their family. This is consistent with the very rare frequency of inactivated H alleles in the whole human population. The birthplaces of the 17 individuals examined in this study are indicated in Table 2 and in the map of Japan of Fig 5, in which we notice that the birthplaces of three Bombay individuals with the h1/h1 and sej/sej genotypes, KK, KaT, and GO, are localized in a restricted area, the Nobi plane. Although they are not aware of any blood relations, they must have common ancestors from the near past. Two h2/h2 homozygotes, AI and MO, who are also not aware of blood relations, were found in Osaka. The G990 deletion in the h2 allele may have occurred in the H gene of their common ancestor from the near past. Fukui, the birthplace of MY and YH with the h3/h3 genotype, has been culturally and geographically isolated by mountains. MY and YH also do not know their blood relations, but we assume that the h3 allele must have appeared in a common ancestor of MY and YH in the Fukui area. The h3 mutant allele seemed to be rather widely distributed in the Japanese population than the other mutant h alleles. The h4 allele found in HA's family may be specific to the Tokushima area in Shikoku island, which is separated by the sea from the other places or may be specific to HA's family because we have not found the h4 allele in anyone except them. As for the h5 allele, we found only one h5/h5 homozygote in Okayama, so it is impossible to make any conjecture on the original place of emergence of the h5 mutant allele. Kanagawa, the birthplace of HN (h3/h3), and Gunma, those of NK (h3/h5) and HiN (h1/h3), are relatively new areas of settlement within Japan's history. Therefore, we conjecture that their ancestors moved from Fukui or the Nobi plane to Kanagawa or Gunma.
The above results suggested that consanguineous marriages in a closed society seemed to give rise to rare homozygous individuals with inactive H genes. H gene inactivation by point mutations must have occurred in the nearer past than that of Le and Se genes.
In a previous report,11 we raised an interesting question as to why a sole inactivated Se allele or only two inactivated Le alleles spread in each ethnic group. In the case of the H gene, which is highly homologous to the Se gene, the mutant H alleles do not seem to have spread in the population. There might be some specific selective advantage on the individuals with the mutant Se and Le alleles, but some selective disadvantage on the individuals with the mutant H alleles.
ACKNOWLEDGMENT
We thank Dr K. Saito for critical reading of this manuscript.
Supported in part by Special Coordination Funds from the Science and Technology Agency of the Japanese Government, and by a Grant-in-Aid for Scientific Research on Priority Areas No. 05274103 from the Ministry of Education, Science, and Culture of Japan.
Address reprint requests to Hisashi Narimatsu, MD, PhD, Division of Cell Biology, Institute of Life Science, Soka University, 1-236 Tangi-cho, Hachioji, Tokyo 192, Japan.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal