Abstract
To provide quantitative information on the sites that promote polymerization of sickle hemoglobin (HbS) after formation of the initial hydrophobic bond involving Val-6(β) [E6V(β)] and also to provide hemoglobins with an enhanced polymerization that could be used in a mouse model for sickle cell anemia, we have expressed recombinant double, triple, and quadruple HbS mutants with substitutions on both the α- and β-chains, E6V(β)/E121R(β), D75Y(α)/E6V(β)/E121R(β) and D6A(α)/D75Y(α)/E6V(β)/E121R(β). These recombinant hemoglobins were extensively characterized by high-performance liquid chromatography analysis, sodium dodecyl sulfate-polyacrylamide gel electrophoresis, isoelectric focusing, amino acid analysis, and mass spectroscopy. They retained the functional properties of the Hb tetramer and polymerized in a linear manner at progressively lower Hb concentration as a function of the degree of substitution, suggesting that these remote sites (αD6A, αD75Y, and βE121R) on the α- and β-chains exhibit additive, enhanced polymerization properties. The quadruple mutant has a polymerization concentration close to that of the purified SAD hemoglobin from transgenic mouse red blood cells consisting of HbS, Hb Antilles, and Hb D-Punjab. Normal mouse Hb increases the polymerization concentration of each mutant. Thus, the general approach of using recombinant Hbs as described here should prove useful in elucidating the quantitative aspects of the mechanism of HbS polymerization and in identifying the contribution of individual sites to the overall process. The strategy described here demonstrates the feasibility of a systematic approach to achieve future recombinant HbS mutants that could provide a new generation of the transgenic mouse model for sickle cell anemia.
THE LACK OF A naturally occurring animal model for sickle cell disease to test the efficacy of pharmacologic agents for treatment of the disease has prompted the development of transgenic mice in which the gene encoding sickle hemoglobin (HbS) is expressed in their red blood cells (RBCs). Several variations of this model have been developed,1-6 ie, mice expressing additional mutant Hbs together with HbS, ie, the SAD mouse model expressing HbS, Hb Antilles, and Hb D-Punjab. However, the continued expression of mouse Hb in such RBCs interferes with the polymerization of HbS or the higher mutants of HbS, thus impeding the formation of morphologically sickled erythrocytes to the extent that they appear in the circulation of sickle cell anemia patients. Attempts to delete the endogenous mouse Hb genes are underway to circumvent this problem. Moreover, the sickle mouse model has proven useful for studying other problems associated with sickle cell anemia.7
Because the strategy of using multiple naturally occurring HbS variants is now an accepted experimental route, we have taken the approach that the presence of higher order recombinant mutant Hbs in mouse RBCs with even more pronounced polymerization properties than HbS itself may be a viable strategy. Furthermore, the study of such mutants in a progressive fashion would provide a comprehensive and quantitative appreciation of the role of various sites in HbS polymerization to complement the x-ray8 and electron microscopic results,9,10 which identify the sites of polymerization but do not address their contribution to polymerization. Studies on the effects of such mutants on the well-known kinetics of polymerization11 may also provide new information. Accordingly, we report here the expression and study of three recombinant HbS mutants with enhanced polymerization properties.
There are a number of natural Hbs that inhibit polymerization, and this information was used initially to identify the contact sites between HbS tetramers in the polymer.12 Currently, information on polymerization enhancing mutants is limited to a small number of naturally occurring Hb double mutants12 that were identified because their presence in the RBC together with HbS led to a more severe clinical state than homozygous sickle cell disease.13 Furthermore, the molecular basis for understanding the enhanced polymerization properties of these mutants is not understood. The availability of recombinant DNA technology greatly expands the possibilities of enlarging that base of information and also to substitute a variety of amino acids at any given site to achieve either an enhanced or a reduced polymerization.
The current transgenic mouse models for sickle cell anemia are based on the enhanced polymerization of certain naturally occurring single and double mutants of sickle Hb.12 In this communication, we have used the identification of these important sites12 as a foundation to initiate a systematic study of higher order recombinant sickle Hb mutants.
MATERIALS AND METHODS
Reagents and plasmids. The restriction endonucleases, alkaline phosphatase, and DNA ligase were from Boehringer Mannheim (Indianapolis, IN) or New England Biolab (Beverley, MA). The DNA sequencing kit and the PCR reagent kit were obtained from US Biochemical Corp (Cleveland, OH). The Geneclean kit was from Bio 101 (La Jolla, CA). Qiagen plasmid midi kit was from Qiagen Inc (Santa Clarita, CA). The 35S-labeled dATP was from Amersham (Arlington Heights, IL). Dextran and 2,3-diphosphoglycerate (DPG) were purchased from Sigma (St Louis, MO). The nucleotides used to make the mutations were synthesized by the Protein Sequencing Facility at the Rockefeller University (New York, NY). The construction of pGS189 and pGS389 plasmids has been described elsewhere.14 15 All the other reagents were of the highest purity available.
Polymerase chain reaction (PCR)-based site-directed mutagenesis of the double HbS mutant E6V(β)/E121R(β). An Xho I fragment of pGS189 containing the sickle β-globin cDNA was inserted into the Xho I site of Bluescript II SK (+) to produce a plasmid with 4.2 kb, BSK-BetaE6V(β).16 17 This plasmid was used as a template in the PCR reactions. The two synthetic oligonucleotides (5′-AC TTT GGC AAA MGR TTC ACC CCA CCA GTG C and 5′-G CAC TGG TGG GGT GAA YCK TTT GCC AAA GT) were used for the PCR amplifications, and the underlined bases were those used to create the desired mutation. M denotes a mixture of A + C and Y denotes a mixture of C + T. These two primers were used in two PCR reactions to generate two PCR fragments. PCR amplifications were performed on a GeneAmp PCR system 2400 (Perkin-Elmer, Norwalk, CT) with 25 cycles of 30 seconds of denaturation at 97°C, 1 minute of primer/template annealing at 55°C, and 1 minute of extension at 72°C. These two fragments were then recombined in a separate PCR reaction, with primer/template annealing at 45°C, to produce recombinant sickle β gene containing the E121R(β) mutation. From this recombinant DNA fragment, the BamHI fragment was excised and used to replace the BamHI fragment of BSKBetaE6V(β). The correct insertional direction of the mutated BamHI fragment in BSK-BetaE6V(β) was verified by restriction enzyme analysis. The recombinant plasmid with desired mutation was identified directly by DNA sequence analysis and the base change encoding the Glu121(β) → Arg was the only mutation of the sickle β-globin gene. The β-globin cDNA with the base change encoding the Glu6(β) → Val and Glu121(β) → Arg was isolated as an Xho I fragment and used to replace the Xho I fragment of pGS189. The resulting plasmid pGS189E6V(β)/E121R(β) was digested with Not I and Bgl I to release the α- and β-gene cassette. The cassette was purified from agarose gel using a Geneclean kit (Bio 101) and inserted into pGS389 previously digested with Not I to give the recombinant plasmid, pGS389E6V(β)/E121R(β).
PCR-based site-directed mutagenesis of the triple HbS mutant D75Y(α)/E6V(β)/E121R(β). The plasmid pAD190, which was constructed by Dr A. Dumoulin in our laboratory as a derivative of pGS189, was used as a template in the PCR reactions to create the recombinant α gene. This plasmid contains the full-length α-globin cDNA under transcriptional control of a pGGAP promoter. The two synthetic oligonucleotides (5′-CAC GTG GAC TAC ATG CCC AAC G and 5′-C GTT GGG CAT GTA GTC CAC GTG) were used for the PCR amplifications, and the underlined bases were those used to create the desired mutation. The PCR amplifications were performed under conditions similar to those described above. From the recombinant α-globin gene containing the mutation, the Nco I and Sal I fragment was excised and used to replace the Nco I and Sal I fragment of pAD190. The recombinant plasmid with desired mutation was identified directly by DNA sequence analysis and the base change encoding the Asp75 (α) → Tyr was the only mutation of the α-globin gene. The Sph I fragment of plasmid pGS189E6V(β)/E121R(β) was excised and inserted into Sph I site of pAD190D75Y(α) to produce a plasmid with α- and β-genes, pAD190D75Y(α)/E6V(β)/E121R(β). This α and β cassette was excised as Not I fragment and similarly inserted into the Not I site of pGS389 to give the recombinant plasmid, pGS389D75Y(α)/E6V(β)/E121R(β).
PCR-based site-directed mutagenesis of the quadruple HbS mutant D6A(α)D75Y(α)/E6V(β)/E121R(β). Two overlapping PCR products were synthesized using the 5′-ATA AAC CAT GGT GCT GTC TCC TGC CGCCAA GAC C and 5′-C GTT GGG CAT GTA GTC CAC GTG and the 5′-CAC GTG GAC TAC ATG CCC AAC G and 5′-GAA CAA AGT CGA CTT AAC GGT A, respectively. The underlined bases were those used to create the desired mutations Asp6 (α) → Ala and Asp75 (α) → Tyr. PCR amplifications were performed as described above to produce the recombinant α-globin gene containing the D6A(α) and D75Y(α) mutations. The recombinant α-globin gene was excised as the Nco I and Sal I fragment and used to replace the Nco I and Sal I fragment of pAD190. The recombinant plasmid with desired mutations was identified directly by DNA sequence analysis and the base change encoding the Asp6 (α) → Ala and Asp75 (α) → Tyr was the only mutation of the α-globin gene. The Sph I fragment of plasmid pGS189E6V(β)/E121R(β) was excised and inserted into Sph I site of pAD190D6A(α)/D75Y(α) to produce a plasmid with α- and β-genes, pAD190D6A(α)/D75Y(α)/E6V(β)/E121R(β). This α and β cassette was excised as Not I fragment and similarly inserted into the Not I site of pGS389 to give the recombinant plasmid, pGS389D6A(α)/D75Y(α)/E6V(β)/E121R(β).
Yeast expression system and purification of Hb mutants. The recombinant pGS389E6V(β)/E121R(β), pGS389D75Y(α)/E6V(β)/E121R(β), and pGS389D6A(α)/D75Y(α)/E6V(β)/E121R(β) were transformed into Saccharomyces cerevisiae GSY112 cir° strain, using the lithium acetate method described previously.15-18 The transformants were selected using a complete medium first without uracil and then without uracil and leucine. To express each recombinant mutant, the yeast strain was grown in 12 L YP medium in a New Brunswick (Edison, NJ) Fermentor Bio IV for 3 to 4 days, using ethanol as the carbon source until an absorbance at 600 nm in the range of 5 to 10 was reached. The promoter controlling the transcription of the globin genes was induced for 20 hours by adding galactose (Pfanstiehl [Waukegan, IL] or US Biochemicals containing <0.1% glucose) to a final concentration of 3%. The collection and breakage of the cells after bubbling with CO gas has been described previously.15-18 The purification of the double mutant Hb was accomplished by chromatography on CM-Cellulose 52, using the linear gradient that consisted of 10 mmol/L potassium phosphate, pH 5.8, and 25 mmol/L potassium phosphate, pH 8.0 (150 mL of each). The purification of the triple mutant was accomplished using the linear gradient that consisted of 10 mmol/L potassium phosphate, pH 5.8, and 50 mmol/L potassium phosphate, pH 8.5 (150 mL of each). The quadruple mutant was initially chromatographed on CM-Cellulose 52 using the linear gradient that consisted of 10 mmol/L potassium phosphate, pH 5.8, and 50 mmol/L potassium phosphate, pH 8.5 (150 mL of each). The main fraction containing Hb was then purified on a Superose-12 HR 10/30 column on a Pharmacia FPLC system (Pharmacia, Uppsala, Sweden) and eluted with 120 mmol/L Tris-Ac buffer, pH 7.5, at a flow rate of 0.4 mL/min. The concentrated Hb sample was applied in 100 μL each, and the absorbance was measured at 280 nm with the Pharmacia on-line mercury lamp detection system with a 5-mm flow cell. The Hb quadruple mutant eluted at 13.6 mL as a tetramer and was collected for all structural and functional study.
Preparation of mouse hemolyzate and purification of Hb SAD. The RBCs were collected by centrifugation of a blood sample from the normal mouse or SAD mouse. Lysis of the cells was achieved by addition of 2 vol of distilled water and freeze/thawing of the suspension. After centrifugation in a microcentrifuge for 20 minutes for removal of RBC debris, the mouse hemolyzate was dialyzed against 0.1 mol/L NaCl and stored at −80°C until further use. Hb SAD was purified from SAD mouse hemolysate on Mono-S (Pharmacia, Piscataway, NJ) using 100 mmol/L phosphate, pH 6.3, and an NaCl gradient from 0 to 30 mmol/L. Its purity was ascertained by the presence of only one band upon isoelectric focusing.
Analytical methods. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of the recombinant mutants was performed on the Phast System from Pharmacia Biotech. The protein bands were stained with Coomassie brilliant blue R-250. Isoelectric focusing was performed on the pH 7-10 Hb-Resolve system from Isolab. The α- and β-globin chains from recombinant Hbs were separated by reverse phase high-performance liquid chromatography (HPLC) on a Vydac C4 column with a gradient of 20% to 60% acetonitrile containing 0.1% trifluoroacetic acid. The effluent was monitored at 220 nm. Amino acid analysis of globin chains isolated by this procedure was performed on a Beckman 6300 instrument with System Gold data handling system (Beckman Instruments, Palo Alto, CA). The spectrum of each mutant was recorded on a Shimadzu (Columbia, MD) 1601 UV-Visible spectrophotometer.
Mass spectrometry analysis. Electrospray mass spectrometric analysis of the purified recombinant mutant Hb tetramers was performed with a Finnigan-MAT TSQ-700 triple quadrupole mass spectrometer (San Jose, CA).19 20 Fifty picomoles of the Hb sample was loaded onto a desalting protein cartridge (Michrom BioResources, Inc, Auburn, CA) and washed with 1 mL of deionized water. The sample was eluted from the cartridge using a solution of water/acetonitrile/acetic acid, 30/67.5/2.5 (vol/vol/vol), and electrosprayed directly into the mass spectrometer. The flow of the eluting solution was maintained at 6 μL/min through a 100-μm inner diameter fused silica capillary.
IEF of natural and recombinant Hbs. Lane 1, natural HbA; lane 2, HbS; lane 3, Hb E6V(β)/E121R(β) double mutant; lane 4, Hb D75Y(α)/E6V(β)/E121R(β) triple mutant; lane 5, D6A(α)/D75Y(α)/E6V(β)/E121R(β); lane 6, HbS. The anode is at the top and the cathode is at the bottom.
IEF of natural and recombinant Hbs. Lane 1, natural HbA; lane 2, HbS; lane 3, Hb E6V(β)/E121R(β) double mutant; lane 4, Hb D75Y(α)/E6V(β)/E121R(β) triple mutant; lane 5, D6A(α)/D75Y(α)/E6V(β)/E121R(β); lane 6, HbS. The anode is at the top and the cathode is at the bottom.
Functional studies. The oxygen binding curves of the Hbs were determined at 37°C on a modified Hem-O-Scan instrument (American Instrument Co, Silver Springs, MD). Before the measurements, the purified Hb sample was dialyzed in 50 mmol/L bis-tris, pH 7.5, and converted to the oxy form.15-18 These samples were concentrated using CentriPrep, Centricon, and MicroCon ultrafiltration devices (10,000 molecular weight cut-off; Amicon, Beverly, MA) to a final concentration of 0.6 mmol/L. To measure the effect of anions on the oxygen affinity of these Hb mutants, an aliquot of a solution of 5.6 mmol/L DPG or 2.5 mol/L NaCl in 50 mmol/L bis-tris, pH 7.5, was added to the Hb sample to achieve the desired final concentration.
Measurement of gelation concentration (Csat ). The gelation concentration of the Hb mutants or Hb mixtures was determined by Dextran-Csat micromethod, as described previously.21,22 The concentrated Hb sample in the oxy form in 50 mmol/L potassium phosphate buffer, pH 7.5, was mixed with dextran at a final concentration of 120 mg/mL. Mineral oil was layered on top and fresh sodium dithionite solution (50 mmol/L final concentration) was added anaerobically below the Hb-dextran mixture using a gas-tight syringe. After stirring and incubation for 30 minutes in a 37°C water bath, the resulting gel under the oil layer was disrupted with a narrow wire loop, and the tubes were centrifuged in a microcentrifuge for 30 minutes. The clear supernatant was carefully separated from the aggregated Hb and its Hb concentration was measured spectrophotometrically and verified by amino acid analysis after acid hydrolysis of an aliquot.22
RESULTS
PCR-based site-directed mutagenesis. During the preparation of the double mutant β gene for E6V(β)/E121R(β), an initial attempt by the method of Kunkel23 using a template of M13mp18 phage containing the sickle β-globin gene and one mutagenic primer failed to create the desired mutation at β121 position. Use of different mutagenic primers and Gene 32 Protein did not improve the mutation frequency of the mutagenic reaction. Using agarose gel electrophoresis, we found that a significant amount of higher molecular weight oligonucleotides was present in the mutagenic primer. Presumably, the oligonucleotide that was used to create a mutation in this region of the β globin gene had a very strong tendency to self-associate so that synthesis of the single strand circular cDNA could not be completed. However, the desired mutation at β-121 position (Glu → Arg) and the mutation at α-75 position (Asp75 → Tyr) and α-6 position (Asp6 → Ala) were obtained by PCR-based site-directed mutagenesis strategy. After the mutagenesis, the sequence of the entire α and β genes of the recombinant mutants was verified by Sanger's dideoxy method.
Expression of the recombinant Hb mutants in yeast. The recombinant Hb double mutant E6V(β)/E121R(β), triple mutant D75Y(α)/E6V(β)/E121R(β), and quadruple mutant D6A(α)/D75Y(α)/E6V(β)/E121R(β) were expressed in yeast, which was grown and collected as described previously.15-18 A fermentor was used for the large production of yeast cells instead of the batch-wise flask method previously used, and it greatly simplified the procedure for medium preparation, ethanol addition during cultivation of the yeast, and induction of Hb mutant by galactose.
Purification of the HbS recombinant mutants. The purification of the recombinant HbS mutants was achieved by CM-Cellulose 52 chromatography as described earlier.15-18 However, these HbS mutants were relatively basic compared with HbS, so a gradient of high pH and increased ionic strength was required to elute them from the cation exchange column. For elution of the double mutant, a gradient of up to 25 mmol/L phosphate at pH 8.0 was used instead of 15 mmol/L phosphate for the purification of HbS. For the triple mutant, the purification was achieved on CM-Cellulose 52 using a gradient of up to 50 mmol/L phosphate at pH 8.5. The quadruple mutant was initially purified on CM 52 using a gradient of up to 50 mmol/L phosphate at pH 8.5 and further purified using Superose-12 gel filtration chromatography to remove several minor nonheme protein components that coeluted with the Hb mutant. The purity of each mutant was verified by SDS-PAGE, isoelectric focusing, globin chain analysis, and mass spectrometric analysis, as described below.
Isoelectric focusing (IEF ). Analysis of the purified recombinant mutants by IEF was performed on the pH 7 to 10 Hb-Resolve system from Isolab (Akron, OH)15-18 because of the basic nature of these mutants (Fig 1). For the double mutant E6V(β)/E121R(β), the loss of one negative charge (Glu → Val) and a substitution equivalent to the loss of two negative charges (Glu → Arg) per subunit was confirmed by the IEF migrations, ie, there is one charge difference between HbA (lane 1) and HbS (lane 2) and two charge differences between HbS and the double mutant (lane 3). The triple mutant D75Y(α)/E6V(β)/E121R(β) has a mutation site in which Asp of its α subunit has been replaced by Tyr. Its migration on IEF was consistent with a loss of a negative charge between the double mutant and triple mutant (lane 4). Compared with the triple mutant, IEF of the quadruple mutant D6A(α)/D75Y(α)/E6V(β)/E121R(β) indicated an additional loss of a negative charge (lane 5), consistent with an additional mutation site in which Asp of its a subunit has been replaced by Ala. Because all four mutations are on the exterior of the Hb, the full effect of these pKa changes is reflected in their electrophoretic behavior.
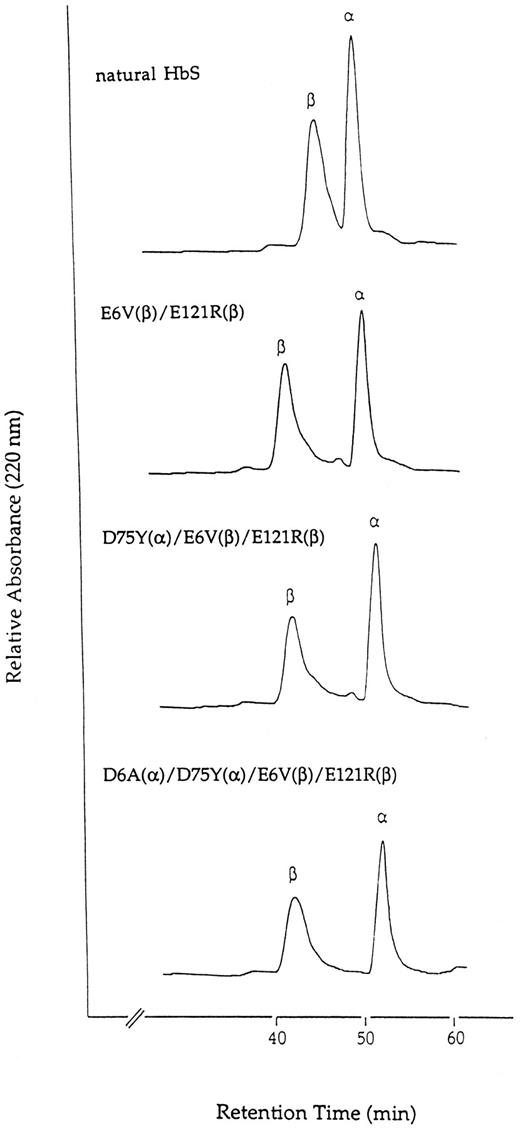
HPLC analysis of globin chains and amino acid analysis. The purified recombinant HbS mutants were analyzed by reverse-phase HPLC using a denaturing solvent to separate the globin chains (Fig 2). The β-chain of the double mutant E6V(β)/E121R(β) eluted at 42.9 minutes, much faster than the normal β-chain from HbS (47.6 minutes), indicating that replacement of Glu by Arg at 121 position significantly changed the chromatographic behavior of the β-chain. For the triple D75Y(α)/E6V(β)/E121R(β) mutant and quadruple mutant D6A(α)/D75Y(α)/E6V(β)/E121R(β), the elution times of their β-chains at 42.8 minutes were the same as that of the double mutant, consistent with the absence of further mutations of doubly mutated β-chain. The elution position of the α-chains from these recombinant mutants coincided with the α-chain from natural HbS (51.8, 52.6, 53.1, and 51.9 minutes, respectively), indicating that the replacement of the hydrophilic residue Asp at the α-chain by nonpolar amino acids Ala and Tyr does not alter their chromatographic behavior significantly on the reverse-phase C4 column.
Separation of α- and β-chains of natural HbS and recombinant HbS double, triple, and quadruple mutants. The samples were applied to a Vydac C4 HPLC column and eluted as described in the text. The amounts of Hb applied were about 100 μg.
Separation of α- and β-chains of natural HbS and recombinant HbS double, triple, and quadruple mutants. The samples were applied to a Vydac C4 HPLC column and eluted as described in the text. The amounts of Hb applied were about 100 μg.
The α- and β-chains of the recombinant mutants isolated by HPLC on Vydac C4 as described above were subjected to amino acid analysis, which permitted their assignments (Tables 1, 2, and 3). For the mutated β-chain of the double mutant (Table 1), the mutated α-chain of the triple mutant (Table 2), and the doubly mutated α-chain of the quadruple mutant (Table 3), the values for Arg, Glu, Asp, and Ala were in reasonable accord with their theoretical values. The values for the other amino acids were also in good agreement with the known composition of the recombinant mutants and confirmed their purity as a basis for the polymerization studies below.
Amino Acid Analysis of the α- and β-Globin Chains of HbS Double Mutant E6V( β)/E121R( β)
| Amino Acid . | Found . | Theory . | ||
|---|---|---|---|---|
| . | α-Chain . | β-Chain . | α-Chain . | β-Chain . |
| Asx | 12.0 | 11.8 | 12 | 13 |
| Thr | 7.5 | 5.9 | 9 | 7 |
| Ser | 10.7 | 4.8 | 11 | 5 |
| Glx | 5.7 | 9.5 | 5 | 9 |
| Gly | 7.7 | 14.1 | 7 | 13 |
| Ala | 21.2 | 16.4 | 21 | 15 |
| Cys | 1.4 | 0.3 | 1 | 2 |
| Val | 10.4 | 14.4 | 13 | 19 |
| Met | 0 | 0 | 2 | 1 |
| Ile | 0 | 0 | 0 | 0 |
| Leu | 18.0 | 18.0 | 18 | 18 |
| Tyr | 1.3 | 0.3 | 3 | 3 |
| Phe | 6.8 | 7.5 | 7 | 8 |
| His | 9.9 | 8.6 | 10 | 9 |
| Lys | 10.5 | 10.6 | 10 | 10 |
| Trp | 0 | 0 | 1 | 2 |
| Arg | 2.4 | 3.5 | 3 | 4 |
| Pro | 6.1 | 6.4 | 7 | 7 |
| Amino Acid . | Found . | Theory . | ||
|---|---|---|---|---|
| . | α-Chain . | β-Chain . | α-Chain . | β-Chain . |
| Asx | 12.0 | 11.8 | 12 | 13 |
| Thr | 7.5 | 5.9 | 9 | 7 |
| Ser | 10.7 | 4.8 | 11 | 5 |
| Glx | 5.7 | 9.5 | 5 | 9 |
| Gly | 7.7 | 14.1 | 7 | 13 |
| Ala | 21.2 | 16.4 | 21 | 15 |
| Cys | 1.4 | 0.3 | 1 | 2 |
| Val | 10.4 | 14.4 | 13 | 19 |
| Met | 0 | 0 | 2 | 1 |
| Ile | 0 | 0 | 0 | 0 |
| Leu | 18.0 | 18.0 | 18 | 18 |
| Tyr | 1.3 | 0.3 | 3 | 3 |
| Phe | 6.8 | 7.5 | 7 | 8 |
| His | 9.9 | 8.6 | 10 | 9 |
| Lys | 10.5 | 10.6 | 10 | 10 |
| Trp | 0 | 0 | 1 | 2 |
| Arg | 2.4 | 3.5 | 3 | 4 |
| Pro | 6.1 | 6.4 | 7 | 7 |
The value found for Leu was set at the theoretical value of 18.0 and used to normalize the values for the other amino acids. The values for Thr, Cys, Met, Tyr, and Trp are very low or absent because they are partially or completely destroyed under the conditions of acid hydrolysis in the presence of oxygen. For the amino acids underlined, there are significant differences between the amounts in the α- and β-chains.
Amino Acid Analysis of the α- and β-Globin Chains of HbS Triple Mutant D75Y(α)/E6V( β)/E121R( β)
| Amino Acid . | Found . | Theory . | ||
|---|---|---|---|---|
| . | α-Chain . | β-Chain . | α-Chain . | β-Chain . |
| Asx | 11.6 | 12.8 | 11 | 13 |
| Thr | 9.0 | 6.6 | 9 | 7 |
| Ser | 9.3 | 5.3 | 11 | 5 |
| Glx | 6.1 | 9.7 | 5 | 9 |
| Gly | 8.0 | 13.2 | 7 | 13 |
| Ala | 22.6 | 16.7 | 21 | 15 |
| Cys | 0 | 0 | 1 | 2 |
| Val | 15.7 | 18.3 | 13 | 19 |
| Met | 1.0 | 0.4 | 2 | 1 |
| Ile | 0.1 | 0.1 | 0 | 0 |
| Leu | 18.0 | 18.0 | 18 | 18 |
| Tyr | 2.6 | 1.8 | 4 | 3 |
| Phe | 7.1 | 7.7 | 7 | 8 |
| His | 10.5 | 9.0 | 10 | 9 |
| Lys | 10.8 | 10.6 | 10 | 10 |
| Trp | 0 | 0 | 1 | 2 |
| Arg | 2.2 | 2.8 | 3 | 4 |
| Pro | 6.9 | 6.7 | 7 | 7 |
| Amino Acid . | Found . | Theory . | ||
|---|---|---|---|---|
| . | α-Chain . | β-Chain . | α-Chain . | β-Chain . |
| Asx | 11.6 | 12.8 | 11 | 13 |
| Thr | 9.0 | 6.6 | 9 | 7 |
| Ser | 9.3 | 5.3 | 11 | 5 |
| Glx | 6.1 | 9.7 | 5 | 9 |
| Gly | 8.0 | 13.2 | 7 | 13 |
| Ala | 22.6 | 16.7 | 21 | 15 |
| Cys | 0 | 0 | 1 | 2 |
| Val | 15.7 | 18.3 | 13 | 19 |
| Met | 1.0 | 0.4 | 2 | 1 |
| Ile | 0.1 | 0.1 | 0 | 0 |
| Leu | 18.0 | 18.0 | 18 | 18 |
| Tyr | 2.6 | 1.8 | 4 | 3 |
| Phe | 7.1 | 7.7 | 7 | 8 |
| His | 10.5 | 9.0 | 10 | 9 |
| Lys | 10.8 | 10.6 | 10 | 10 |
| Trp | 0 | 0 | 1 | 2 |
| Arg | 2.2 | 2.8 | 3 | 4 |
| Pro | 6.9 | 6.7 | 7 | 7 |
The value found for Leu was set at the theoretical value of 18.0 and used to normalize the values for the other amino acids. The values for Thr, Cys, Met, Tyr, and Trp are very low or absent because they are partially or completely destroyed under the conditions of acid hydrolysis in the presence of oxygen. For the amino acids underlined, there are significant differences between the amounts in the α- and β-chains.
Amino Acid Analysis of the α- and β-Globin Chains of HbS Quadruple Mutant D6A(α)/D75Y(α)/E6V( β)/E121R( β)
| Amino Acid . | Found . | Theory . | ||
|---|---|---|---|---|
| . | α-Chain . | β-Chain . | α-Chain . | β-Chain . |
| Asx | 10.7 | 13.8 | 10 | 13 |
| Thr | 8.7 | 6.9 | 9 | 7 |
| Ser | 9.9 | 5.3 | 11 | 5 |
| Glx | 5.9 | 10.0 | 5 | 9 |
| Gly | 7.3 | 12.9 | 7 | 13 |
| Ala | 21.4 | 15.1 | 22 | 15 |
| Cys | 0 | 0 | 1 | 2 |
| Val | 12.0 | 16.4 | 13 | 19 |
| Met | 0.2 | 0 | 2 | 1 |
| Ile | 0 | 0 | 0 | 0 |
| Leu | 18.0 | 18.0 | 18 | 18 |
| Tyr | 2.9 | 0.9 | 4 | 3 |
| Phe | 6.7 | 7.7 | 7 | 8 |
| His | 9.5 | 9.1 | 10 | 9 |
| Lys | 10.5 | 10.5 | 10 | 10 |
| Trp | 0 | 0 | 1 | 2 |
| Arg | 2.6 | 3.2 | 3 | 4 |
| Pro | 6.9 | 7.5 | 7 | 7 |
| Amino Acid . | Found . | Theory . | ||
|---|---|---|---|---|
| . | α-Chain . | β-Chain . | α-Chain . | β-Chain . |
| Asx | 10.7 | 13.8 | 10 | 13 |
| Thr | 8.7 | 6.9 | 9 | 7 |
| Ser | 9.9 | 5.3 | 11 | 5 |
| Glx | 5.9 | 10.0 | 5 | 9 |
| Gly | 7.3 | 12.9 | 7 | 13 |
| Ala | 21.4 | 15.1 | 22 | 15 |
| Cys | 0 | 0 | 1 | 2 |
| Val | 12.0 | 16.4 | 13 | 19 |
| Met | 0.2 | 0 | 2 | 1 |
| Ile | 0 | 0 | 0 | 0 |
| Leu | 18.0 | 18.0 | 18 | 18 |
| Tyr | 2.9 | 0.9 | 4 | 3 |
| Phe | 6.7 | 7.7 | 7 | 8 |
| His | 9.5 | 9.1 | 10 | 9 |
| Lys | 10.5 | 10.5 | 10 | 10 |
| Trp | 0 | 0 | 1 | 2 |
| Arg | 2.6 | 3.2 | 3 | 4 |
| Pro | 6.9 | 7.5 | 7 | 7 |
The value found for Leu was set at the theoretical value of 18.0 and used to normalize the values for the other amino acids. The values for Cys, Met, Tyr, and Trp are very low or absent because they are partially or completely destroyed under the conditions of acid hydrolysis in the presence of oxygen. For the amino acids underlined, there are significant differences between the amounts in the α- and β-chains.
Mass spectrometry. The results of the molecular masses of the purified recombinant HbS mutants are shown in Table 4. Previous measurements on HbA and HbS have shown the correct values within experimental error.15-18 The molecular mass obtained for the β-chain of these three mutants (15,868.0, 15,868.0, and 15,867.0, respectively) by the ESI-MS method agreed with the theoretical value of 15,864.2 for a β-chain having the two substitutions (Glu → Val, Glu → Arg). The difference of about 29 Daltons from the sickle β-chain (15,838.3 mass units) is within experimental error of the expected difference of 27 Daltons between a Glu (129 mass units) and an Arg (156 mass units) residue. The measured molecular mass for the α-chain of E6V(β)/E121R(β) double mutant (15,130.0 Daltons) is consistent with the calculated value (15,126.4 Daltons) for the native α-chain within experimental error. The difference of 48.0 mass units between the mass of the α-chain of D75Y(α)/E6V(β)/E121R(β) triple mutant (15,178.0) and of the E6V(β)/E121R(β) double mutant (15,130.0) is in good accord with the calculated difference of 48 Daltons between an Asp (115 mass units) and a Tyr (163 mass units) residue, suggesting the presence of the desired substitution on the α-chain (Asp → Tyr) of the triple mutant. The difference of 6.6 mass units from the native α-chain (15,126.4) is in good accord with the combined differences for the substitutions Asp → Ala and Asp → Tyr (44.0 and 48.1 mass units difference), consistent with the presence of two desired substitutions on the α-chain of the quadruple mutant. These results together with amino acid analysis established that the recombinant mutants were the desired ones.
Mass Spectrometric Analysis of Recombinant Hbs
| Hb . | Theory . | Measured . | ||
|---|---|---|---|---|
| . | α-Chain . | β-Chain . | α-Chain . | β-Chain . |
| E6V( β)/E121R( β) | 15,126.4 | 15,864.2 | 15,130 | 15,868 |
| D75Y(α)/E6V( β)/E121R( β) | 15,174.0 | 15,864.2 | 15,178 | 15,868 |
| D6A(α)/D75Y(α)/E6V( β)/E121R( β) | 15,130.4 | 15,864.2 | 15,133 | 15,867 |
| Hb . | Theory . | Measured . | ||
|---|---|---|---|---|
| . | α-Chain . | β-Chain . | α-Chain . | β-Chain . |
| E6V( β)/E121R( β) | 15,126.4 | 15,864.2 | 15,130 | 15,868 |
| D75Y(α)/E6V( β)/E121R( β) | 15,174.0 | 15,864.2 | 15,178 | 15,868 |
| D6A(α)/D75Y(α)/E6V( β)/E121R( β) | 15,130.4 | 15,864.2 | 15,133 | 15,867 |
There is an error of 3 or 4 mass units in the measured mass of both α- and β-chains. It is a systematic error, so we can safely conclude that mutations were successful and that the Hbs were expressed properly.
Functional studies. The oxygen binding properties of the recombinant mutants were determined at a Hb concentration of 0.6 mmol/L (Table 5). The oxygen affinity of the double and triple mutants showed an average P50 value of 9 mmHg, with a Hill coefficient of 3.0 and 3.1, indicating that they retained full cooperativity. The quadruple mutant showed a high oxygen affinity with a P50 value of 5 mmHg, consistent with the reported value of natural Hb Sawara,24 and it was also cooperative with a Hill coefficient of 2.8. The effects of DPG (at 2:1 ratio to Hb) on both double and triple mutants were similar to those on natural HbS. The effect of DPG (at 2:1 ratio to Hb) on quadruple mutant was also comparable to its effect on natural HbS under the same conditions, with a higher value of P50 by about twofold. In the presence of 500 mmol/L of chloride, oxygen affinity of the double and triple mutants was lowered twofold to threefold similar to that of HbS. However, the quadruple mutant shows a somewhat decreased response to chloride. The reasons for this behavior are under investigation.
Functional Properties of Natural HbS and Recombinant HbS Mutants
| Hb . | Additive . | P50 . | n . |
|---|---|---|---|
| Natural HbS | 0 | 9 | 3.1 |
| 1.2 mmol/L DPG | 22 | 2.8 | |
| 0.5 mmol/L NaCl | 22 | 3.1 | |
| E6V( β)/E121R( β) | 0 | 9 | 3.1 |
| 1.2 mmol/L DPG | 22 | 3.1 | |
| 0.5 mmol/L NaCl | 21 | 2.9 | |
| D75Y(α)/E6V( β)/E121R( β) | 0 | 9 | 3.0 |
| 1.2 mmol/L DPG | 21 | 2.8 | |
| 0.5 mmol/L NaCl | 19 | 2.8 | |
| D6A(α)/D75Y(α)/E6V( β)/E121R( β) | 0 | 5 | 2.8 |
| 1.2 mmol/L DPG | 9 | 2.8 | |
| 0.5 mmol/L NaCl | 7 | 2.8 |
| Hb . | Additive . | P50 . | n . |
|---|---|---|---|
| Natural HbS | 0 | 9 | 3.1 |
| 1.2 mmol/L DPG | 22 | 2.8 | |
| 0.5 mmol/L NaCl | 22 | 3.1 | |
| E6V( β)/E121R( β) | 0 | 9 | 3.1 |
| 1.2 mmol/L DPG | 22 | 3.1 | |
| 0.5 mmol/L NaCl | 21 | 2.9 | |
| D75Y(α)/E6V( β)/E121R( β) | 0 | 9 | 3.0 |
| 1.2 mmol/L DPG | 21 | 2.8 | |
| 0.5 mmol/L NaCl | 19 | 2.8 | |
| D6A(α)/D75Y(α)/E6V( β)/E121R( β) | 0 | 5 | 2.8 |
| 1.2 mmol/L DPG | 9 | 2.8 | |
| 0.5 mmol/L NaCl | 7 | 2.8 |
The procedures used to calculate the P50 and n values are described in the text, and the Hb concentration for oxygen dissociation curves was 0.6 mmol/L in 50 mmol/L bis-Tris, pH 7.5.
Polymerization of the recombinant mutants and of the purified Hb SAD. A new micromethod21,22 was used to measure the Csat (Table 6). The gelation concentration of the HbS was 35.3 mg/mL, consistent with the value of HbS (34 mg/mL) reported previously.22 Under the same conditions, the double mutant E6V(β)/E121R(β) polymerizes at a concentration of 24.3 mg/mL, indicating a one third lowering of the gelation of HbS itself. The triple mutant shows a doubling of this enhancement of gelation, ie, it gels at 11.8 mg/mL, which is a two thirds lower concentration than that of HbS itself. The average Csat of the quadruple mutant was 7.0 mg/mL, which is close to that of the purified SAD Hb (6.1 mg/mL). This value is one fifth the polymerization concentration of HbS itself. These results show that the polymerization can be significantly enhanced by selected amino acid substitution on various regions of the HbS tetramer. Substitution at these widely separated sites (αD6A, αD75Y, and βE121R) on the α- and β-chains gives rise to linearly additive effects on the overall process of HbS polymerization (Fig 3). The importance of this observation as a basis for further studies is described in the Discussion.
Gelation Concentrations of Natural HbS and Recombinant HbS Mutants
| Hb . | Csat (mg/mL) . |
|---|---|
| Natural HbS | 34.4, 34.9, 36.5 (35.3) |
| E6V( β)/E121R( β) | 24.0, 24.6 (24.3) |
| D75Y(α)/E6V( β)/E121R( β) | 11.4, 12.2 (11.8) |
| D6A(α)/D75Y(α)/E6V( β)/E121R( β) | 7.6, 6.8, 6.6 (7.0) |
| Purified Hb SAD | 6.2, 6.1, 6.1 (6.1) |
| Hb . | Csat (mg/mL) . |
|---|---|
| Natural HbS | 34.4, 34.9, 36.5 (35.3) |
| E6V( β)/E121R( β) | 24.0, 24.6 (24.3) |
| D75Y(α)/E6V( β)/E121R( β) | 11.4, 12.2 (11.8) |
| D6A(α)/D75Y(α)/E6V( β)/E121R( β) | 7.6, 6.8, 6.6 (7.0) |
| Purified Hb SAD | 6.2, 6.1, 6.1 (6.1) |
All measurements were performed in the presence of dextran in 50 mmol/L potassium phosphate buffer, pH 7.5, at 37°C, as described in the text. The average values are in parentheses.
Gelation equilibrium concentrations (Csat ) of recombinant HbS mutants. The Csat is given in milligrams per milliliter. All measurements were performed in the presence of dextran in 50 mmol/L potassium phosphate buffer, pH 7.5, at 37°C, as described in the text. (1) HbS; (2) double HbS mutant E6V(β)/E121R(β); (3) triple HbS mutant D75Y(α)/E6V(β)/E121R(β); and (4) quadruple HbS mutant D6A(α)/D75Y(α)/E6V(β)/E121R(β).
Gelation equilibrium concentrations (Csat ) of recombinant HbS mutants. The Csat is given in milligrams per milliliter. All measurements were performed in the presence of dextran in 50 mmol/L potassium phosphate buffer, pH 7.5, at 37°C, as described in the text. (1) HbS; (2) double HbS mutant E6V(β)/E121R(β); (3) triple HbS mutant D75Y(α)/E6V(β)/E121R(β); and (4) quadruple HbS mutant D6A(α)/D75Y(α)/E6V(β)/E121R(β).
Polymerization of the mixture of recombinant mutants and normal mouse hemolysate. To determine the effects of normal mouse Hb on the polymerization tendency of these recombinant mutants, we have measured gelation concentration of the mixture of these mutants with mouse hemolysate in different ratios (Table 7). The data show clearly the effect of the normal mouse Hb in preventing polymerization of sickle Hb, which was already appreciated but is presented here in quantitative terms. When the triple mutant was mixed with two equivalents of mouse hemolysate, the mixture polymerizes at a Hb concentration close to that of HbS alone. The gelation concentration of equimolar mixture of the triple mutant with normal mouse Hb is much lower than that of sickle Hb. For the quadruple mutant, equimolar mixture with mouse hemolysate has a gelation concentration of 12.4 mg/mL, which is one third the polymerization concentration of HbS itself.
Gelation Concentrations of Mixture of Recombinant HbS Mutants With Mouse Hemolysate
| Hb . | HbS Mutants/Mouse Hemolysate . | |
|---|---|---|
| . | 1/1 . | 1/2 . |
| Natural HbS | 79.1 | 95.0 |
| E6V( β)/E121R( β) | 48.2 | 72.6 |
| D75Y(α)/E6V( β)/E121R( β) | 28.0 | 39.6 |
| D6A(α)/D75Y(α)/E6V( β)/E121R( β) | 12.4 | — |
| Hb . | HbS Mutants/Mouse Hemolysate . | |
|---|---|---|
| . | 1/1 . | 1/2 . |
| Natural HbS | 79.1 | 95.0 |
| E6V( β)/E121R( β) | 48.2 | 72.6 |
| D75Y(α)/E6V( β)/E121R( β) | 28.0 | 39.6 |
| D6A(α)/D75Y(α)/E6V( β)/E121R( β) | 12.4 | — |
All measurements were performed in the presence of dextran in 50 mmol/L potassium phosphate buffer, pH 7.5, at 37°C, as described in the text.
DISCUSSION
Using recombinant DNA techniques, we have previously shown that inhibition of polymerization of HbS was dependent on the site of the amino acid substitution.17,18,22,25 Furthermore, we found that substitution at one particular site on the exterior of the HbS tetramer inhibited polymerization17 more than a substitution at the initial hydrophobic interaction itself between Val-6(β) and Leu-88(β) on adjacent tetramer25 and that they acted independently.22 Those results clearly demonstrated in a quantitative manner that some sites are more important than others in inhibiting polymerization of HbS. In the present report, we used the same recombinant DNA methodology but to study enhanced polymerization. Although it is possible to understand the inhibition of polymerization based on the x-ray and electron microscopic studies by interference with contact sites between tetramers, the mechanism of the polymerization-enhancing effects by many mutants has not yet reached that level of understanding. The present results represent the initial attempt in that direction.
In choosing the replacement sites for the double, triple, and quadruple mutants of HbS described in this report, we used the naturally occurring single and double mutants that lead to increased polymerization12 as a guide to select the mutation sites and to achieve the additive effect. Hb D-Los Angeles (Hb D-Punjab), which is part of the SAD mouse construct, has a Gln substitution for Glu at β121. Its presence in the RBC together with HbS led to a more severe clinical state than homozygous sickle cell disease.26 Hb O-Arab with a Lys substitution at the same site promotes the gelation of HbS to an even greater extent.27 A natural double mutant HbS-Oman (Glu6 → Val, Glu121 → Lys) gels at about 30% lower Hb concentration than HbS.28 We reasoned that the substitution for Glu at β121 by an even larger basic amino acid could stabilize the polymer structure to an even greater extent. Therefore, Arg was selected to achieve this effect. Our results show that the double mutant E6V(β)/E121R(β) polymerizes at about one third lower Hb concentration than HbS. This value is comparable to that from natural double mutant HbS-Oman, suggesting that the enhancing effect is retained by the introduction of a more basic amino acid residue and the size of the residue at this site does not play a significant role in the polymerization process of HbS.
In choosing the third and fourth replacement site for the triple and quadruple mutants, we selected the sites on the α-chain distant from the substitution in the β-chain anticipating that an additive effect on the enhanced polymerization may be obtained. We therefore selected Hb Winnipeg (Asp75 → Tyr)29 and Hb Sawara (Asp6 → Ala).24 The HbS double mutant αWinnipeg2βS2 and αSawara2βS2 , prepared from hybridization of the Hb single mutant and sickle β-chain, were found to polymerize at about 50% and 20% lower Hb concentration, respectively.30 In this instance, we saw no advantage in substituting an amino acid different than that occurring naturally. This strategy has been borne out, because the triple mutant D75Y(α)/E6V(β)/E121R(β) has doubled the enhanced polymerization of the mutant E6V(β)/E121R(β) and it gels at a Hb concentration that is two thirds less than HbS itself. For the quadruple mutant, the polymerization concentration (7.0 mg/mL) is about 40% lower than that of triple mutant D75Y(α)/E6V(β)/E121R(β). Thus, the substitution of Ala for Asp at α6 further decreased the solubility of the HbS triple mutant by about 40%. These results not only show the overall enhanced effects but also the individual contribution of different sites on HbS polymerization.
The findings demonstrate the feasibility of expressing multiple HbS mutants in the yeast system to understand the sites that lead to an enhanced polymerization as well as those that give a decreased polymerization.17,22 25 The comparison of the polymerization concentration of the recombinant mutants with that of the purified SAD Hb indicates a progressive tendency towards the latter (Fig 3). Because the polymerization concentration of the purified SAD Hb has not yet been reported at physiologic concentrations to our knowledge, it is useful to have this information as a point of reference.
An important conclusion of these studies is that recombinant higher order sickle Hb mutants can be selected beginning with the fundamental knowledge of the gelation concentration of the natural HbS double mutants as a guide.12 Thus, the finding that the polymerization concentration of the quadruple mutant is virtually the same as that of the triple SAD mutant purified from the transgenic mouse indicates that the experimental approach is logical. This result also suggests that some sites are more effective than others in promoting polymerization. The correlation of the extent of polymerization enhancement with increasing order of mutation, ie, the linearity in Fig 3, represents a concept that we plan to extend. The absence of any saturation effect in this profile suggests that recombinant mutants with even lower polymerization concentration could be designed.
ACKNOWLEDGMENT
The authors thank Drs Antoine Dumoulin and Vincenzo Nardi-Dei for their continual assistance with the techniques, Dr Peter W. van Ophem for carrying out the amino acid analysis, and William Fowle for the photography. We are also grateful to Dr Marie Trudel and to Dr Alvin Head for providing blood samples from the SAD mouse.
Supported in part by National Institutes of Health Grant No. HL-18819 (J.M.M.).
Address reprint requests to James M. Manning, PhD, Northeastern University, Department of Biology, 414 Mugar Bldg, 360 Huntington Ave, Boston, MA 02115.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal