Abstract
A French kindred with autosomal dominant hereditary renal amyloidosis was found to have a novel mutation in the fibrinogen Aα-chain gene. In this kindred, renal disease appeared early in life and led to terminal renal failure at an early age. Renal transplantation resulted in rapid destruction of the allograft by amyloid deposition within 2 years. Amyloid fibril protein isolated from a transplanted kidney was found to contain a novel, hybrid peptide of 49 residues whose N-terminal 23 amino acids were identical to residues 499 to 521 of normal fibrinogen Aα-chain. The remainder of the peptide (26 residues) represented a completely new sequence for mammalian proteins. DNA sequencing documented that the new sequence was the result of a single nucleotide deletion at position 4897 of the fibrinogen Aα-chain gene that gives a frame-shift at codon 522 and premature termination at codon 548. The contributions toward fibrillogenesis of the two portions of the amyloid fibril protein, ie, N-terminal fibrinogen sequence and C-terminal novel sequence, are presently unknown. However, the early onset and rapid reoccurrence of amyloid in renal transplants is unlike the clinical course with other amyloid proteins having single amino acid substitutions that give hereditary renal amyloidosis. Liver transplantation to stop synthesis of this abnormal hepatic derived protein should be considered early in the course of the disease.
HEREDITARY AMYLOIDOSIS is an autosomal dominant trait characterized by progressive extracellular deposition of amyloid protein in various organs. Systemic forms of hereditary amyloidosis have been associated with mutants of several plasma proteins: transthyretin, gelsolin, apolipoprotein A1, lysozyme, and fibrinogen Aα-chain.1
Most transthyretin (TTR)1 associated amyloidoses have peripheral neuropathy and cardiomyopathy as major clinical manifestations. Patients may present with gastrointestinal dysfunction, nephropathy, sexual impotence, vitreous opacification, and cerebral hemorrhage. The disease usually progresses over 10 to 20 years after a delayed onset of symptoms in the third to seventh decades. To date, more than 50 amyloidogenic TTR variants have been identified by protein and DNA analysis.2 All are the results of point mutations except for one in which there is a trinucleotide deletion.3
The first manifestation of amyloidosis related to variant gelsolin is a dystrophic change of the cornea due to amyloid deposition. Over several decades, thickening of the skin on the forehead and back occurs, and patients may develop facial paralysis caused by cranial neuropathies. Kindreds have been reported in Finland, the United States, Denmark, Canada, and Japan. Two mutations at position 187 of the gelsolin protein have been identified in patients with this disease.1
In a number of kindreds, hereditary renal amyloidosis (HRA) is the major clinical finding. Since a German kindred with HRA was originally described by Ostertag in 1932,4 several additional kindreds have been reported. More recently, three proteins associated with HRA have been identified: apolipoprotein A1, lysozyme, and fibrinogen Aα-chain. Nephrotic syndrome and renal insufficiency may result from deposition of variant forms of apolipoprotein A1, all of which are the result of single nucleotide mutations except for one reported deletion/insertion mutation in exon 4 of the apolipoprotein A1 gene.1,5,6 Several English families have been reported with hereditary non-neuropathic systemic amyloidosis with renal failure in which lysozyme is the major fibril protein and two mutations, Thr-56 and His-67, have been described.7
In 1993, amyloid fibrils were isolated from a transplanted kidney of a patient with HRA and found to contain a fragment of fibrinogen Aα-chain with an amino acid substitution of Leu for Arg at position 554.8 Subsequently, two additional mutations in the fibrinogen Aα-chain gene were shown to cosegregate with the disease.9,10 One of these mutations was a nucleotide deletion, resulting in a frame shift and early termination of the fibrinogen Aα-chain after amino acid residue 547.10 However, no amyloid-laden tissue was available for study and, therefore, existence of the postulated novel protein and its involvement in amyloid fibril formation could not be confirmed. In the present study, we describe the first French kindred with a fibrinogen mutation causing HRA, and we report histologic, biochemical, and molecular biologic characterization of a novel amyloid protein.
CASE DESCRIPTION
The propositus (case no. 1) was a 31-year-old man who developed nephrotic syndrome in 1984. Immunoelectrophoresis of blood and urine showed no free Ig light chains. A renal biopsy showed glomerular amyloid deposits that were birefringent in polarized light after Congo red staining and were resistant to oxidation by potassium permanganate. Hepatic, bone marrow, and rectal biopsies showed no amyloid deposits. Amyloid was considered to be of the Ig (AL) type and the patient was treated with plasmaphoresis, cyclophosphamide, melphalan, and methyl prednisone. At the age of 35 in 1988, the patient received a renal transplant. Two years later, recurrence of amyloidosis in the renal graft was documented by biopsy. The renal graft was removed in 1995 and was used for extraction of amyloid fibrils. Liver biopsy showed a slight vascular amyloid infiltrate. Echocardiography showed no signs of amyloid heart involvement. A new immunohistochemical study of amyloid deposits showed reaction with antifibrinogen (anti-residue 518-584 of fibrinogen Aα-chain), anti-SAP, anti-IgG, and anti-κ light chain.
In 1992, renal amyloidosis was diagnosed in the propositus' son (case no. 2), who presented with nephrotic syndrome at the age of 12. Renal biopsy disclosed amyloid deposits and immunohisto-chemistry favored the diagnosis of AL type. However, there was no circulating monoclonal protein and bone marrow examination was normal. Renal failure progressed rapidly with severe hypertension, and peritoneal dialysis was begun 1 year after the occurrence of the nephrotic syndrome. Renal transplantation was performed at the age of 15. Repeat immunohistochemistry of the renal biopsy showed positive staining with anti-fibrinogen Aα, anti-IgM, and anti-λ light chain localized to glomerular deposits. Immunohistochemistry with antibodies to gelsolin, apolipoprotein A1, lysozyme, and transthyretin showed no staining of glomerular deposits. One year after the transplantation, proteinuria of 1 g/d and hypertension recurred and biopsy of the graft showed the presence of marked amyloid deposits. Plasma fibrinogen concentration as well as activated partial thromboplastin and reptilase times were normal in both patients.
MATERIALS AND METHODS
Materials. Sepharose CL6B was obtained from Pharmacia (Uppsala, Sweden) and Ultrasphere-ODS high-performance liquid chromatography (HPLC) column was from Beckman Instruments, Inc (Fullerton, CA). Polyvinyldifluoride (PVDF; Problott) membranes were from Applied Biosystem (Foster City, CA). GeneAmp polymerase chain reaction (PCR) reagents were from Perkin-Elmer Cetus Corp (Norwalk, CT). Nusieve GTG agarose was from FMC Bioproducts (Rockland, ME). Centricon-30 microconcentrator was from Amicon (Beverly, MA). Sequenase Version 2.0 and Pst I were from US Biochemical Corp (Cleveland, OH). α 32P-labeled dCTP was obtained from Amersham Corp (Arlington Heights, IL).
Histology. Sections (6 μm) of formalin-fixed, paraffin-embedded kidney were stained with hematoxylin and eosin or with alkaline Congo red using standard techniques. Congo red-stained sections were viewed by polarized light.
Isolation of amyloid fibrils. Amyloid fibrils were isolated from 9.5 g of rejected transplanted kidney tissue of case no. 1 using repeated homogenization and centrifugation, as previously described.11 Briefly, tissues were repeatedly homogenized in 0.9% sodium chloride and centrifuged, and the supernatant was discarded until its absorbance at 280 nm was less than 0.075. The remaining pellet was then homogenized in water and centrifuged. The procedure was repeated three more times. Water washes and water resuspended pellet were dialyzed against water and lyophilized. Smears of lyophilized water washes and final pellet stained with Congo red were positive for amyloid fibrils.
Isolation of amyloid protein. Amyloid fibrils were suspended in 6 mol/L guanidine hydrochloride, 0.5 mol/L Tris, pH 8.5, containing 1 mg EDTA/mL, reduced with dithiotreitol, and alkylated with iodoacetic acid as previously described.12 After centrifugation at 12,000 rpm for 30 minutes at 4°C, the supernatant was fractionated on a Sepharose CL6B column (2.6 × 90 cm), equilibrated, and eluted with 4 mol/L guanidine hydrochloride, 0.05 mol/L Tris, pH 8.2. Eluant was monitored by absorbance at 280 nm. Fractions in the 15-kD and lower elution area were pooled, dialysed against water, and lyophilized.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Protein samples were analyzed on SDS-PAGE using the Tricine system of Schagger and Von Jagow.13 Separated proteins were electrophoretically transferred to PVDF membrane for 2 hours at 70 V using 25 mmol/L Tris, 192 mmol/L glycine, pH 8.3, buffer containing 10% methanol.
Trypsin digestion and peptide fractionation. CL6B fractionated protein was digested with trypsin (2% wt/wt) in 0.1 mol/L ammonium bicarbonate for 18 hours at pH 8.0. Digestion was stopped by lyophilization. The sample was then dissolved in 50% acetic acid and the supernatant was fractionated on an Ultrasphere-ODS HPLC column (0.46 × 25 cm) equilibrated with 0.1% trifluoroacetic acid TFA in water and eluted with a 0% to 60% acetonitrile in 0.1% TFA linear gradient over 120 minutes. Eluant was monitored by absorbance at 215 nm. Fractions were dried in a Savant Speed Vac Concentrator (Savant Instruments, Farmingdale, NY).
Amino acid sequence determination. Samples were sequentially degraded in an Applied Biosystems Model 473A Protein Sequencer using standard cycle programs provided by the manufacturer.
DNA isolation. Total genomic DNA was isolated from peripheral blood cells of the propositus and his son using the conventional method.14
Single-strand conformation polymorphism (SSCP) analysis. A region of exon 5 of the fibrinogen Aα-chain gene (nucleotides 4832-5051) that codes for residues 500 to 573 of the carboxyl terminus was enzymatically amplified using Fib3 primer (5′-CTTCGACACTGCCTCAACTG-3) and Fib2 primer (5′-TCCTCTGTTGTAACTCGTGCT-3′). PCR was performed using primers, Gene Amp PCR reagents, and α 32P-labeled dCTP. PCR conditions were 35 cycles of denaturing at 94°C for 30 seconds, annealing at 62°C for 30 seconds, and extension at 72°C for 1 minute. PCR products were diluted with 100 vol of buffer containing 50% formamide, 0.05% SDS, 0.02% xylene cyanol FF, 0.02% bromophenol blue, and 10 mmol/L EDTA, heated at 95°C for 5 minutes, and loaded onto a 5% nondenaturing polyacrylamide gel (40 × 20 × 0.04 cm). The gel was electrophoresed at 4°C for 20 hours at 300 V, dried, and exposed to Kodak X-Omat film (Eastman Kodak, Rochester, NY).
Direct DNA sequence analysis. Portions of the fibrinogen Aα-chain gene were amplified as described above and separated by electrophoresis through 2% Nusieve GTG agarose gel. The band was excised and melted in 100 μL of water. Asymmetric PCR was performed using 1 μL of the gel-purified template and the same set of primers (primer ratio, 1:30). The sample was extracted with chloroform and subjected to spin dialysis with a Centricon-30 concentrator. Seven microliters of the retentate was directly used for dideoxynucleotide DNA sequencing by Sequenase version 2.0, as suggested by the manufacturer. Samples were electrophoresed on an 8% polyacrylamide gel at 2,000 V for 3 hours.
Restriction fragment length polymorphism (RFLP) analysis. A PCR-induced mutation restriction analysis was performed using a new primer (Fib9) corresponding to nucleotides 4874 to 4896 of the fibrinogen Aα gene (5′-CTCACCTATGTTAGGAGAGTCTG-3′). This primer with a C instead of T at the third position from the 3′ end creates a recognition site for Pst I only in the mutant gene PCR products. PCR was performed with primers Fib9 and Fib2 under the same conditions as described above. PCR-amplified DNA fragments were digested with the restriction enzyme Pst I at 37°C overnight, electrophoresed through 3% Nusieve GTG agarose gel, stained with ethidium bromide, and photographed under UV light.
RESULTS
Serial sections of the renal transplant stained with alkaline Congo red showed glomerular deposits that were weakly birefringent but gave a green color characteristic of amyloid.
Chromatography on Sepharose CL6B of guanidine hydrochloride solubilized reduced and alkylated amyloid fibrils yielded 2.3 mg of lyophilized material from the pooled fractions in the 15-kD and lower molecular weight elution area. SDS-PAGE analysis of the pool showed major bands at 4-5, 11, and 13 kD and several intermediate minor bands. Amino acid sequence analysis of the pool identified the majority of the first 35 residues of the amyloid subunit protein (Fig 1). Residues 1 to 23 were identical to residues 499-521 of fibrinogen Aα-chain, but the remaining sequence was completely different. However, residues 27-35 were identical to the predicted sequence of residues 525-533 in an Aα-chain resulting from the 4904delG mutation previously identified in the Aα-chain gene.10 The other 3 residues, Ala-Val-Arg in positions 24-26, could be derived from the Aα gene by a nucleotide deletion at the second base of codon 522 (4897delT). The resulting frame shift would yield the observed sequence for residues 24-35 in the amyloid protein and would predict a premature termination at codon 548 of Aα-chain or after residue 49 of the amyloid protein.
Sequence of amyloid fibril protein. The carboxyl-terminal portion of the fibrinogen Aα-chain amino acid and gene sequence are shown. The abnormal amino acid sequence resulting from deletion of the second nucleotide in the codon for Val522 (delT4897) is shown in the boxed area. The arrows indicate the peptides used to determine the amino acid sequence of the amyloid protein. The parentheses within the arrow for intact protein indicate residues tentatively identified, and dots at the end indicate that the protein continued but was not sequenced further. Peptides labeled with T were isolated from HPLC fractionation of a trypsin digest of the subunit protein (Fig 2). Dashed arrows indicate peptides recovered at ≤5% the molar amount of the other peptides.
Sequence of amyloid fibril protein. The carboxyl-terminal portion of the fibrinogen Aα-chain amino acid and gene sequence are shown. The abnormal amino acid sequence resulting from deletion of the second nucleotide in the codon for Val522 (delT4897) is shown in the boxed area. The arrows indicate the peptides used to determine the amino acid sequence of the amyloid protein. The parentheses within the arrow for intact protein indicate residues tentatively identified, and dots at the end indicate that the protein continued but was not sequenced further. Peptides labeled with T were isolated from HPLC fractionation of a trypsin digest of the subunit protein (Fig 2). Dashed arrows indicate peptides recovered at ≤5% the molar amount of the other peptides.
The complete sequence of the amyloid subunit protein was obtained from analyses of peptides after reverse-phase HPLC separation of a trypsin digest of the CL6B fractionated low molecular weight protein (Fig 2). These analyses confirmed the identity of the first 35 residues and identified 14 additional C-terminus residues (Fig 1). The sequence of the last 26 residues was identical to that predicted by a 4897delT mutation in the Aα gene. No normal sequence past residue 521 of Aα-chain was found. Partial tryptic peptides T2a and T2b resulting from cleavage between Met517 and Leu518 were recovered in molar amounts equal to intact T2. Also, peptides T2c and T2d resulting from cleavage between Phe521 and Ala522 were recovered in yields ≤10% that of the other T2 peptides. These peptides probably resulted from trypsin proteolysis, because no sequence starting with positions 518 or 522 of fibrinogen Aα-chain was evident in the sequence analysis of the Sepharose CL6B low molecular weight pool. Although trypsin normally cleaves after Arg and Lys residues, cleavage after other residues such as Met or Phe has been observed.
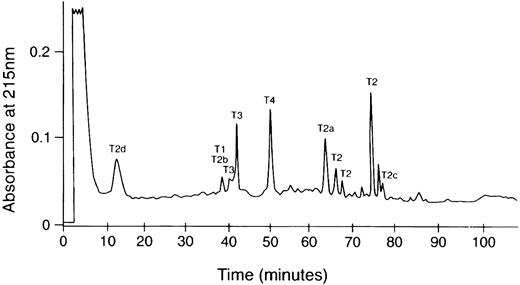
HPLC chromatogram of tryptic digest of the amyloid subunit protein. The trypsin digestion product was dissolved in 50% acetic acid and fractionated on an Ultrasphere ODS column (0.46 × 25 cm) equilibrated with 0.1% TFA in water and eluted with a 0% to 60% acetonitrile linear gradient in 120 minutes. The labeled peaks yielded the sequences in Fig 1. Multiple peaks containing the T2 sequence may be the result of changes in hydrophobicity due to cis-trans isomerization of Pro residues.
HPLC chromatogram of tryptic digest of the amyloid subunit protein. The trypsin digestion product was dissolved in 50% acetic acid and fractionated on an Ultrasphere ODS column (0.46 × 25 cm) equilibrated with 0.1% TFA in water and eluted with a 0% to 60% acetonitrile linear gradient in 120 minutes. The labeled peaks yielded the sequences in Fig 1. Multiple peaks containing the T2 sequence may be the result of changes in hydrophobicity due to cis-trans isomerization of Pro residues.
The recovered amounts of T2, T3, and T4 peptides were approximately equimolar, whereas that of T1 was 10% compared with the others. Because only the sequence starting with residue 499 of Aα chain was identified in the intact protein, these results would suggest that some of the amyloid protein (possibly the majority) may have an N-terminus blocked to Edman degradation analysis starting between residue 494 to 508 of Aα-chain. The blocked N-terminal tryptic peptide from the protein would also have been undetected by Edman analysis. This possibility is presently under investigation. Some of the smaller HPLC peaks such as that eluting at 76 minutes between T2 and T2c (Fig 2) had sequences consistent with the V10 subunit of vitronectin and histones 2, 3, and 4. The molar yields of these were less than 5% that of Aα-chain tryptic peptides, suggesting that these are probably not part of the fibrils, but rather low-level contaminants from the fibril extraction from the kidney.
The low molecular weight pool from Sepharose CL6B was fractionated by SDS-PAGE and electrophoretically transferred to PVDF membrane, and individual bands were excised and analyzed by Edman degradation. Aα-chain sequence starting with residue 499 was present in the approximately 4-5–kD band. This is close to the calculated mass of the residue 499-547 peptide from the 4897delT Aα gene. The 11- and 13-kD bands contained sequence consistent with the V10 peptide of vitronectin starting at residue 399 of the vitronectin precursor. The 13-kD band had sequence starting with residue 27 of histone 3. No sequence was detected in the other minor bands.
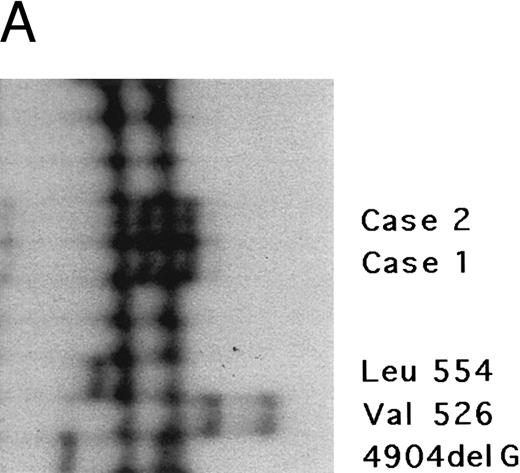
The fibrinogen Aα gene of two affected members of this kindred was examined by SSCP analysis. DNAs with previously described mutations in the fibrinogen Aα-chain gene associated with renal amyloidosis were included for comparison. SSCP analysis of exon 5 of the fibrinogen Aα gene in the DNA from the two affected individuals showed abnormally migrating bands in the PCR products compared with normal control subjects and gave a different pattern than the bands observed in patients with known amyloid mutations in the fibrinogen Aα-chain gene, suggesting a new fibrinogen Aα-chain mutation (Fig 3A).
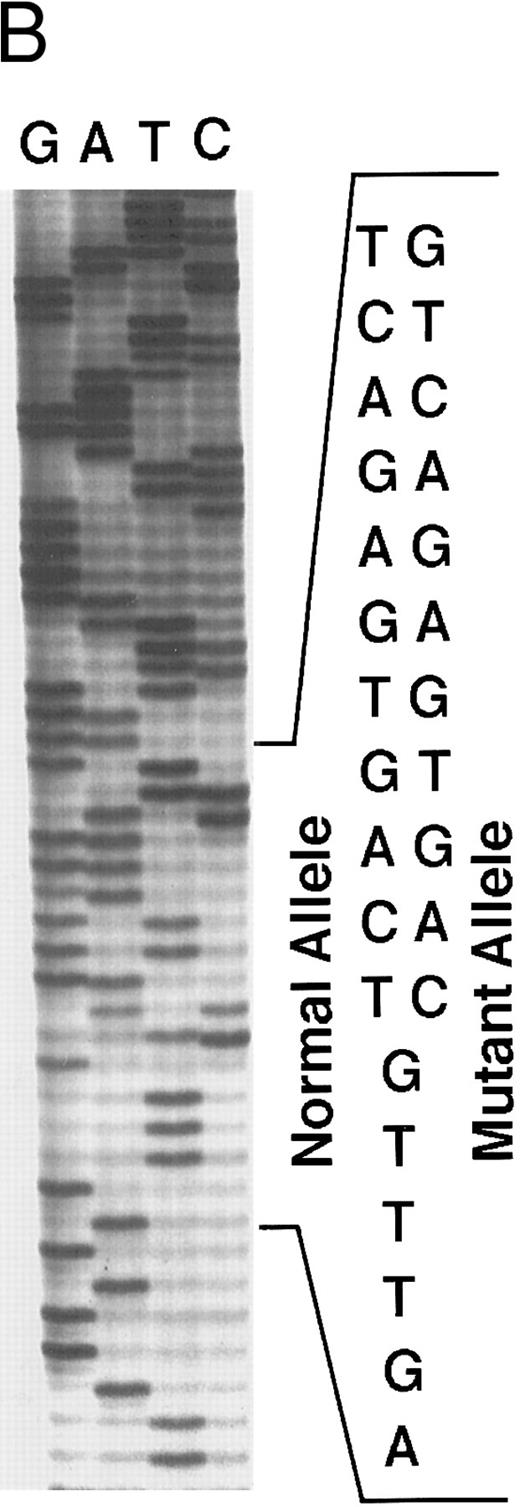
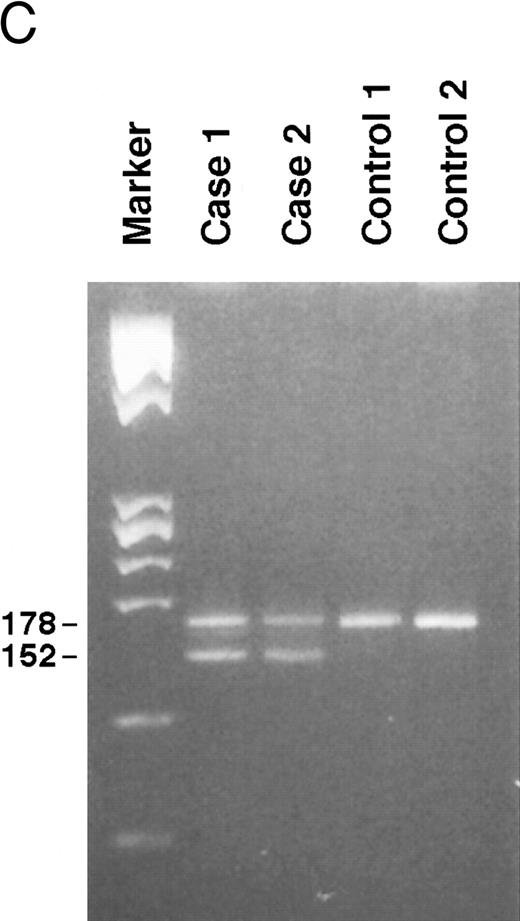
Genomic analysis of mutant fibrinogen Aα-chain gene. (A) Autoradiography of SSCP gel for exon 5 of fibrinogen Aα-chain gene. Cases no. 1 (affected father) and 2 (affected son) presented abnormal migrating bands in addition to the normal ones. DNAs with the known amyloidogenic fibrinogen Aα gene mutations Leu554, Val526 and 4904delG, were analyzed on the same gel for comparison. Unlabeled lanes are normal controls. (B) Autoradiography of DNA sequencing gel of exon 5 of the propositus. The propositus is heterozygous with the normal fibrinogen Aα-chain gene and the mutant gene that is missing T at the second base of codon 522. (C) Ethidium bromide-stained agarose gel of PCR-induced mutation restriction analysis for the detection of the 4897delT mutant gene. Case no. 1, affected father; case no. 2, affected son; controls no. 1 and 2, normal subjects; marker, DNA size marker (ΦX 174 DNA-HaeIII digest). The numbers on the left denote the sizes of the bands in basepairs.
Genomic analysis of mutant fibrinogen Aα-chain gene. (A) Autoradiography of SSCP gel for exon 5 of fibrinogen Aα-chain gene. Cases no. 1 (affected father) and 2 (affected son) presented abnormal migrating bands in addition to the normal ones. DNAs with the known amyloidogenic fibrinogen Aα gene mutations Leu554, Val526 and 4904delG, were analyzed on the same gel for comparison. Unlabeled lanes are normal controls. (B) Autoradiography of DNA sequencing gel of exon 5 of the propositus. The propositus is heterozygous with the normal fibrinogen Aα-chain gene and the mutant gene that is missing T at the second base of codon 522. (C) Ethidium bromide-stained agarose gel of PCR-induced mutation restriction analysis for the detection of the 4897delT mutant gene. Case no. 1, affected father; case no. 2, affected son; controls no. 1 and 2, normal subjects; marker, DNA size marker (ΦX 174 DNA-HaeIII digest). The numbers on the left denote the sizes of the bands in basepairs.
Direct DNA sequencing of exon 5 of the fibrinogen Aα-chain gene15 in the propositus and his son showed both a normal gene sequence and a mutant gene sequence with a nucleotide deletion at position 4897 (4897delT) that corresponded to the second base of codon 522 (Fig 3B). The mutant gene coded for a completely abnormal peptide sequence after codon 521 of the fibrinogen Aα-chain and was identical to the amino acid sequence found in isolated amyloid fibrils. There is only a difference of 3 amino acids at residues 522-524 between this case and the predicted protein sequence for the amyloid-associated, previously described deletion at the third base of codon 524.10 Termination at position 548 is the same for both.
PCR-RFLP analysis showed that both affected subjects had the Pst I recognition site associated with the deletion and therefore had a digestion band of 152 bp in addition to a normal band of 178 bp (Fig 3C).
DISCUSSION
Fibrinogen is a 340-kD plasma protein that plays a major role in platelet aggregation and blood clot formation.16 The fibrinogen molecule consists of two identical ensembles each containing an Aα, Bβ, and γ polypeptide chain. The Aα is the largest of the three chains with 610 amino acids and a molecular weight of 66 kD.17 The fibrinogen Aα-chain gene spans over 7 kb and has 6 exons.18 The gene has been assigned to 4q28 in humans.19 Three mutations in the carboxyl terminal region of the fibrinogen Aα-chain responsible for hereditary amyloidosis with dominant inheritance have been reported.8-10 In affected subjects, the disease has several common features. First, it is relatively early in onset. Second, the principal manifestation of the amyloidosis is nephropathy often presenting with hypertension and proteinuria. Third, there is no peripheral neuropathy.
The first Aα-chain mutation was found in three members of a Peruvian family with renal amyloidosis who shared a guanine to thymine transversion at position 4993 of this gene, resulting in a Leu for Arg substitution at residue 554 in the protein.8 A fragment of fibrinogen Aα-chain, residues 500 to 580, with a Leu at position 554 was isolated from renal amyloid deposits. No normal fibrinogen peptide with Arg554 was found. The second fibrinogen Aα-chain mutation associated with renal amyloidosis was originally found in two Irish-American kindreds, but other kindreds of European origin have now been described.9,20 DNA analysis showed an adenine to thymine transversion at the second base of codon 526 that is responsible for a Val for Glu substitution. The plasma fibrinogen values of mutant gene carriers were within normal range, and sequence analysis of their plasma fibrinogen showed about equal amounts of normal and variant protein. The third mutation in this protein associated with renal amyloidosis was found in a Caucasian-American kindred.10 DNA sequencing showed a single nucleotide deletion at the third base of codon 524 of the fibrinogen Aα-chain gene (4904delG) that resulted in a frame shift and predicted premature termination of the protein at codon 548. Sequence analysis of plasma fibrinogen failed to detect the presence of this predicted variant Aα-chain. However, plasma fibrinogen levels in gene carriers were lower than normal, suggesting that the abnormal Aα-chain may be degraded much more rapidly than normal Aα-chain.
We present here a novel mutation of the fibrinogen Aα-chain in a kindred with HRA. The mutation was first characterized in the peptide purified from renal amyloid and then confirmed at the DNA level. The fragment of the fibrinogen Aα-chain purified from renal amyloid has 49 amino acid residues and a molecular weight of approximately 5,000. The first 23 amino acid residues of the peptide are identical to the normal Aα-chain residues 499-521, whereas the C-terminal 26 amino acid residues correspond to a completely different sequence due to a nucleotide deletion and frame shift at codon 522 that results in a stop codon at 548.
In the previously described family with HRA in which a 1-base deletion resulted in a frame shift in the fibrinogen Aα-chain coding region, a new sequence was predicted from codon 525 to codon 548, where a premature termination would appear.10 However, attempts to identify the putative abnormal Aα-chain in plasma fibrinogen were unsuccessful, probably due to its low plasma levels and rapid clearance, and no amyloid-laden tissue from an affected subject was available to purify amyloid protein. However, we have successfully isolated the amyloid protein in the present family and shown that it is a hybrid of normal fibrinogen Aα-chain sequence and a completely novel peptide.
Although the family is small and linkage analysis is not feasible, the results strongly support a pathogenic role for the mutation: (1) the mutation is present in the propositus and in his affected son, (2) amyloid deposits reacted with antiserum raised against fibrinogen Aα-chain, (3) to date all mutations of the fibrinogen Aα-chain associated with HRA have been localized in this portion of the gene, and (4) deletion of the thymidine nucleotide at position 4897 resulted in a frame shift and predicted a new and shortened Aα-chain sequence that was identical to that found in the amyloid protein purified from renal tissue.
The age of onset in this kindred is particularly early, as the propositus developed nephrotic syndrome at the age of 31 and the disease began much earlier in his son at age 12. This onset age is earlier than that of patients with other mutations of the fibrinogen Aα-chain and resulted in terminal renal failure at a young age. The propositus received a renal transplant at age 35, but amyloid recurred within 2 years. The disease progressed more rapidly in the propositus' son, who reached terminal renal failure at age 13 and received a renal transplant at age 15. Unfortunately, histologic and clinical signs of recurrence of amyloid disease were present 1 year after renal transplantation. Although kidney transplantation is a possible therapeutic option in patients with fibrinogen Aα amyloidosis, the rapid reoccurrence of amyloid deposition in subjects with the 4897delT gene is a relative contraindication for this form of treatment. Other alternatives such as liver transplantation should be considered. Fibrinogen, like transthyretin, is synthesized in the liver. In transthyretin-associated amyloidosis, liver transplantation has been shown to lower the availability of mutated transthyretin and the results to date suggest that liver transplantation is an effective therapy.21 The fact that fibrinogen synthesis is essentially limited to the liver suggests that liver or combined liver/kidney transplantation should be considered for this form of amyloidosis.
In the present case, the purified amyloid protein was shown to be derived from a mutated fibrinogen Aα-chain and no trace of normal Aα-chain past residue 521 was found. This peptide fragment has two main characteristics. First, its size is relatively small compared with previously identified amyloid proteins associated with hereditary amyloidosis that range from 8 to 14 kD. This protein, therefore, belongs to a family of small amyloid subunit proteins that includes proteins of localized amyloid such as atrial natriuretic factor (3-4 kD) and the Aβ peptide (4-5 kD) of Alzheimer disease. The second characteristic is that the sequence of the protein in the C-terminal portion is completely different from the normal fibrinogen Aα-chain sequence and, indeed, lacks homology with any other mammalian-derived protein. This raises the question of the relative contributions of the different regions of the Aα-chain to amyloid-forming potential. Comparison of this amyloid protein with the protein isolated from amyloid deposits in fibrinogen Aα-chain Leu554 shows that the common portion of 22 amino acids, residues 500-521, could contribute to amyloid fibril formation. However, another fibrinogen variant with an Arg to Cys mutation at residue 554 (Dusart) has not been associated with amyloid formation, suggesting that residues 500-521 alone are not sufficient to produce fibril formation.22 Comparison with the amyloid subunit peptide in fibrinogen Aα Val526, which is similar in size to the Leu554 amyloid peptide (Liepnieks and Benson, unpublished observations), suggests that size of the fragment may be important to fibril formation. It has been documented that the carboxyl terminal portion of fibrinogen Aα-chain normally undergoes proteolytic cleavage at a number of sites, generating several early catabolic intermediates of different molecular size.23 Mutations in this part of the molecule may change either the tertiary structure or proportion of one or more degradation peptides and these may have increased amyloid-forming potential. Because early proteolysis of fibrinogen Aα-chain occurs intravascularly, this may explain the targeting of the kidney for this type of amyloid fibril formation. A factor in favor of the increased amyloidogenicity of the novel sequence of the carboxyl terminal portion of the Aα 4897delT peptide is that the 23 C-terminal residue sequence is homologous to the predicted sequence for the fibrinogen Aα 4904delG mutant that is also associated with HRA. Renal amyloidosis in both kindreds with these deletion mutations has a particularly early onset and rapid progression when compared with the amyloid in kindreds with the Val526 or Leu554 single amino acid substitution. Studies of tertiary structure of these variant peptides and their metabolism may help to define the pathogenesis of this condition. Certainly metabolic studies using isotopic markers such as those performed with the apolipoprotein A1Iowa variant,24 which has a significantly increased plasma clearance, may help to show how alterations and degradation of the different fibrinogen variants may contribute to amyloid fibril formation.
ACKNOWLEDGMENT
Much gratitude is offered to D. Dian Dillon (3D) for the use of her word-processing expertise.
Supported by Association Paulette Ghiron-Bistagne contre l'amylose, Association Française contre les Myopathies, a contrat de recherche clinique from Assistance Publique-Hôpitaux de Paris (No. 1869 for year 1992), Veterans Affairs Medical Research (MIS 583-0888), grants from RR-00750 (GCC), The US Public Health Service (Grants No. AG10608, AR20582, DK42111, and DK49596), the Marion E. Jacobson Fund, and the Machado Family Research Fund.
Address reprint requests to Merrill D. Benson, MD, c/o RLR VAMC (583), 1481 W 10th St, Room No. C7154, Indianapolis, IN 46202.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal