Abstract
Although abnormalities involving the short arm of chromosome 12 (12p) are one of the most frequently observed rearrangements in childhood acute lymphoblastic leukemia (ALL), little is known about the frequency of different structural abnormalities and their relationship to the status of the ETV6 (also named TEL) gene in this region. Of 815 children with newly diagnosed ALL, 94 (11.5%) had a total of 104 cytogenetic 12p abnormalities. Loss of genetic material was observed in 67 (64%) of these abnormalities. Cases with 12p alterations had a much lower frequency of hyperdiploidy greater than 50 (7%) than did the ALL population in general, but these cases had a similar distribution of immunophenotype and similar 5-year event-free survival (70% ± 5% SE v 64% ± 2%, P = .64). Rearrangement of the ETV6 gene was identified in 36 (56%) of 64 cases evaluated. The ETV6-CBFA2 (TEL-AML1) fusion transcript was found in 25 (66%) of 38 cases evaluated, and all but one of these showed ETV6 rearrangement. Importantly, ETV6 rearrangement was associated with a favorable prognosis (5-year event-free survival: 89% ± 6% v 60% ± 1%, P < .01). We conclude that most but not all 12p cytogenetic abnormalities in childhood ALL involve ETV6, and that rearrangement of ETV6 is associated with a favorable treatment outcome.
CHROMOSOME 12p abnormalities are identified by conventional cytogenetics studies in 8% to 11% of acute lymphoblastic leukemia (ALL) cases.1-3 Most 12p abnormalities involve translocations, such as dic(9; 12) (p11; p11),4-6 dic(7; 12)(p11; p11-12),6,7 t(12; 13)(p13; q14),6-8 t(2; 12)(q14; p13),1,6 and t(12; 17)(p13; q21).9 Approximately one fourth of the 12p abnormalities are deletions.3
Most ALL cases with a 12p abnormality have a B-lineage immunophenotype, although a few T-lineage cases have been reported.1,2 Abnormalities of 12p, most notably the dic(9; 12)(p11; p12), are generally associated with a favorable prognosis,5,10,11 although one report described an increased risk of central nervous system (CNS) relapse.2
ETV6, also known as TEL, is one of the gene(s) involved in 12p abnormalities, and has recently drawn intense investigation.12,13 This gene encodes an ETS-like putative transcription factor and was initially identified by its fusion with the platelet-derived growth factor receptor beta (PDGFRβ) in a case of chronic myelomonocytic leukemia with a t(5; 12) (q33; p13).12 The gene was subsequently found to be involved in a number of translocations, and fusion transcripts have been identified for ETV6-MDS1/EVI1, t(3; 12) (q26; p13)14,15; ETV6-ABL, t(9; 12)(q34; p13)16; MN1-ETV6, t(12; 22)(p13; q11)17; ETV6-?, t(10; 12)(q24; p13)18; and ETV6-CBFA2 (also termed TEL-AML1 ), t(12; 21)(p13; q22).19-23 The t(12; 21), a cryptic translocation rarely observed by conventional cytogenetics, was first identified by fluorescence in situ hybridization (FISH).20,24 In ALL blasts, this translocation fuses the 5′ part of the ETV6 gene with almost the entire CBFA2 gene, producing the chimeric transcript ETV6-CBFA2.19 Recently, the t(12; 21) has been identified as the most frequent chromosomal abnormality in childhood ALL, affecting 20% to 25% of B-lineage cases.20-23
The frequency of ETV6 involvement in pediatric ALL with 12p abnormalities has not been described, nor has its effect, if any, on clinical characteristics and prognosis. To address these issues, we studied the largest cohort of patients with 12p abnormalities reported to date.
MATERIALS AND METHODS
Patients
Between December 1979 and August 1994, 1,090 children with newly diagnosed ALL were enrolled in four consecutive front-line clinical trials (Total Therapy Studies X-XIII) at St Jude Children's Research Hospital.10,25,26 The diagnosis of ALL was based on the morphologic criteria of the French-American-British (FAB) Cooperative Group and on negative myeloperoxidase and esterase stains.27 Cytogenetic analysis was successfully performed in 815 cases; most of the unsuccessful cases were enrolled during the 1979 to 1983 era, when chromosome banding was generally suboptimal. Of the 815 evaluable cases, abnormalities of chromosome 12p11-p13 were identified in 94 (11.5%), including 23 cases previously reported.1 Earlier we have described the ETV6 status of two overlapping subsets of our study group, one comprising 18 patients22 and the other 22 patients.28 Informed consent was obtained from all patients or their guardians, and the investigations were approved by the institution's clinical trials review committee.
Cytogenetic Evaluation
Blast Cell Phenotyping
Blast cell surface antigens were detected by standard indirect immunofluorescence assays with monoclonal antibodies to lymphoid-associated antigens. Blast cells were also tested for surface and cytoplasmic immunoglobulins (sIg and cIg) and for formation of heat-stable rosettes with sheep erythrocytes. Depending on their reactivity patterns, cells were classified as T (CD7+, CD5+, E-rosette±), B (sIg+), pre-B (cIg+), early pre-B (cIg−, sIg−, T−, HLA-DR+, CD19+, CD10±), or common B-lineage (sIg−, CD19+, DR+, CD10+, but unknown cIg status).31
Southern Blot Analysis
DNA was extracted from frozen marrow samples obtained at diagnosis, using standard techniques.22 For Southern analysis, 5 to 10 μg of DNA were digested with the restriction enzymes BamHI and HindIII, size-fractionated on a 0.8% agarose gel, and blotted onto nylon membranes. All blots were hybridized with a 32P-labeled probe consisting of the 0.6-kb Sac I/BamHI fragment of the ETV6 cDNA.22
Reverse Transcriptase Polymerase Chain Reaction (RT-PCR)
RNA was extracted (Purescript kit; Gentra, Research Triangle, NC) and RT-PCR performed as previously described.32 Samples were analyzed for the presence of both derivative fusion products using previously reported primer pairs.22 Following amplification, PCR products were separated by electrophoresis, transferred to nylon membrane, and hybridized to 32P–end-labeled oligonucleotide probes specific for CBFA2 or ETV6. All cases apparently negative for the der(21) products were reamplified using a more 5′ oligonucleotide, as described.22
FISH
Of the 25 cases with germline ETV6 by Southern analysis, only 11 cases (Nos. 11, 12, 16, 26, 45, 49, 57, 58, 68, 74, and 83) had frozen cells/cytogenetics pellets available for FISH analysis; 4 (Nos. 45, 57, 58, and 68) exhibited cytogenetic evidence of 12p13 involvement. Three cases with ETV6 rearrangement (Nos. 42, 46, and 69) were also subjected to the FISH assay as positive controls, and four normal donors were subjected as negative controls.
Two P1 clones containing the 5′ (exon 1; 25,000 bp) and 3′ (exon 8; 24,000 bp) ends of the ETV6 gene were used for the FISH analysis. The probes were dual-color labeled with biotin or digoxigenin by Nick translation (GIBCO-BRL, Life Technologies, Gaithersburg, MD) according to the manufacturer's protocol. The hybridizations were performed on either fixed cell pellets after cytogenetic analysis (n = 8) or cytospin slides from frozen marrow samples (n = 6) obtained at diagnosis. The probes were hybridized to the target slides as previously described.33 All hybridizations were repeated with reverse labeling to check the consistency of the results. Screening and analysis were done using an Olympus fluorescence microscope (Olympus America Inc, Melville, NY) attached to the VYSIS imaging system (VYSIS Inc, Downers Grove, IL). In each case, a minimum of 100 to 200 interphase nuclei were evaluated by two independent investigators.
Statistical Analysis
Event-free survival was estimated by the method of Kaplan and Meier,34 with standard errors calculated by the method of Peto and coworkers.35 Statistical comparisons of event-free survival distributions were made by the log-rank test, stratified by study.36 The distributions of categorically defined presenting features were compared between the groups with and without ETV6 rearrangement using Fisher's exact test.
RESULTS
Cytogenetic Findings
Table 1 lists the clinical and cytogenetic features of the 94 cases with 12p abnormalities. The modal number was 45 in 15 cases (16%), 46 in 54 (57%), 47 to 50 in 18 (19%), and greater than 50 in 7 (8%). Aberrations affecting 12p were the sole chromosomal abnormality in 21 of the cases (22%). The other 73 cases had additional aberrations comprising mainly random numerical and structural chromosomal abnormalities; the few recurrent aberrations included deletions in 6q (n = 7), 9p (n = 5), 11q (n = 5), and 13q (n = 4). Four cases had common recurrent translocations other than those involving the 12p region: a pre-B case with a der(19)t(1; 19)(q21; p13) (patient 56), an early pre-B case with a t(17; 19)(q22; p13) (patient 88), and two T-cell cases, one with a t(10; 14)(q24; q11) (patient 11) and one with a t(7; 14)(p15; q32) (patient 83).
Clinical and Laboratory Features of 94 ALL Patients With 12p Abnormality
| Patient No. . | Race . | Age . | WBC . | Immuno- . | Karyotype . | ETV6 Status . | ETV6/CBFA2 . | Remission Duration (mo) . | Type of Failure . |
|---|---|---|---|---|---|---|---|---|---|
| . | . | (yr) . | (×109/L) . | phenotype . | . | . | . | . | . |
| ONE 12P HOMOLOG ABNORMAL | |||||||||
| Deletion (n = 26) | |||||||||
| 1 | W | 4.1 | 5.7 | Pre-B | 46,XX,del(12)(p12),−13,+mar/47,idem,+mar* | NA | NA | 142+ | |
| 2 | W | 8.3 | 154.8 | Early pre-B | 46,XX,del(9)(p21),del(12)(p12)* | NA | NA | 5 | CNS |
| 3 | W | 5.6 | 8.9 | Pre-B | 46,XY,del(8)(q21.2q21.3),del(9)(p22),del(12)(p12),add(16)(q24)/46,idem,−del(12)* | R | NA | 126+ | |
| 4 | B | 3.3 | 11.3 | Early pre-B | 46,XX,del(12)(p11)/46,XX,t(7; 12)(p22; q12)* | R | NA | 125+ | |
| 5 | W | 2.2 | 34.3 | Early pre-B | 47,XX,del(12)(p12),+16/47,idem,del(13)(q12q21-22)* | G | NA | 81+ | |
| 6 | W | 4.3 | 2.1 | Early pre-B | 46,XY,del(12)(p12)* | R | NA | 116+ | |
| 7 | W | 13.1 | 73.7 | Early pre-B | 46,XX,del(12)(p12),−22,+der(?)t(?; 22)(?; q11)/46,XX,?del(8)(p22), del(12)/47,XX,del(12),+21 | R | + | 99+ | |
| 8 | W | 10.1 | 4.7 | Early pre-B | 53,XX,+5,+6,+del(6)(q21q23),+del(12)(p12),+18,+21,+21* | G | NA | 72+ | |
| 9 | B | 7.6 | 24.5 | Common ALL | 46,XY,t(10; 11)(p14-15; q22-23),del(12)(p11)/47,idem,+19* | NA | NA | 9 | H |
| 10 | W | 2.6 | 15.9 | Early pre-B | 46,XX,del(12)(p12) | R† | + | 78+ | |
| 11 | B | 12.8 | 16.1 | T-cell | 46,XY,t(10; 14)(q24; q11),del(12)(p12)* | G | NA | 89+ | |
| 12 | W | 4.6 | 128 | Early pre-B | 48,XY,+X,del(12)(p11),+21 | G† | − | 77+ | |
| 13 | W | 4 | 5 | Pre-B | 45,X,add(X)(p22),del(9)(p22),del(12)(p12),−13/45,idem,−del(12)* | R† | + | 72+ | |
| 14 | W | 18.2 | 50.3 | Pre-B | 46,XX,dup(1)(q21q?41),del(12)(p11) | G† | − | 0 | IF |
| 15 | W | 2.2 | 3.4 | Early pre-B | 46,XY,del(12)(p11) | R† | + | 53+ | |
| 16 | W | 13.2 | 12 | T-cell | 47,XY,+8,del(9)(p21),del(10)(q24q26),del(12)(p11) | G† | − | 17 | H + T |
| 17 | W | 9 | 21.1 | Early pre-B | 46,XX,der(6)t(4; 6)(q21; q21),del(12)(p11) | G† | − | 38 | H |
| 18 | W | 6.8 | 13.7 | Early pre-B | 45,X,−X,del(12)(p11) | R† | + | 20+ | |
| 19 | W | 2.9 | 18 | Pre-B | 46,XX,t(6; 19)(p21; q13),del(12)(p11) | R† | + | 50+ | |
| 20 | W | 5.1 | 1.9 | Early pre-B | 65,XX,+X,dup(2)(q21q35),+3,+4,+5,+6,+6,+7,+8,+9,+10,+10,+11, +del(12)(p11),+14,+15,+17,+18,+21,+21 | NA | NA | 20 | H |
| 21 | W | 3.2 | 38.7 | Pre-B | 46,XY,del(12)(p12)/46,XY,−C,+mar | R† | + | 40+ | |
| 22 | W | 13.4 | 2.3 | T-cell | 46,XX,t(5; 6)(q13; q23),del(7)(p15-p21),del(12)(p12) | G† | NA | 0 | IF |
| 23 | W | 2.8 | 94.8 | Pre-B | 46,XX,add(5)(q35),del(12)(p12) | R† | + | 30+ | |
| 24 | W | 3.3 | 8.9 | Pre-B | 46,XY,del(12)(p11) | R† | + | 19+ | |
| 25 | W | 6.8 | 1.2 | Early pre-B | 46,XX,del(11)(q22-23),del(12)(p12),der(21)t(1; 21)(q21; q22)/46,XX,t(3; 5)(p25; q13),der(21)t(1; 21) | G | NA | 16+ | |
| 26 | W | 16.5 | 6.9 | T-cell | 53,XY,+7,+8,+13,+18,+19,+19,+22/51,idem,−7,−19/51,idem,−7,del(12)(p11),−19/52,idem,−7,del(12)(p11)* | G† | NA | 13+ | |
| Inversion (n = 3) | |||||||||
| 27 | W | 16.5 | 3.4 | T-cell | 46,XX,t(11; 17)(p13; q21),inv(12)(p13q22)/46,XX,inv(12) | NA | NA | 28 | H |
| 28 | B | 4.6 | 357 | Early pre-B | 46,XY,del(3)(p23),del(7)(p13p15),inv(12)(p11p13) | NA | NA | 7 | H |
| 29 | W | 9.3 | 2.4 | Early pre-B | 46,X,add(Y)(q12),del(11)(q23),inv(12)(p13q12) | R | NA | 17+ | |
| Translocation-balanced simple (n = 20) | |||||||||
| 30 | W | 6.5 | 4.1 | Early pre-B | 47,XX,t(12; 17)(p12; q12),+21* | NA | NA | 11 | H |
| 31 | W | 9.9 | 13.8 | Early pre-B | 46,XX,?del(4)(q?25),t(5; 12)(q22; p13) | NA | NA | 134+ | |
| 32 | W | 2.3 | 20.7 | Early pre-B | 46,XY,t(12; 21)(p13; q11) | NA | NA | 151+ | |
| 33 | W | 5.5 | 79 | Common ALL | 46,XY,t(1; 12)(q25; p12)* | NA | NA | 136+ | |
| 34 | W | 2.3 | 41.4 | Pre-B | 46,XY,t(9; 12)(p21; p12)* | NA | NA | 24 | CNS |
| 35 | W | 11.5 | 2.2 | Early pre-B | 46,XY,t(3; 12)(q12; p12),del(6)(q15)/46,idem,−t(3; 12)* | NA | NA | 127+ | |
| 36 | W | 5.2 | 8.2 | Early pre-B | 47,XX,t(10; 12)(q22; p13),+21* | NA | NA | 126+ | |
| 37 | W | 4.6 | 39.2 | Early pre-B | 46,XX,t(8; 12)(p21; p13)* | R | NA | 115+ | |
| 38 | W | 7.7 | 7.5 | Technic. Inc | 47,XX,t(12; 14)(p11; q11),+21* | NA | NA | 101+ | |
| 39 | W | 2.4 | 3.3 | Early pre-B | 47,XY,+10/47,idem,t(7; 12)(q22; p12)* | R | NA | 102+ | |
| 40 | W | 6 | 7.8 | T-cell | 46,XY,add(1)(p36),del(2)(p22),del(8)(q11q22),del(9)(q11q22),del(11) (q22-23),add(11)(q23-24),t(12; 13)(p13; q14)/92,idem,del(6)(q16)* | NA | NA | 42+ | |
| 41 | B | 3.3 | 1.8 | Technic. Inc | 46,XY,t(11; 12)(q23; p13)* | NA | NA | 40 | Other |
| 42 | W | 7.1 | 3.4 | Early pre-B | 48,XY,t(6; 12)(q21; p13),+16,+add(19)(p13) | R | NA | 40 | Other |
| 43 | W | 4.8 | 7.9 | Early pre-B | 46,XY,t(12; 13)(p13; q14),add(19)(q13) | R | + | 92+ | |
| 44 | W | 3.8 | 9.3 | Early pre-B | 46,XX,t(9; 12)(q13; p12) | R† | + | 44 | H + CNS |
| 45 | W | 4.5 | 112.6 | Early pre-B | 46,XY,t(5; 12)(q31; p13) | G† | − | 21 | H |
| 46 | W | 2.9 | 11.6 | Early pre-B | 47,XY,t(8; 12)(p21; p13),+10 | R† | + | 33.1 | Died CR |
| 47 | W | 10 | 18.4 | Pre-B | 46,X,Y,t(2; 12)(q21; p13),der(11)t(3; 11)(p11; q23),+mar/47,idem,+X, add(1)(q32) | G | − | 19+ | |
| 48 | W | 3.4 | 5.3 | Early pre-B | 46,XY,t(12; 13)(p13; q14) | R† | + | 13+ | |
| 49 | W | 11 | 2.3 | T-cell | 47,XX,+4,t(10; 12)(q22; p13) | G | − | 0 | IF |
| Translocation-balanced complex (n = 3) | |||||||||
| 50 | W | 2.3 | 43.4 | Early pre-B | 47,XY,+X,t(7; 12; 14)(p15; p13; q22)/48,idem,+19* | G | − | 118+ | |
| 51 | W | 4.2 | 10.3 | Early pre-B | 46,XY,t(3; 12; 11)(q21; p13; q23),del(6)(p22) | R† | + | 65+ | |
| 52 | W | 2.8 | 7 | Early pre-B | 46,XX,t(12; 11)(11; 14)(p1?2; q14→q2?4; q22) | R† | − | 46+ | |
| Translocation-unbalanced: known origin (n = 6) | |||||||||
| 53 | W | 3.2 | 8.4 | Pre-B | 46,XY,der(12)t(1; 12)(q22-23; p13)* | NA | NA | 35.5 | 2° AML |
| 54 | W | 5 | 17 | Early pre-B | 49,XX,+X,der(12)t(2; 12)(q21; p13),+del(16)(q13)x2/49,idem,del(7)(p11)* | NA | NA | 6 | H |
| 55 | W | 2.6 | 18.2 | Pre-B | 45,XX,−4,der(12)t(4; 12)(q21; p13)* | NA | NA | 109+ | |
| 56 | B | 7.8 | 28.6 | Pre-B | 49,XY,+Y,6,+10,der(12)t(6; 12)(p21; p13),der(19)t(1; 19)(q23; p13),+21,+21/49,XY,+Y,+7,der(15)t(1; 15)(q11; p11),der(19)t(1; 19),+21/50,XY,+Y,+17,der(19)t(1; 19),+20,+21* | G | NA | 90+ | |
| 57 | W | 4.2 | 2 | Early pre-B | 46,XX,der(12)t(12; 17)(p13; q21),−17,+mar | G† | − | 34+ | |
| 58 | W | 1.6 | 186 | Early pre-B | 45,XX,del(1)(q32),−7,der(12)t(7; 12)(q11; p13) | G† | − | 34+ | |
| Translocation-unbalanced: unknown origin (n = 11) | |||||||||
| 59 | W | 12 | 22.4 | Early pre-B | 46,X,X,del(9)(p21),add(12)(p12),+16/46,idem,del(1)(q42)* | NA | NA | 134+ | |
| 60 | W | 7.8 | 40.4 | Early pre-B | 46,XY,add(12)(p12),add(19)(p13)/46,XX,?del(8)(p21),add(12)(p12)* | R | + | 119+ | |
| 61 | W | 2.7 | 11.8 | Pre-B | 54,XX,+6,+10,+11,+add(12)(p13),+14,+17,+21,+21* | NA | NA | 32 | H |
| 62 | W | 14.7 | 14.7 | Pre-B | 47,XY,add(12)(p13),−9,add(17)(p13),+2mar* | NA | NA | 25 | T |
| 63 | W | 15.5 | 3 | Pre-B | 94,XXYY,+del(2)(p21),+3,−7,add(12)(p13)x2,+mar* | NA | NA | 102+ | |
| 64 | W | 2.8 | 18.8 | Early pre-B | 46,XY,add(12)(p13) | G | NA | 89+ | |
| 65 | W | 6 | 18.5 | Early pre-B | 46,Y,−X,del(2)(q21q24),del(6)(q21q25),add(12)(p12),+mar* | R | + | 99.7 | H |
| 66 | W | 4.6 | 6.7 | Early pre-B | 46,XY,add(12)(p13),del(13)(q12q21-22) | NA | NA | 77+ | |
| 67 | W | 2.4 | 4.7 | Pre-B | 59,XY,+X,+4,+5,+6,+9,+10,+11,+14,+15,+17,+18,+21,+21/59,idem,add(12)(p13)* | G† | − | 46+ | |
| 68 | W | 13.5 | 2.3 | Pre-B | 46,XX,t(1; 9)(p36; q13),add(1)(p32),add(12)(p13),?del(14)(q?)/46,idem,−?del(14)* | G† | NA | 21 | CNS |
| 69 | W | 4 | 48.7 | Early pre-B | 48,XY,+10,add(12)(p13),+21 | R† | + | 26+ | |
| Dicentric dic(9; 12) (n = 8) | |||||||||
| 70 | B | 4.6 | 132 | Early pre-B | 45,XY,del(X)(q26),dic(9; 12)(p11; p12)* | NA | NA | Off Study | IF |
| 71 | W | 15.4 | 19.6 | Early pre-B | 45,XY,dic(9; 12)(p11; p12) | NA | NA | 101+ | |
| 72 | W | 5.9 | 12.7 | Early pre-B | 45,XY,dic(9; 12)(p11; p12)* | R | NA | 102+ | |
| 73 | W | 7 | 22.8 | Early pre-B | 45,XX,dic(9; 12)(p11; p12)* | R | + | 89+ | |
| 74 | W | 2.3 | 210 | Pre-B | 45,XX,dic(9; 12)(p11; p12)* | G† | + | 71+ | |
| 75 | W | 3.3 | 36.4 | Early pre-B | 46,XY,dic(9; 12)(p11; p12),+mar/45,idem,−mar | R† | + | 66+ | |
| 76 | W | 0.64 | 6 | Pre-B | 45,XY,dic(9; 12)(p11; p12) | R† | + | 36+ | |
| 77 | W | 14 | 3.6 | Pre-B | 45,XY,del(6)(q15q24),dic(9; 12)(p11; p12) | R | NA | 20+ | |
| Dicentric (7; 12) (n = 4) | |||||||||
| 78 | W | 2.3 | 30 | Early pre-B | 46,XY,del(6)(q14q23),dic(7; 12)(p11; p12),+mar/45,idem,−mar* | G† | NA | 28 | H |
| 79 | W | 2.6 | 107.2 | Early pre-B | 46,XY,del(1)(q32),dic(7; 12)(p11; p12),+21* | G | NA | 72 | CNS |
| 80 | B | 10.9 | 68.5 | T-cell | 46,XY,dic(7; 12)(p11; p12),+?18/45,XY,dic(7; 12)/90,XXYY,dic(7; 12)x2* | NA | NA | 70 | H |
| 81 | W | 8.7 | 4 | Pre-B | 47,XX,dic(7; 12)(p11; p12),+dic(7; 12),t(11; 15)(q23; q15-21),+20* | NA | NA | 89+ | |
| Dicentric-random (n = 3) | |||||||||
| 82 | W | 5.7 | 2.1 | Pre-B | 45,XX,t(2; 14)(q24q31; q32),dic(12; 15)(p11; p11)* | NA | NA | 102+ | |
| 83 | W | 4.1 | 5.2 | T-cell | 45,XX,t(1; 2)(p22; p14),t(7; 14)(p15; q32),dic(12; 18)(p11; p11)* | G | NA | 12 | H |
| 84 | W | 14.8 | 85.7 | T-cell | 45,XX,inv(5)(p13q14),del(10)(q22),del(11)(q22-23),dic(12; 17)(p11; p11)* | G | − | 0 | IF |
| BOTH HOMOLOGS 12P ABNORMAL (n = 10) | |||||||||
| 85 | W | 4 | 14 | Pre-B | 47,XX,inv(12)(p13q22),del(12)(p12),del(13)(q12q32),+21/46,XX,inv(12)* | G† | NA | 72+ | |
| 86 | W | 1.8 | 80.6 | Pre-B | 46,XY,del(12)(p11),add(12)(p12) | NA | NA | 65+ | |
| 87 | W | 5.5 | 19.7 | Early pre-B | 46,XY,inv(12)(p13q22),add(21)(q22)/46,idem,t(6; 12)(q21; p13)/46,XX,t(6; 12),inv(12) | R | NA | 106+ | |
| 88 | W | 2.2 | 16.1 | Early pre-B | 46,XY,t(1; 12)(q22-23; p13),add(1)(p36),−7,dic(12; 16)(p11; p13),t(17; 19) (q22; p13),+21,+add(22)(q13)* | G | NA | 111+ | |
| 89 | W | 4.4 | 46.8 | Early pre-B | 46,XX,t(12; 15)(p13; q21)/46,XX,t(7; 12)(p13; p13) | R | NA | 94+ | |
| 90 | B | 6.1 | 15.1 | Early pre-B | 47,XX,t(1; 12)(p22; p13),t(4; 12)(q21; p13),+10,del(11)(q23)/45,X,−X,t(4; 12) | R† | + | 46+ | |
| 91 | W | 13.7 | 3.3 | Early pre-B | 66,XX,+add(X)(p22),+1,+1,+add(2)(p22)x2,add(3)(q21),+add(3)(p13), +4,+4,+6,+6,+8,+9,+10,+11,+i(12)(q10),+add(12)(p11),+14,+18, +22,+mar | G | NA | 16.1 | 2° AML |
| 92 | W | 2.3 | 19.5 | Pre-B | 45,XX,t(3; 12)(p13; p13),dic(12; 22)(p11; p11) | R† | + | 12+ | |
| 93 | W | 3.4 | 61.6 | Pre-B | 46,XY,t(1; 12)(q22-23; p13),del(6)(q14q21),t(12; 17)(p11; p13)/47,idem,+X | R† | + | 6+ | |
| 94 | W | 4.9 | 22.9 | Early pre-B | 46,XY,dup(3)(p23p25),add(11)(q23)/46,idem,del(12)(p12)/45,XY,t(8; 17)(p21; p13),dic(12; 15)(p11; p11) | R† | + | 15+ | |
| Patient No. . | Race . | Age . | WBC . | Immuno- . | Karyotype . | ETV6 Status . | ETV6/CBFA2 . | Remission Duration (mo) . | Type of Failure . |
|---|---|---|---|---|---|---|---|---|---|
| . | . | (yr) . | (×109/L) . | phenotype . | . | . | . | . | . |
| ONE 12P HOMOLOG ABNORMAL | |||||||||
| Deletion (n = 26) | |||||||||
| 1 | W | 4.1 | 5.7 | Pre-B | 46,XX,del(12)(p12),−13,+mar/47,idem,+mar* | NA | NA | 142+ | |
| 2 | W | 8.3 | 154.8 | Early pre-B | 46,XX,del(9)(p21),del(12)(p12)* | NA | NA | 5 | CNS |
| 3 | W | 5.6 | 8.9 | Pre-B | 46,XY,del(8)(q21.2q21.3),del(9)(p22),del(12)(p12),add(16)(q24)/46,idem,−del(12)* | R | NA | 126+ | |
| 4 | B | 3.3 | 11.3 | Early pre-B | 46,XX,del(12)(p11)/46,XX,t(7; 12)(p22; q12)* | R | NA | 125+ | |
| 5 | W | 2.2 | 34.3 | Early pre-B | 47,XX,del(12)(p12),+16/47,idem,del(13)(q12q21-22)* | G | NA | 81+ | |
| 6 | W | 4.3 | 2.1 | Early pre-B | 46,XY,del(12)(p12)* | R | NA | 116+ | |
| 7 | W | 13.1 | 73.7 | Early pre-B | 46,XX,del(12)(p12),−22,+der(?)t(?; 22)(?; q11)/46,XX,?del(8)(p22), del(12)/47,XX,del(12),+21 | R | + | 99+ | |
| 8 | W | 10.1 | 4.7 | Early pre-B | 53,XX,+5,+6,+del(6)(q21q23),+del(12)(p12),+18,+21,+21* | G | NA | 72+ | |
| 9 | B | 7.6 | 24.5 | Common ALL | 46,XY,t(10; 11)(p14-15; q22-23),del(12)(p11)/47,idem,+19* | NA | NA | 9 | H |
| 10 | W | 2.6 | 15.9 | Early pre-B | 46,XX,del(12)(p12) | R† | + | 78+ | |
| 11 | B | 12.8 | 16.1 | T-cell | 46,XY,t(10; 14)(q24; q11),del(12)(p12)* | G | NA | 89+ | |
| 12 | W | 4.6 | 128 | Early pre-B | 48,XY,+X,del(12)(p11),+21 | G† | − | 77+ | |
| 13 | W | 4 | 5 | Pre-B | 45,X,add(X)(p22),del(9)(p22),del(12)(p12),−13/45,idem,−del(12)* | R† | + | 72+ | |
| 14 | W | 18.2 | 50.3 | Pre-B | 46,XX,dup(1)(q21q?41),del(12)(p11) | G† | − | 0 | IF |
| 15 | W | 2.2 | 3.4 | Early pre-B | 46,XY,del(12)(p11) | R† | + | 53+ | |
| 16 | W | 13.2 | 12 | T-cell | 47,XY,+8,del(9)(p21),del(10)(q24q26),del(12)(p11) | G† | − | 17 | H + T |
| 17 | W | 9 | 21.1 | Early pre-B | 46,XX,der(6)t(4; 6)(q21; q21),del(12)(p11) | G† | − | 38 | H |
| 18 | W | 6.8 | 13.7 | Early pre-B | 45,X,−X,del(12)(p11) | R† | + | 20+ | |
| 19 | W | 2.9 | 18 | Pre-B | 46,XX,t(6; 19)(p21; q13),del(12)(p11) | R† | + | 50+ | |
| 20 | W | 5.1 | 1.9 | Early pre-B | 65,XX,+X,dup(2)(q21q35),+3,+4,+5,+6,+6,+7,+8,+9,+10,+10,+11, +del(12)(p11),+14,+15,+17,+18,+21,+21 | NA | NA | 20 | H |
| 21 | W | 3.2 | 38.7 | Pre-B | 46,XY,del(12)(p12)/46,XY,−C,+mar | R† | + | 40+ | |
| 22 | W | 13.4 | 2.3 | T-cell | 46,XX,t(5; 6)(q13; q23),del(7)(p15-p21),del(12)(p12) | G† | NA | 0 | IF |
| 23 | W | 2.8 | 94.8 | Pre-B | 46,XX,add(5)(q35),del(12)(p12) | R† | + | 30+ | |
| 24 | W | 3.3 | 8.9 | Pre-B | 46,XY,del(12)(p11) | R† | + | 19+ | |
| 25 | W | 6.8 | 1.2 | Early pre-B | 46,XX,del(11)(q22-23),del(12)(p12),der(21)t(1; 21)(q21; q22)/46,XX,t(3; 5)(p25; q13),der(21)t(1; 21) | G | NA | 16+ | |
| 26 | W | 16.5 | 6.9 | T-cell | 53,XY,+7,+8,+13,+18,+19,+19,+22/51,idem,−7,−19/51,idem,−7,del(12)(p11),−19/52,idem,−7,del(12)(p11)* | G† | NA | 13+ | |
| Inversion (n = 3) | |||||||||
| 27 | W | 16.5 | 3.4 | T-cell | 46,XX,t(11; 17)(p13; q21),inv(12)(p13q22)/46,XX,inv(12) | NA | NA | 28 | H |
| 28 | B | 4.6 | 357 | Early pre-B | 46,XY,del(3)(p23),del(7)(p13p15),inv(12)(p11p13) | NA | NA | 7 | H |
| 29 | W | 9.3 | 2.4 | Early pre-B | 46,X,add(Y)(q12),del(11)(q23),inv(12)(p13q12) | R | NA | 17+ | |
| Translocation-balanced simple (n = 20) | |||||||||
| 30 | W | 6.5 | 4.1 | Early pre-B | 47,XX,t(12; 17)(p12; q12),+21* | NA | NA | 11 | H |
| 31 | W | 9.9 | 13.8 | Early pre-B | 46,XX,?del(4)(q?25),t(5; 12)(q22; p13) | NA | NA | 134+ | |
| 32 | W | 2.3 | 20.7 | Early pre-B | 46,XY,t(12; 21)(p13; q11) | NA | NA | 151+ | |
| 33 | W | 5.5 | 79 | Common ALL | 46,XY,t(1; 12)(q25; p12)* | NA | NA | 136+ | |
| 34 | W | 2.3 | 41.4 | Pre-B | 46,XY,t(9; 12)(p21; p12)* | NA | NA | 24 | CNS |
| 35 | W | 11.5 | 2.2 | Early pre-B | 46,XY,t(3; 12)(q12; p12),del(6)(q15)/46,idem,−t(3; 12)* | NA | NA | 127+ | |
| 36 | W | 5.2 | 8.2 | Early pre-B | 47,XX,t(10; 12)(q22; p13),+21* | NA | NA | 126+ | |
| 37 | W | 4.6 | 39.2 | Early pre-B | 46,XX,t(8; 12)(p21; p13)* | R | NA | 115+ | |
| 38 | W | 7.7 | 7.5 | Technic. Inc | 47,XX,t(12; 14)(p11; q11),+21* | NA | NA | 101+ | |
| 39 | W | 2.4 | 3.3 | Early pre-B | 47,XY,+10/47,idem,t(7; 12)(q22; p12)* | R | NA | 102+ | |
| 40 | W | 6 | 7.8 | T-cell | 46,XY,add(1)(p36),del(2)(p22),del(8)(q11q22),del(9)(q11q22),del(11) (q22-23),add(11)(q23-24),t(12; 13)(p13; q14)/92,idem,del(6)(q16)* | NA | NA | 42+ | |
| 41 | B | 3.3 | 1.8 | Technic. Inc | 46,XY,t(11; 12)(q23; p13)* | NA | NA | 40 | Other |
| 42 | W | 7.1 | 3.4 | Early pre-B | 48,XY,t(6; 12)(q21; p13),+16,+add(19)(p13) | R | NA | 40 | Other |
| 43 | W | 4.8 | 7.9 | Early pre-B | 46,XY,t(12; 13)(p13; q14),add(19)(q13) | R | + | 92+ | |
| 44 | W | 3.8 | 9.3 | Early pre-B | 46,XX,t(9; 12)(q13; p12) | R† | + | 44 | H + CNS |
| 45 | W | 4.5 | 112.6 | Early pre-B | 46,XY,t(5; 12)(q31; p13) | G† | − | 21 | H |
| 46 | W | 2.9 | 11.6 | Early pre-B | 47,XY,t(8; 12)(p21; p13),+10 | R† | + | 33.1 | Died CR |
| 47 | W | 10 | 18.4 | Pre-B | 46,X,Y,t(2; 12)(q21; p13),der(11)t(3; 11)(p11; q23),+mar/47,idem,+X, add(1)(q32) | G | − | 19+ | |
| 48 | W | 3.4 | 5.3 | Early pre-B | 46,XY,t(12; 13)(p13; q14) | R† | + | 13+ | |
| 49 | W | 11 | 2.3 | T-cell | 47,XX,+4,t(10; 12)(q22; p13) | G | − | 0 | IF |
| Translocation-balanced complex (n = 3) | |||||||||
| 50 | W | 2.3 | 43.4 | Early pre-B | 47,XY,+X,t(7; 12; 14)(p15; p13; q22)/48,idem,+19* | G | − | 118+ | |
| 51 | W | 4.2 | 10.3 | Early pre-B | 46,XY,t(3; 12; 11)(q21; p13; q23),del(6)(p22) | R† | + | 65+ | |
| 52 | W | 2.8 | 7 | Early pre-B | 46,XX,t(12; 11)(11; 14)(p1?2; q14→q2?4; q22) | R† | − | 46+ | |
| Translocation-unbalanced: known origin (n = 6) | |||||||||
| 53 | W | 3.2 | 8.4 | Pre-B | 46,XY,der(12)t(1; 12)(q22-23; p13)* | NA | NA | 35.5 | 2° AML |
| 54 | W | 5 | 17 | Early pre-B | 49,XX,+X,der(12)t(2; 12)(q21; p13),+del(16)(q13)x2/49,idem,del(7)(p11)* | NA | NA | 6 | H |
| 55 | W | 2.6 | 18.2 | Pre-B | 45,XX,−4,der(12)t(4; 12)(q21; p13)* | NA | NA | 109+ | |
| 56 | B | 7.8 | 28.6 | Pre-B | 49,XY,+Y,6,+10,der(12)t(6; 12)(p21; p13),der(19)t(1; 19)(q23; p13),+21,+21/49,XY,+Y,+7,der(15)t(1; 15)(q11; p11),der(19)t(1; 19),+21/50,XY,+Y,+17,der(19)t(1; 19),+20,+21* | G | NA | 90+ | |
| 57 | W | 4.2 | 2 | Early pre-B | 46,XX,der(12)t(12; 17)(p13; q21),−17,+mar | G† | − | 34+ | |
| 58 | W | 1.6 | 186 | Early pre-B | 45,XX,del(1)(q32),−7,der(12)t(7; 12)(q11; p13) | G† | − | 34+ | |
| Translocation-unbalanced: unknown origin (n = 11) | |||||||||
| 59 | W | 12 | 22.4 | Early pre-B | 46,X,X,del(9)(p21),add(12)(p12),+16/46,idem,del(1)(q42)* | NA | NA | 134+ | |
| 60 | W | 7.8 | 40.4 | Early pre-B | 46,XY,add(12)(p12),add(19)(p13)/46,XX,?del(8)(p21),add(12)(p12)* | R | + | 119+ | |
| 61 | W | 2.7 | 11.8 | Pre-B | 54,XX,+6,+10,+11,+add(12)(p13),+14,+17,+21,+21* | NA | NA | 32 | H |
| 62 | W | 14.7 | 14.7 | Pre-B | 47,XY,add(12)(p13),−9,add(17)(p13),+2mar* | NA | NA | 25 | T |
| 63 | W | 15.5 | 3 | Pre-B | 94,XXYY,+del(2)(p21),+3,−7,add(12)(p13)x2,+mar* | NA | NA | 102+ | |
| 64 | W | 2.8 | 18.8 | Early pre-B | 46,XY,add(12)(p13) | G | NA | 89+ | |
| 65 | W | 6 | 18.5 | Early pre-B | 46,Y,−X,del(2)(q21q24),del(6)(q21q25),add(12)(p12),+mar* | R | + | 99.7 | H |
| 66 | W | 4.6 | 6.7 | Early pre-B | 46,XY,add(12)(p13),del(13)(q12q21-22) | NA | NA | 77+ | |
| 67 | W | 2.4 | 4.7 | Pre-B | 59,XY,+X,+4,+5,+6,+9,+10,+11,+14,+15,+17,+18,+21,+21/59,idem,add(12)(p13)* | G† | − | 46+ | |
| 68 | W | 13.5 | 2.3 | Pre-B | 46,XX,t(1; 9)(p36; q13),add(1)(p32),add(12)(p13),?del(14)(q?)/46,idem,−?del(14)* | G† | NA | 21 | CNS |
| 69 | W | 4 | 48.7 | Early pre-B | 48,XY,+10,add(12)(p13),+21 | R† | + | 26+ | |
| Dicentric dic(9; 12) (n = 8) | |||||||||
| 70 | B | 4.6 | 132 | Early pre-B | 45,XY,del(X)(q26),dic(9; 12)(p11; p12)* | NA | NA | Off Study | IF |
| 71 | W | 15.4 | 19.6 | Early pre-B | 45,XY,dic(9; 12)(p11; p12) | NA | NA | 101+ | |
| 72 | W | 5.9 | 12.7 | Early pre-B | 45,XY,dic(9; 12)(p11; p12)* | R | NA | 102+ | |
| 73 | W | 7 | 22.8 | Early pre-B | 45,XX,dic(9; 12)(p11; p12)* | R | + | 89+ | |
| 74 | W | 2.3 | 210 | Pre-B | 45,XX,dic(9; 12)(p11; p12)* | G† | + | 71+ | |
| 75 | W | 3.3 | 36.4 | Early pre-B | 46,XY,dic(9; 12)(p11; p12),+mar/45,idem,−mar | R† | + | 66+ | |
| 76 | W | 0.64 | 6 | Pre-B | 45,XY,dic(9; 12)(p11; p12) | R† | + | 36+ | |
| 77 | W | 14 | 3.6 | Pre-B | 45,XY,del(6)(q15q24),dic(9; 12)(p11; p12) | R | NA | 20+ | |
| Dicentric (7; 12) (n = 4) | |||||||||
| 78 | W | 2.3 | 30 | Early pre-B | 46,XY,del(6)(q14q23),dic(7; 12)(p11; p12),+mar/45,idem,−mar* | G† | NA | 28 | H |
| 79 | W | 2.6 | 107.2 | Early pre-B | 46,XY,del(1)(q32),dic(7; 12)(p11; p12),+21* | G | NA | 72 | CNS |
| 80 | B | 10.9 | 68.5 | T-cell | 46,XY,dic(7; 12)(p11; p12),+?18/45,XY,dic(7; 12)/90,XXYY,dic(7; 12)x2* | NA | NA | 70 | H |
| 81 | W | 8.7 | 4 | Pre-B | 47,XX,dic(7; 12)(p11; p12),+dic(7; 12),t(11; 15)(q23; q15-21),+20* | NA | NA | 89+ | |
| Dicentric-random (n = 3) | |||||||||
| 82 | W | 5.7 | 2.1 | Pre-B | 45,XX,t(2; 14)(q24q31; q32),dic(12; 15)(p11; p11)* | NA | NA | 102+ | |
| 83 | W | 4.1 | 5.2 | T-cell | 45,XX,t(1; 2)(p22; p14),t(7; 14)(p15; q32),dic(12; 18)(p11; p11)* | G | NA | 12 | H |
| 84 | W | 14.8 | 85.7 | T-cell | 45,XX,inv(5)(p13q14),del(10)(q22),del(11)(q22-23),dic(12; 17)(p11; p11)* | G | − | 0 | IF |
| BOTH HOMOLOGS 12P ABNORMAL (n = 10) | |||||||||
| 85 | W | 4 | 14 | Pre-B | 47,XX,inv(12)(p13q22),del(12)(p12),del(13)(q12q32),+21/46,XX,inv(12)* | G† | NA | 72+ | |
| 86 | W | 1.8 | 80.6 | Pre-B | 46,XY,del(12)(p11),add(12)(p12) | NA | NA | 65+ | |
| 87 | W | 5.5 | 19.7 | Early pre-B | 46,XY,inv(12)(p13q22),add(21)(q22)/46,idem,t(6; 12)(q21; p13)/46,XX,t(6; 12),inv(12) | R | NA | 106+ | |
| 88 | W | 2.2 | 16.1 | Early pre-B | 46,XY,t(1; 12)(q22-23; p13),add(1)(p36),−7,dic(12; 16)(p11; p13),t(17; 19) (q22; p13),+21,+add(22)(q13)* | G | NA | 111+ | |
| 89 | W | 4.4 | 46.8 | Early pre-B | 46,XX,t(12; 15)(p13; q21)/46,XX,t(7; 12)(p13; p13) | R | NA | 94+ | |
| 90 | B | 6.1 | 15.1 | Early pre-B | 47,XX,t(1; 12)(p22; p13),t(4; 12)(q21; p13),+10,del(11)(q23)/45,X,−X,t(4; 12) | R† | + | 46+ | |
| 91 | W | 13.7 | 3.3 | Early pre-B | 66,XX,+add(X)(p22),+1,+1,+add(2)(p22)x2,add(3)(q21),+add(3)(p13), +4,+4,+6,+6,+8,+9,+10,+11,+i(12)(q10),+add(12)(p11),+14,+18, +22,+mar | G | NA | 16.1 | 2° AML |
| 92 | W | 2.3 | 19.5 | Pre-B | 45,XX,t(3; 12)(p13; p13),dic(12; 22)(p11; p11) | R† | + | 12+ | |
| 93 | W | 3.4 | 61.6 | Pre-B | 46,XY,t(1; 12)(q22-23; p13),del(6)(q14q21),t(12; 17)(p11; p13)/47,idem,+X | R† | + | 6+ | |
| 94 | W | 4.9 | 22.9 | Early pre-B | 46,XY,dup(3)(p23p25),add(11)(q23)/46,idem,del(12)(p12)/45,XY,t(8; 17)(p21; p13),dic(12; 15)(p11; p11) | R† | + | 15+ | |
Abbreviations: W, white; B, black; R, rearranged; G, germline; NA, not applicable; H, hematologic; CNS, central nervous system; IF, induction failure; AML, acute myelogenous leukemia; T, testicular.
Karyotypes previously reported.
Molecular analyses previously reported.
The 12p region was affected in one homolog in 84 cases (Table 1), with abnormalities including deletion (n = 26), inversion (n = 3), balanced translocation (n = 20), complex translocation (n = 3), unbalanced translocation with exchanged DNA material of known (n = 6) or unknown (n = 11) partner chromosomes, dic(9; 12) (n = 8), dic(7; 12) (n = 4), and random dicentric (n = 3). In the 10 cases with both 12p homologs affected, there were 14 translocations, 3 deletions, 2 inversions, and 1 isochromosome of the 12q.
Overall, 67 (64%) of the 104 12p abnormalities had loss of 12p resulting from deletion, unbalanced translocation, dicentric, or isochromosome of 12q. When all 104 12p abnormalities were examined, the recurrent aberrations included: del(12)(p11) in 13 cases; del(12)(p12) in 16; dic(9; 12) (p11; p12) in 8; dic(7; 12)(p11; p12) in 4; t(1; 12)(q22∼23; p12) and t(12; 13)(p13; q14) in 3 cases each; and t(2; 12)(q21; p13), t(4; 12)(q21; p13), t(8; 12)(p21; p13), and t(10; 12)(q22; p13) in 2 cases each.
Detection of ETV6 Rearrangements and ETV6-CBFA2 Fusion
ETV6 rearrangements were detected by Southern blot analysis in 36 (56%) of the 64 cases with 12p abnormalities for which frozen cells were available for testing. Of the 38 available cases evaluated by RT-PCR, 25 (66%) expressed the ETV6-CBFA2 chimeric transcript. Tables 1 and 2 list the frequency and correlation of ETV6 gene rearrangements and ETV6-CBFA2 fusion transcripts among the subgroups of 12p abnormalities. ETV6-CBFA2 chimeric transcript was undetected in only one (No. 52) of the 25 cases with ETV6 rearrangement that were tested. Notably, 12 of the 25 tested cases with an assigned breakpoint at 12p13 showed no ETV6 rearrangements.
ETV6 Gene Rearrangement as Detected by Southern Blotting and ETV6-CBFA2 as Detected by RT-PCR in Acute Lymphoblastic Leukemia Cases with 12p Abnormalities
| Cytogenetic Abnormality . | ETV6 Rearrangement . | ETV6-CBFA2 Transcript . | ||
|---|---|---|---|---|
| . | Total Evaluated . | No. Rearranged . | Total Evaluated . | No. Positive . |
| One homolog | ||||
| Deletion | 22 | 12 | 13 | 9 |
| Inversion | 1 | 1 | NA | NA |
| Translocation balanced | ||||
| Simple | 10 | 7 | 7 | 4 |
| Complex | 3 | 2 | 3 | 1 |
| Translocation unbalanced | ||||
| Known origin | 3 | 0 | 2 | 0 |
| Unknown origin | 6 | 3 | 4 | 3 |
| Dicentric | ||||
| dic(9; 12) | 6 | 5 | 4 | 4 |
| dic(7; 12) | 2 | 0 | NA | NA |
| dic(random) | 2 | 0 | 1 | 0 |
| Two homologs | ||||
| t/del/inv/i | 9 | 6 | 4 | 4 |
| Total | 64 | 36 (56%) | 38 | 25 (66%) |
| No. of treatment failures | 16 | 4 | 9 | 3 |
| Cytogenetic Abnormality . | ETV6 Rearrangement . | ETV6-CBFA2 Transcript . | ||
|---|---|---|---|---|
| . | Total Evaluated . | No. Rearranged . | Total Evaluated . | No. Positive . |
| One homolog | ||||
| Deletion | 22 | 12 | 13 | 9 |
| Inversion | 1 | 1 | NA | NA |
| Translocation balanced | ||||
| Simple | 10 | 7 | 7 | 4 |
| Complex | 3 | 2 | 3 | 1 |
| Translocation unbalanced | ||||
| Known origin | 3 | 0 | 2 | 0 |
| Unknown origin | 6 | 3 | 4 | 3 |
| Dicentric | ||||
| dic(9; 12) | 6 | 5 | 4 | 4 |
| dic(7; 12) | 2 | 0 | NA | NA |
| dic(random) | 2 | 0 | 1 | 0 |
| Two homologs | ||||
| t/del/inv/i | 9 | 6 | 4 | 4 |
| Total | 64 | 36 (56%) | 38 | 25 (66%) |
| No. of treatment failures | 16 | 4 | 9 | 3 |
Abbreviations: NA, not available; t, translocation; del, deletion; inv, inversion; i, isochromosome.
One 12p homolog abnormal. As shown in Table 2, cases with one abnormal 12p homolog included 12 with ETV6 gene rearrangement out of 22 that had cytogenetically visible 12p deletions (54.5%). Nine of the 13 cases (69%) evaluated by RT-PCR expressed the ETV6-CBFA2 fusion transcript. Likewise, 54.5% (12 out of 22) of all the translocations had ETV6 rearranged, and 50% (8 out of 16) were positive for the ETV6-CBFA2 fusion transcript. Only 1 case (patient 52, with a complex translocation) had ETV6 rearranged but was negative for the ETV6-CBFA2 fusion transcript. Patient 56, who had the t(1; 19) [E2A-PBX1 fusion], had normal ETV6 genes, but no material was available for RT-PCR evaluation. Among cases with a dic(9; 12), 5 of 6 tested had the ETV6 locus rearranged, and all 4 evaluated had an ETV6-CBFA2 fusion transcript. However, the single dic(9; 12) was the only case (No. 74) in this study in which we did not detect an ETV6 rearrangement, although it was positive for the ETV6-CBFA2 fusion transcript.
Two homologs abnormal. Of 9 cases that had two abnormal homologs (Table 2), a rearranged ETV6 was found in 6, of which all 4 tested had an ETV6-CBFA2 fusion transcript.
FISH
All 11 cases with germline ETV6 as well as 4 normal controls showed two red/green signals by FISH, indicating the lack of ETV6-CBFA2 fusion. Notably, case No. 74, who was shown to be positive for the fusion transcript by RT-PCR but lacked rearrangement by Southern, was among the negative cases by FISH analysis. Three positive controls (Nos. 42, 46, and 69) showed the ETV6 rearrangement by all three methods (Southern, RT-PCR, and FISH [split signals]).
Clinical Characteristics and Outcome
In the 94 children with 12p abnormalities, the median age was 4.7 years (range, 0.64 to 18.2 years) and the median initial leukocyte count was 14.3 × 109/L (range, 1.2 to 357 × 109/L). The only infant studied (patient 76) presented with a dic(9; 12), a 6 × 109/L leukocyte count, a pre-B phenotype, and ETV6-CBFA2 fusion transcript; this patient is alive, 36 months postdiagnosis.
According to the NCI risk classification for B-lineage ALL,37 57 cases with abnormal 12p had a standard risk (age ≥ 1 and ≤ 10, leukocyte count ≤ 50 × 109/L) and 25 cases had a worse risk. Two cases had incomplete phenotypic data and were not classified, and the remaining 10 patients had a T-cell immunophenotype. No differences were identified in age, leukocyte count, or immunophenotype among the different 12p cytogenetic subgroups (data not shown). The 5-year event-free survival rate of the 94 cases with a 12p abnormality did not differ from that of the 557 patients without 12p abnormalities (70% ± 5% v 64% ± 2%, respectively; P = .64). The 12p subgroups were too small for meaningful statistical comparisons, but patients with a dic(9; 12) or involvement of both 12p homologs had better outcomes (5-year event-free survival: 88% ± 13% and 90% ± 12%, respectively). Among the 30 patients who had an adverse event, 18 had low-risk and 12 had high-risk or T-cell ALL. Five of the failures were CNS relapses.
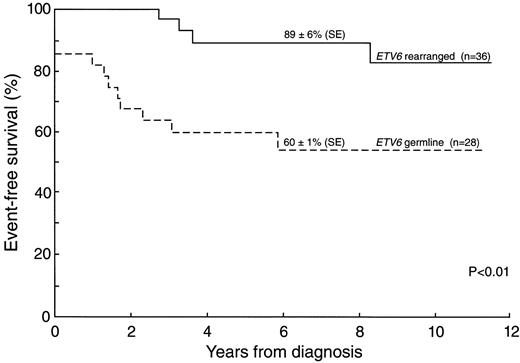
Table 3 lists presenting features of cases according to ETV6 status. Although no significant differences were found in the distribution of leukocyte count, race, sex, or ploidy, patients with ETV6 gene rearrangement were more likely to be between and including ages 1 and 10 years and have a B-lineage immunophenotype (early pre-B or pre-B); all 7 cases with a T-cell immunophenotype had a germline ETV6. Importantly, as shown in Fig 1, patients with ETV6 rearrangements had a better prognosis than those without the rearrangement (5-year event-free survival: 89% ± 6% v 60% ± 1%, respectively, P < .01). Furthermore, the event-free survival data was further analyzed according to ETV6-CBFA2 fusion, instead of ETV6 rearrangements, to evaluate which one may be relevant to outcome. We have performed the exact log-rank test stratified by study, which indicates no significant difference value (P = .13) according to the presence or absence of ETV6-CBFA2 fusion. However, this analysis should be interpreted with caution because only about half of the patients evaluated for ETV6 status by Southern had material available for the RT-PCR analysis, which limited the statistical power.
Presenting Features and ETV6 Gene Status of 64 Patients With ALL and 12p Cytogenetic Abnormalities
| Feature . | Rearranged ETV6 . | Germline ETV6 . | P Value3-150 . |
|---|---|---|---|
| . | (n = 36) . | (n = 28) . | . |
| Age (yr) | |||
| ≥1-≤10 | 33 | 17 | <0.01 |
| Other | 3 | 11 | |
| WBC count (×109/L) | |||
| ≤50 | 33 | 21 | 0.07 |
| >50 | 3 | 7 | |
| Race | |||
| White | 34 | 26 | 0.79 |
| Other | 2 | 2 | |
| Sex | |||
| Male | 22 | 14 | 0.24 |
| Female | 14 | 14 | |
| Immunophenotype | |||
| Non-T | 36 | 21 | <0.01 |
| T-lineage | 0 | 7 | |
| Ploidy | |||
| Hypodiploid | 7 | 4 | 0.06 |
| Pseudodiploid | 24 | 13 | |
| Hyperdiploid (47-50) | 5 | 7 | |
| Hyperdiploid (>50) | 0 | 4 |
| Feature . | Rearranged ETV6 . | Germline ETV6 . | P Value3-150 . |
|---|---|---|---|
| . | (n = 36) . | (n = 28) . | . |
| Age (yr) | |||
| ≥1-≤10 | 33 | 17 | <0.01 |
| Other | 3 | 11 | |
| WBC count (×109/L) | |||
| ≤50 | 33 | 21 | 0.07 |
| >50 | 3 | 7 | |
| Race | |||
| White | 34 | 26 | 0.79 |
| Other | 2 | 2 | |
| Sex | |||
| Male | 22 | 14 | 0.24 |
| Female | 14 | 14 | |
| Immunophenotype | |||
| Non-T | 36 | 21 | <0.01 |
| T-lineage | 0 | 7 | |
| Ploidy | |||
| Hypodiploid | 7 | 4 | 0.06 |
| Pseudodiploid | 24 | 13 | |
| Hyperdiploid (47-50) | 5 | 7 | |
| Hyperdiploid (>50) | 0 | 4 |
Abbreviation: WBC, white blood cell.
P values obtained by Fisher exact test.
Kaplan-Meier estimates of 5-year event-free survival for ALL patients with 12p abnormalities comparing blast cells showing rearranged or germline ETV6 gene.
Kaplan-Meier estimates of 5-year event-free survival for ALL patients with 12p abnormalities comparing blast cells showing rearranged or germline ETV6 gene.
DISCUSSION
Our pediatric ALL population had an 11.5% rate of leukemic cell 12p rearrangement by conventional cytogenetics. The frequency and heterogeneity of these chromosomal abnormalities suggest that the 12p region is subjected to diverse genetic alterations that contribute to the leukemogenic process in childhood ALL. In the 94 cases that had 12p changes, a total of 104 structural chromosomal abnormalities were observed in the 12p11-p13 region, including translocations (n = 69), deletions (n = 29), inversions (n = 5), and isochromosome 12q (n = 1). The 69 translocations involved 20 different reciprocal partner chromosomes and 43 distinct breakpoints. The most frequent exchanges involved 9p11 (n = 8); 7p11 (n = 4); 1q22-23 and 13q14 (n = 3); and 2q21, 4q21, 8p21, and 10q22 (n = 2 each). The sex chromosomes and chromosomes 19 and 20 were not involved.
A cryptic t(12; 21)(p13; q22) that is rarely observed cytogenetically has recently been a focus of interest in ALL.19-23 Molecular studies have shown that the ETS-family gene (ETV6 ) (12p13) is fused to the CBFA2 gene (21q22) encoding the alpha 2 subunit of the core binding factor.19 The chimeric ETV6-CBFA2 gene is present in 20% to 25% of childhood B-lineage ALL cases.20-23 Romana et al20 identified ETV6 gene rearrangements in 8 of 46 children diagnosed with ALL, but only 1 case had a 12p deletion visible by light microscopy. In all 8 cases, the abnormality was further defined as a t(12; 21) by FISH or RT-PCR. In a previous study, we described ETV6 rearrangements and ETV6-CBFA2 transcripts in 24% and 22%, respectively, of 160 cases of childhood B-precursor ALL, which included 18 cases with a cytogenetic 12p aberration.22 Our most recent study found 48 (25.5%) ETV6 rearrangements in 188 patients with B-lineage leukemia; 22 patients had a 12p abnormality.28 We have observed in an earlier study that polymorphisms of ETV6 do not contribute to the high frequency of detected ETV6 abnormalities.22 In that study, 19 nonleukemic control individuals and 50 acute myeloid leukemia (AML) cases did not show any cases with ETV6 rearrangements with the restriction endonuclease/probe combination used. In the present study, we showed that the subgroup with a cytogenetically determined 12p abnormality has an even higher frequency of ETV6 rearrangement or ETV6-CBFA2 chimeric transcript than previously reported. Overall, these molecular abnormalities were detected in 56% and 66%, respectively, of the cases tested. The frequency of the molecular abnormality did not appear to differ among the subtypes of 12p structural abnormalities, except for a higher frequency in cases with a dic(9; 12) or two abnormal homologs.
In general, the FISH results were in agreement with the results obtained by Southern. All the cases with the germline ETV6 by Southern showed normal ETV6 constitution by FISH. However, a discrepancy in the results of RT-PCR versus Southern and FISH was observed in one case (No. 74), and will require other molecular techniques to resolve.
The precision of cytogenetic breakpoint assignments in 12p is limited. In this study, cases with a break at p13, where the ETV6 locus has been mapped, did not have a higher frequency of the evaluated genetic abnormalities than did cases with other 12p breakpoints. Few other cases have been reported to have a cytogenetically abnormal 12p that did not reveal a t(12; 21).38,39 Thus, other gene(s) critical to leukemogenesis may be affected in this region. Until recently, CDKN1B (cyclin-dependent kinase inhibitor 1B), also named p27kip1, was considered a possible candidate. This gene has been localized to 12p13 by FISH, and one allele is deleted in a variety of hematologic disorders.13,40 However, extensive sequence analysis has failed to detect mutations in the retained allele.40-43 Assuming that hemizygosity for p27kip1 is insufficient to cause growth alterations, as is suggested by p27kip1 knockout studies in mice,44 the gene is unlikely to be the potential target for 12p alterations.
The presence of a 12p cytogenetic abnormality did not significantly influence outcomes. The only possible exceptions are cases with a dic(9; 12) or both 12p homologs affected who had a higher rate of 5-year event-free survival than other 12p cytogenetic subgroups; however, the number of cases was too small for meaningful statistical analysis. Recently, van der Plas et al2 reported CNS relapse in 7 of 12 children with ALL and a 12p aberration and suggested that a 12p abnormality increases the risk of CNS relapse. In contrast, only 5 of our 94 patients had such an adverse event. Whether this disparity reflects the small number of cases in their study or a difference in treatment efficacy is uncertain.
Most of the 94 cases with a cytogenetic 12p abnormality (78%) had additional chromosomal abnormalities, but with the exception of one with a t(1; 19)(q23; p13), none had the known common recurrent translocations. This finding is consistent with that of Cayuela et al,45 who evaluated 35 cases of childhood ALL for the BCR-ABL, E2A-PBX1, MLL-AF4, and ETV6-CBFA2 chimeric transcripts and failed to find more than one chimeric transcript in any. We have evaluated more than 100 newly diagnosed ALL cases for these transcripts with similar results (unpublished data, September 1996), suggesting that ETV6-CBFA2 is an exclusive genetic fusion and a probable instigating factor in leukemogenesis.
Among the one third of cases with 12p abnormality that had no ETV6-CBFA2 chimeric transcript, we found no distinguishing presenting clinical feature except a higher frequency of T-cell immunophenotype. A clinically relevant leukemic subset is likely to be identified among these cases as molecular data accumulate.
In summary, the ETV6 gene was rearranged in 56% and the ETV6-AML1 fusion transcript was expressed in 66% of the 94 ALL cases with visible 12p abnormalities. Because of the potential importance of recognizing the t(12; 21) for proper treatment stratification, the recommended test to identify patients with an ETV6-CBFA2 fusion transcript is RT-PCR because of its specificity and sensitivity. Southern analysis for the ETV6 gene using DNA probes shows an excellent correlation with the presence of a ETV6-CBFA2 fusion transcript but lacks the sensitivity of RT-PCR. FISH with whole chromosome painting probes requires metaphases to detect the t(12; 21), which can be a limiting factor when few metaphases are available in the bone marrow preparations. At present there are no commercially available probes for the detection of the ETV6 and CBFA2 in interphase nuclei. Conventional cytogenetic techniques do not detect the cryptic t(12; 21), but they would facilitate the selection of cases in which FISH analysis is unnecessary to detect the fusion; that is, hyperdiploidy (51+) (25% to 30% of ALL cases overall), cases with a t(4; 11), t(9; 22), t(1; 19) (approximately 15%), and T-cell lineage (15%).
ACKNOWLEDGMENT
We thank the cytogenetic technologists for their excellent technical assistance; M. Griffith, W.P. Conn, W.K. Williams, and A. Curcio-Brint for technical expertise; R. Seshadri for statistical programming; and S. Naron for editorial assistance.
Supported in part by Grants No. CA 20180 and CA 21765 from the National Cancer Institute and by the American Lebanese Syrian Associated Charities (ALSAC).
Address reprint requests to S.C. Raimondi, PhD, Department of Pathology and Laboratory Medicine, St Jude Children's Research Hospital, 332 N Lauderdale, Memphis, TN 38105.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal