Abstract
The mannose receptor (MR) is a transmembrane protein that functions primarily as a phagocytic receptor for a wide range of microorganisms. Its expression appears to be restricted to tissue macrophages and Langerhans cells. To gain an understanding of the regulation of the gene, we have isolated the 5′ flanking sequence of the murine MR gene and have analyzed a 536-bp sequence upstream of the ATG start site for transcriptional activity. This sequence lacks a TATA box but contains an initiator (Inr) consensus element overlapping the single transcriptional start site. Transcription factor binding sites contained within this sequence include PU.1, Sp1, ETS, GATA, and MYB motifs. Serial 100-bp deletions of this promoter fragment fused to a luciferase reporter gene showed various patterns of activity when transfected into different cell types. In myeloid cells, sequence elements upstream of bp −300 appeared to have a silencing effect on promoter activity. Of the four potential PU.1 binding sites contained within the fragment, one site (at −164) bound the PU.1 factor most strongly, whereas the adjacent PU.1 site (at −177 bp) bound PU.1 to a lesser degree. Mutations of these sites decreased transcriptional activity but did not abolish it. However, promoter activity was abrogated when both the −164 bp PU.1 site and the adjacent −177 bp PU.1 site were mutated. In addition, mutation of the Sp1 site also significantly reduced promoter activity. Cotransfection studies in Drosophila Schneider cells indicated that PU.1 and Sp1 may function synergistically in transactivating the murine MR. This study indicates that MR gene expression is regulated in part by the interaction between the ubiquitously expressed factor Sp1 and the lymphoid/myeloid factor PU.1 and provides a basis for studying the regulation of this gene.
THE MACROPHAGE MANNOSE receptor (MR) is a 175-kD transmembrane protein expressed on the surface of mature tissue macrophages1 and Langerhans cells.2 The MR recognizes a pattern of apparently disparate carbohydrate ligands that coat a wide array of microorganisms. The MR recognizes gram-positive and gram-negative bacteria,3 yeast,4 fungi,5 mycobacteria,6 and certain parasites such as Leishmania donovani7 and Trypanosoma cruzi.8 This broad range of recognition is mediated by the receptor ectodomain that contains an amino terminal cysteine-rich domain, a fibronectin type II region, and a tandem array of eight carbohydrate recognition domains (CRDs).9
MR levels are upregulated by α and β interferon,10 1,25-dihydroxyvitamin D3,11 and interleukin-412 and are downregulated by interferon-γ10 and phorbol ester.13 Circulating monocytes do not express surface MRs, but high levels of expression are observed within hours after adherence of monocytes in vitro. However, whereas something is known about the biology of the receptor, the regulation of the gene and the structure and function of its promoter have not been well studied.
The PU.1 transcription factor was first identified in mice and is expressed mainly in B lymphocytes and in immature myeloid and erythroid cells. It is a member of the ETS family of transcription factors that includes a number of other factors important in hematopoiesis, such as ETS-1 and Fli-1.14-16 Several members of the ETS family of transcription factors are expressed in a tissue-specific manner and may be critical in regulating tissue-specific gene expression.17
A number of studies have described PU.1 as a transactivator of transcription. For instance, PU.1 appears to transactivate the Ig λ 2-4 enhancer18 as well as the Ig κ 3′ enhancer to which it binds after recruiting a second factor, NF-EM5. But PU.1 has also been reported to function as a transrepressor. Thus, in B lymphocytes, it is thought to transrepress the IgH enhancer by competing with a second factor for DNA binding.19
PU.1 appears to play an important role in macrophage differentiation. Ross et al19 have postulated that it may be part of a switch contributing to lineage divergence between B lymphocytes and macrophages. More recently, gene targeting experiments have shown that PU.1 is a critical factor for the expression of genes associated with terminal myeloid differentiation, such as CD11b, the receptor for macrophage colony-stimulating factor (M-CSFR), and CD64.20 In the CD64 promoter, our own work has shown a critical PU.1 binding site 20-bp upstream of the transcription start site that plays an integral role both in basal transcription as well as in the interferon-γ inducibility of the gene.21
Like PU.1, Sp1 has been shown to be capable of interacting with components of the basal transcription machinery. The two glutamine-rich activation domains of Sp1 have been shown to interact with the TBP-associated factor dTAFII1022 as well as with the basal factor YY1 that binds the adeno-associated virus P5 promoter.23 Other studies have found that Sp1 functioned most effectively in the context of a core promoter containing an initiator element.24-26 It has been suggested that Sp1 may be involved in bringing distally bound protein into the presence of the proximal promoter through DNA looping.27
Sp1 has furthermore been shown to be involved in the regulation of the tissue-specific expression of certain genes, eg, CD14 gene expression in monocytic cells.28 Sp1 has been reported to be involved in determining tissue specificity of some erythroid genes by recruiting the factor GATA-1 to the promoter.29 On the other hand, in the murine thrombospondin I gene, there appears to be an element of redundancy in the function of Sp1. The Sp1 binding sites overlap Egr-1 sites but mutation and small deletions in these sites did not markedly inhibit transcriptional activity.30
That some form of synergistic interaction between PU.1 and Sp1 might occur in certain promoters has been suggested by a number of studies. For example, in both CDIIb and the Bruton's tyrosine kinase promoters, it has been hypothesized that PU.1 binds first and then induces a change in chromatin structure that provides access for Sp1 to its binding site.31 32
We previously described the cDNA cloning and intron-exon structure of the murine MR gene and showed that downregulation of the receptor by interferon-γ occurs at the level of transcription.33 34 In this study, we set out to isolate the 5′ flanking region of the murine MR gene and to characterize some of the regulatory mechanisms involved in its promoter function. We have determined that PU.1 and Sp1 are major regulators of transcription in this promoter.
MATERIALS AND METHODS
Cell culture. The cell lines U937 (human promyelomonocytic), RAW264 (murine macrophage cell line), HeLa (human epithelial carcinoma), and Jurkat (human T lymphocyte) were obtained from the American Type Culture Collection (ATCC; Rockville, MD). The cells were cultured at 37°C in humidified air with 5% CO2 in RPMI 1640 medium supplemented with heat-inactivated fetal calf serum (HyClone, Logan, UT) and containing penicillin and streptomycin.
Cloning and characterization of the MR promoter. The murine MR promoter was identified by screening two λ-FIX genomic libraries, one derived from NIH/3T3 cells (#946301; Stratagene, La Jolla, CA) and the other from mouse liver (strain 129, #946305; Stratagene) with a radiolabeled murine mannose receptor cDNA. Three clones (the longest of which contained 536 bp of 5′ flanking sequence) were isolated from the murine libraries and sequenced by the dideoxy Sanger procedure using Sequenase DNA polymerase enzyme (US Biochemical, Cleveland, OH). No sequence mismatches were detected between the murine clones.
In vitro transcription and translation. In vitro transcribed and translated PU.1 cDNA (a gift from Dr R. Maki, La Jolla Cancer Research Foundation, La Jolla, CA) was made using the TNT Coupled Reticulocyte Lysate System supplied by Promega Corp (Madison, WI).
Electrophoretic mobility assay (EMSA). EMSAs were performed as previously described.21 Oligonucleotides were annealed (50 ng each) and end-labeled with [γ-32P]-dATP using polynucleotide kinase, and the unincorporated label was separated through a DNA grade Sephadex G-50 column (Pharmacia LKB Biotechnology, Piscataway, NJ). Nuclear extracts (∼5 mg), prepared by the method of Dignam et al,35 were incubated on ice with 0.5 ng of probe at a specific activity of 4 × 108 cpm/mg. The binding assays with in vitro-translated PU.1 protein and nuclear extracts were performed in a 30 μL reaction mixture containing 12 mmol/L HEPES (pH 7.9), 0.5 mmol/L EDTA, 5 mmol/L MgCl2 , 10% (vol/vol) glycerol, 1 mmol/L dithiothreitol, 60 mmol/L KCl, 0.05% nonidet P40, and 2 μg of poly (dI-dC) (Pharmacia LKB Biotechnology). Unlabeled competitor oligonucleotides were added to the incubation mixtures 15 minutes preceding the addition of the radioactive probe. Reactions were separated on a 4% polyacrylamide gel at 4°C (acrylamide:bisacrylamide, 29:1). DNA binding assays with recombinant Sp1 transcription factor (Promega) were performed for 15 minutes on ice in a 20-μL binding buffer containing 10 mmol/L HEPES, pH 7.5, 50 mmol/L KCl, 5 mmol/L MgCl2 , 1 mmol/L dithiothreitol, 1 mmol/L EDTA, and 5% glycerol.
Antibody supershift assays were performed by preincubating the relevant protein for 15 minutes with specific antibody or preimmune serum before the addition of the probe for an additional 15 minutes. The anti-PU.1 serum, directed against the NH2 -terminal domain of the protein, was a gift from Dr R. Maki. The anti-Sp1 antibody was obtained from Santa Cruz Biotechnology (Santa Cruz, CA).
Plasmid constructs. Development of the pGLO luciferase reporter gene vector has been previously described.21 The 100-bp serial deletion constructs of the murine 5′ flanking region were made by polymerase chain reaction (PCR) using primers containing Not1 and Spe1 restriction sites for directional cloning into the Not1 and Spe1 sites of pGLO. The site-directed mutation constructs were developed by PCR overlap extension. The 5′-end outer oligonucleotide for priming of each construct contained a Not1 site, and the 3′-end contained a Spe1 site for directional cloning into the pGLO vector. All constructs were sequenced on an automated Applied Biosystem 373 DNA Sequencer using Dye Taq flourescein. Another set of constructs were developed in which either 2-bp or 6-bp mutations were introduced into each of the four PU.1 sites either separately or in combination with other sites. Similarly, constructs were developed in which mutations were introduced into the Sp1 site. A construct was developed with mutations in two PU.1 sites and an Sp1 site. The Sp1 expression vectors have been previously described.36
Transient transfections. Transient transfections of luciferase reporter gene constructs into mammalian cells were performed by electroporation as previously described.21
The following conditions were used for electroporation of different cell lines: U937 (300 V; 960 μF ), RAW264 (300 V; 960 μF ), HeLa (150 V, 960 μF ), and Jurkat (250 V; 1,600 μF ). To control for efficiency of transfection, 2 μg of a plasmid containing a human growth hormone (hGH) gene sequence linked to the thymidine kinase (tk) promoter was cotransfected into the cells. Luciferase activity was normalized to 1 μg of hGH as measured by radioimmunoassay (Nichols Institute, San Juan Capistrano, CA). Luciferase activity was measured as described21 and was expressed in millivolts per microgram of growth hormone produced per milliliter in the transfection medium (luc/GH). Between three and six transfections in duplicate were performed for each construct.
Sp1 and PU.1 transactivation of promoter in Schneider SL cells.Drosophila Schneider SL2 cells were transfected by the calcium phosphate coprecipitation method37 with 10 μg of reporter plasmid, in the presence or absence of 1 μg of the Sp1 expression vector pPACSp1.37 Schneider SL cells were similarly transfected with the PU.1 expression vector PUpECE. In other assays, pPACSp1 and PUpECE were cotransfected together with various mutated MR constructs. As an internal control for transfection efficiency, 100 ng of the tkgH growth hormone expressing plasmid was included in all cases. Sonicated double-stranded salmon sperm DNA to a total of 10 μg was included as carrier. Luciferase assays were performed 48 hours after transfection.
RESULTS
Sequence analysis of MR 5′ upstream region. Sequence analysis of the 5′ flanking fragment identified four potential binding sites for the ETS family transcription factor PU.1 (GAGGAA)38 located around −22, −32, −164, −177, and an ETS-1 consensus sequence around −204,15,39 as shown in Fig 1. An Sp1 site (CCGCCC) is located around −104.40,41 Other transcription factor binding sites of potential relevance to hematopoiesis are a MYB site located around +2542 and a GATA site around −90.43 There is no TATA box −25 to −35 bp upstream of the transcription start site (designated +1), but there is an initiator consensus sequence (PyPyA + 1 NT/APyPy) overlapping the transcription start site.44 45
Murine MR 5′ flanking region. Both strands of the flanking region were sequenced by the dideoxy method. The translation start site ATG is indicated in bold letters, followed by some translated sequence in small letters. The transcription start site is indicated by a large arrow. The boxes indicate potential and experimentally verified transcription factor binding sites (see Results). The location of the four potential PU.1 sites (P177, P164, P32, and P22) and the single Sp1 site as well as the initiator (Inr) element are portrayed.
Murine MR 5′ flanking region. Both strands of the flanking region were sequenced by the dideoxy method. The translation start site ATG is indicated in bold letters, followed by some translated sequence in small letters. The transcription start site is indicated by a large arrow. The boxes indicate potential and experimentally verified transcription factor binding sites (see Results). The location of the four potential PU.1 sites (P177, P164, P32, and P22) and the single Sp1 site as well as the initiator (Inr) element are portrayed.
Transfection of wild-type MR promoter constructs into myeloid and nonmyeloid cells. Serial 100-bp deletion constructs of the wild-type MR receptor fused to a luciferase reporter gene were transfected into myeloid (U937 and RAW264) and nonmyeloid cells (HeLa and Jurkat) and assayed for activity. Promoter activity (luc/GH) was measured in millivolts (mV) and normalized to 1 μg of growth hormone produced by a cotransfected tk-GH plasmid per milliliter of medium (see the Materials and Methods). Values of the transfected vector backbone (pGLO) are included. Bars represent the average value obtained from between three and six transfections performed on each construct.
Transfection of wild-type MR promoter constructs into myeloid and nonmyeloid cells. Serial 100-bp deletion constructs of the wild-type MR receptor fused to a luciferase reporter gene were transfected into myeloid (U937 and RAW264) and nonmyeloid cells (HeLa and Jurkat) and assayed for activity. Promoter activity (luc/GH) was measured in millivolts (mV) and normalized to 1 μg of growth hormone produced by a cotransfected tk-GH plasmid per milliliter of medium (see the Materials and Methods). Values of the transfected vector backbone (pGLO) are included. Bars represent the average value obtained from between three and six transfections performed on each construct.
Gel-shift analyses using labeled oligonucleotides representing the MYB consensus (and including adjacent basepairs) failed to bind either purified MYB protein (a gift from Dr Peter Dini, Harvard Medical School, Boston, MA) or any other factor in nuclear extracts of U937 cells.
Functional analysis of the 5′ upstream sequence shows positive and negative regulatory regions. The 5′ flanking region of the gene was examined for promoter activity. One-hundred–basepair serial deletion fragments were cloned into the pGLO luciferase reporter gene vector. Promoter activity was ascertained by measuring luciferase activity after transfection of these constructs into promyelomonocytic U937 cells and macrophage RAW264 cells as well as into Jurkat T cells and HeLa cells.
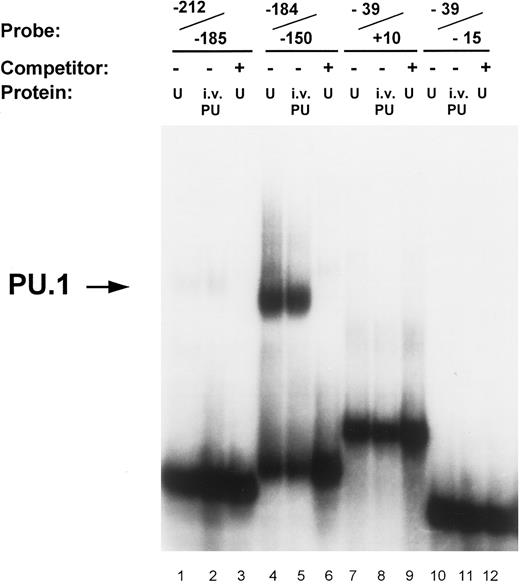
Identification of PU.1 binding to the MR promoter by gel mobility shift analysis. Oligonucleotide probes representing promoter segments that included each of the four potential PU.1 sites were prepared as described in the Materials and Methods. Binding reactions were performed on ice for 30 minutes by incubating the respective probes with either 5 μg of nuclear extract (U) or 1 μL of in vitro-translated PU.1 protein (i.v. PU) in the presence of 2 μg of double-stranded poly (dI:dC). The source of nuclear extract in the assay portrayed was from U937 cells. The probe −212/−185 contains the ETS-1 site at −204 (lanes 1 through 3); the probe −184/−150 contains the pair of PU.1 sites at −164 and −177 (lanes 4 through 6); the probe −39/−15 contains the pair of PU.1 sites at −32 and −22 (lanes 7 through 9); similarly, the probe −39/+10 also contained the −32 and −22 PU.1 sites but in addition included the initiator element (Inr) overlapping the transcription start site (lanes 10 through 12; see Fig 1). Unlabeled double-stranded competitor oligonucleotides of each promoter segment were added at 50-molar excess over the probe oligo (lanes 3, 6, 9, and 12). The arrow on the left indicates the complex of PU.1 binding to the −184/−150 oligonucleotide.
Identification of PU.1 binding to the MR promoter by gel mobility shift analysis. Oligonucleotide probes representing promoter segments that included each of the four potential PU.1 sites were prepared as described in the Materials and Methods. Binding reactions were performed on ice for 30 minutes by incubating the respective probes with either 5 μg of nuclear extract (U) or 1 μL of in vitro-translated PU.1 protein (i.v. PU) in the presence of 2 μg of double-stranded poly (dI:dC). The source of nuclear extract in the assay portrayed was from U937 cells. The probe −212/−185 contains the ETS-1 site at −204 (lanes 1 through 3); the probe −184/−150 contains the pair of PU.1 sites at −164 and −177 (lanes 4 through 6); the probe −39/−15 contains the pair of PU.1 sites at −32 and −22 (lanes 7 through 9); similarly, the probe −39/+10 also contained the −32 and −22 PU.1 sites but in addition included the initiator element (Inr) overlapping the transcription start site (lanes 10 through 12; see Fig 1). Unlabeled double-stranded competitor oligonucleotides of each promoter segment were added at 50-molar excess over the probe oligo (lanes 3, 6, 9, and 12). The arrow on the left indicates the complex of PU.1 binding to the −184/−150 oligonucleotide.
Figure 2A indicates that, in the U937 cells, promoter activity is concentrated between −100 and −300 bp upstream of the transcription start site. The luciferase activity of the construct containing this region was up to 35-fold higher than that of the promoterless vector. The inclusion of the additional 200 bp of upstream sequence resulted in diminished reporter gene activity in the myeloid cell lines U937 and RAW264 (Fig 2). Further analysis is required to determine whether this region contains a true repressor.
Transcription factor PU.1 binds to the MR promoter. The proximal murine MR promoter contains four PU.1 consensus binding sites (GAGGAA) located on the noncoding strand (Fig 1). These purine-rich sites appear to be located in pairs, between −19 and −34 (separated by 4 bp) and between −160 and −179 (separated by 7 bp). In gel mobility shift assays, we tested whether each of these potential PU.1 sites bound in vitro translated PU.1 or PU.1 contained in nuclear extracts isolated from U937 and RAW264 cells.
In vitro-translated PU.1 and a comobile factor in U937 nuclear extracts bound to the oligonucleotide −184/−150, which contains the −164 and −177 PU.1 sites (Fig 3). The intensity of PU.1 binding to this region is clearly greater than binding to the oligonucleotides −39/+10 and −39/−15 that encompass the PU.1 sites at −22 and −32 (Fig 3). Nuclear extracts of U937 cells consistently demonstrated more intense binding activity to the −164 PU.1 site, which was abrogated upon mutation of this site (Fig 4). The mobility of the DNA-protein complex was not retarded in the presence of anti-PU.1 antisera, but rather the intensity of the band was reduced by the addition of the specific antisera (results not shown). This suggests that PU.1 antisera may inhibit binding of protein to the complex.
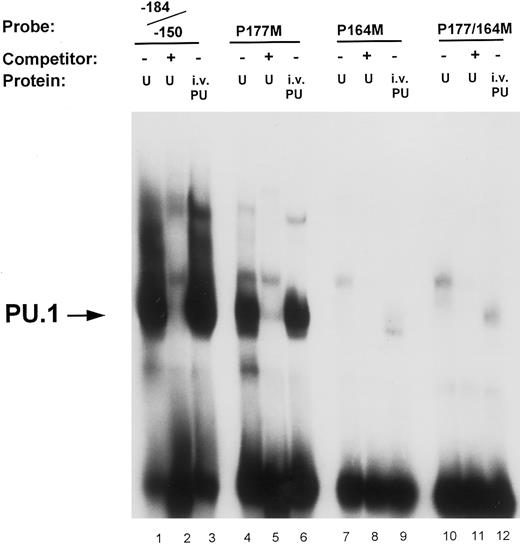
Mutational analysis of the −164 and −177 PU.1 sites. Oligonucleotide probes were prepared in which the PU.1 sites at −164 and −177 were mutated (GAGGAA to GACTAA) either separately (P177M, lanes 4 through 6; and P164M, lanes 7 through 9) or in combination (P177/164M, lanes 10 through 12). Binding assays of nuclear extract (U) and in vitro-translated PU.1 (i.v. PU) to these probes were performed as described in the Materials and Methods and compared with binding to the unmutated wild-type oligonucleotide probe (−184/−150, lanes 1 through 3). Unlabeled double-stranded competitor oligonucleotides were added at 50-molar excess over the probe oligo (lanes 2, 5, 8, and 11). The arrow on the left indicates PU.1 binding to the oligonucleotide probes.
Mutational analysis of the −164 and −177 PU.1 sites. Oligonucleotide probes were prepared in which the PU.1 sites at −164 and −177 were mutated (GAGGAA to GACTAA) either separately (P177M, lanes 4 through 6; and P164M, lanes 7 through 9) or in combination (P177/164M, lanes 10 through 12). Binding assays of nuclear extract (U) and in vitro-translated PU.1 (i.v. PU) to these probes were performed as described in the Materials and Methods and compared with binding to the unmutated wild-type oligonucleotide probe (−184/−150, lanes 1 through 3). Unlabeled double-stranded competitor oligonucleotides were added at 50-molar excess over the probe oligo (lanes 2, 5, 8, and 11). The arrow on the left indicates PU.1 binding to the oligonucleotide probes.
Site-directed mutational analyses of PU.1 sites. To ascertain the functional contribution of the various potential PU.1 sites to promoter activity, we analyzed and compared the effects of mutating these sites in gel shift and reporter gene transfection assays. Results shown in Fig 4 demonstrate that the binding to the −164 PU.1 site is abrogated when the central guanine (GG) residues of the consensus sequences were mutated. Lanes 1 and 3 indicate that both in vitro-translated PU.1 and a factor in U937 nuclear extracts bind to an oligonucleotide (−184/−150) that contains the PU.1 sites at −177 and −164. This binding is competed off with cold competitor oligonucleotide (lane 2). An oligonucleotide in which the −177 PU.1 was mutated retained binding of PU.1, albeit to a lesser extent (lanes 4 through 6), whereas one in which the −164 PU.1 site was mutated exhibited loss of binding (lanes 7 through 9). Mutation of both PU.1 sites in this region abolished binding (lanes 10 through 12). The −164 PU.1 site consistently demonstrated the strongest PU.1 binding activity.
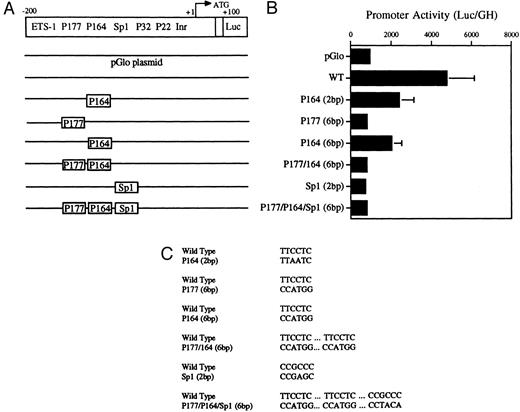
Decreased transcriptional activity of promoter activity of promoter fragments containing mutated PU.1 and/or Sp1 sites. (A) Diagramatic representation of reporter gene constructs of the murine MR 5′ flanking region. The wild-type (WT) MR construct depicting the four PU.1 sites (P177, P164, P32, and P22) as well as the single Sp1 site and the initiator element (Inr) overlapping the transcription start site at +1 (large arrow) are presented. The translation start site (ATG) at +100 and the fused luciferase reporter gene (luc) are also indicated. (B) The mutations introduced at the various transcription factor sites are portrayed above the wild-type sequence in the diagrams of each construct. (C) Constructs containing 300 bp of upstream promoter sequence fused to a luciferase reporter gene were developed in which either of the PU.1 sites at −177 or −164, respectively, were mutated or the single Sp1 site at −104 was mutated (see Fig 2). In the P177 (6 bp) and the P164 (6 bp) constructs, 6 bp of the PU.1 consensus were mutated (GAGGAA to CCATGG), whereas in the P164 (2 bp) construct, 2 bp of the PU.1 consensus were mutated (GAGGAA to GACTAA). In the Sp1 (2 bp) construct, 2 bp of the consensus sequence were mutated (CCGCCC to CCGAGC). In the P177/P164/Sp1 (6 bp) construct, 6 bp of the PU.1 consensus were mutated (GAGGAA to CCTGG) and 2 bp of the Sp1 consensus were mutated (GC to TA). The constructs were transiently transfected by electroporation and the luciferase activity was determined as described in the Materials and Methods.
Decreased transcriptional activity of promoter activity of promoter fragments containing mutated PU.1 and/or Sp1 sites. (A) Diagramatic representation of reporter gene constructs of the murine MR 5′ flanking region. The wild-type (WT) MR construct depicting the four PU.1 sites (P177, P164, P32, and P22) as well as the single Sp1 site and the initiator element (Inr) overlapping the transcription start site at +1 (large arrow) are presented. The translation start site (ATG) at +100 and the fused luciferase reporter gene (luc) are also indicated. (B) The mutations introduced at the various transcription factor sites are portrayed above the wild-type sequence in the diagrams of each construct. (C) Constructs containing 300 bp of upstream promoter sequence fused to a luciferase reporter gene were developed in which either of the PU.1 sites at −177 or −164, respectively, were mutated or the single Sp1 site at −104 was mutated (see Fig 2). In the P177 (6 bp) and the P164 (6 bp) constructs, 6 bp of the PU.1 consensus were mutated (GAGGAA to CCATGG), whereas in the P164 (2 bp) construct, 2 bp of the PU.1 consensus were mutated (GAGGAA to GACTAA). In the Sp1 (2 bp) construct, 2 bp of the consensus sequence were mutated (CCGCCC to CCGAGC). In the P177/P164/Sp1 (6 bp) construct, 6 bp of the PU.1 consensus were mutated (GAGGAA to CCTGG) and 2 bp of the Sp1 consensus were mutated (GC to TA). The constructs were transiently transfected by electroporation and the luciferase activity was determined as described in the Materials and Methods.
Transient transfections of the various PU.1-mutated constructs into U937 cells (Fig 5A, B, and C) showed that mutation of the −177 PU.1 site abrogated transcriptional activity. Mutation of the −164 PU.1 site also greatly reduced transcriptional activity. These results apparently contradict the gel-shift binding studies shown in Fig 4. However, the transactivation experiments shown in Fig 7 (see below) show a clear role for the −164 PU.1 site. Mutations of both the −164 and −177 PU.1 sites as well as an Sp1 site resulted in reduced transcriptional activity (Fig 5B). We conclude that both the −177 PU.1 site and the −164 PU.1 site act together with the Sp 1 site to regulate the expression of this gene.
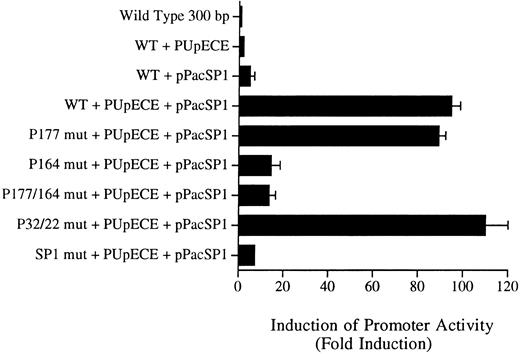
Sp1 and PU.1 act synergistically in cotransactivating transcription of the murine MR promoter. Fragments of the murine MR promoter, in which the Sp1 site or each of the four PU.1 sites (separately or in combination) were mutated, were transfected into Drosophila SL cells (Sp1 and PU.1 negative) by the calcium phosphate precipitation method of Chen and Okayama.37 Ten micrograms of the mutated MR plasmid construct was transfected along with 1 μg of each expression plasmid (PUpECE and pPacSP1) and luciferase activity was measured as described in the Materials and Methods. Wild-type (WT) plasmid alone or together with PUpECE and/or pPacSp1 was transfected as a control. The figure portrays the luciferase reporter gene results as a fold increase over activity obtained from transfection of the wild-type (WT) construct.
Sp1 and PU.1 act synergistically in cotransactivating transcription of the murine MR promoter. Fragments of the murine MR promoter, in which the Sp1 site or each of the four PU.1 sites (separately or in combination) were mutated, were transfected into Drosophila SL cells (Sp1 and PU.1 negative) by the calcium phosphate precipitation method of Chen and Okayama.37 Ten micrograms of the mutated MR plasmid construct was transfected along with 1 μg of each expression plasmid (PUpECE and pPacSP1) and luciferase activity was measured as described in the Materials and Methods. Wild-type (WT) plasmid alone or together with PUpECE and/or pPacSp1 was transfected as a control. The figure portrays the luciferase reporter gene results as a fold increase over activity obtained from transfection of the wild-type (WT) construct.
Transcription factor Sp1 binds to the murine MR promoter. Having established that the putative PU.1 sites at −164 and −177 may be necessary for promoter activity, we next evaluated whether the Sp1 site was functional and whether indeed Sp1 might synergize with PU.1.
The Sp1 consensus binding site (CCGCCC) is located around −104 bp (Fig 1). A factor contained in U937 (and RAW264) nuclear extracts bound specifically to this site (Fig 6, lanes 1 and 2) and was supershifted to a higher molecular weight complex by addition of Sp1 antibody (Fig 6, lane 3). A binding complex of identical mobility using purified Sp1 protein (Promega) was similarly competed off using unlabeled oligonucleotide in lanes 4 and 5.
Identification of Sp1 binding to the MR promoter by electrophoretic mobility shift analysis. A double-stranded MR promoter oligonucleotide (bp −90 to −125) that included the single Sp1 site was labeled with [γ-32P]-dATP and incubated with either nuclear extract (U) or with purified Sp1 protein (Promega) in the presence of 2 μg of double-stranded poly (dI:dC) under conditions described in the Materials and Methods. Unlabeled double-stranded competitor oligonucleotides of each promoter segment were added at a 50-molar excess over the probe oligonucleotide (lanes 2 and 5). The antibody supershift assay was performed by preincubating the binding protein for 15 minutes with anti-Sp1 antibody (Santa Cruz Biotechnology) before the addition of the probe for an additional 20 minutes. The arrows on the left of the diagram indicate the positions of the Sp1 binding complex (lanes 1 and 4) and the antibody supershifted complex (lane 3).
Identification of Sp1 binding to the MR promoter by electrophoretic mobility shift analysis. A double-stranded MR promoter oligonucleotide (bp −90 to −125) that included the single Sp1 site was labeled with [γ-32P]-dATP and incubated with either nuclear extract (U) or with purified Sp1 protein (Promega) in the presence of 2 μg of double-stranded poly (dI:dC) under conditions described in the Materials and Methods. Unlabeled double-stranded competitor oligonucleotides of each promoter segment were added at a 50-molar excess over the probe oligonucleotide (lanes 2 and 5). The antibody supershift assay was performed by preincubating the binding protein for 15 minutes with anti-Sp1 antibody (Santa Cruz Biotechnology) before the addition of the probe for an additional 20 minutes. The arrows on the left of the diagram indicate the positions of the Sp1 binding complex (lanes 1 and 4) and the antibody supershifted complex (lane 3).
Mutations were introduced into the Sp1 site to determine the functional significance of this site in the murine MR promoter. Luciferase activity was measured after transfection of these constructs into U937 cells. Results shown in Fig 5 indicate that mutation of the Sp1 site alone causes a drastic reduction in luciferase activity in U937 cells.
PU.1 and Sp1 transactivate the MR promoter in Drosophila SL cells. To show that PU.1 and Sp1 are critical regulators of MR promoter activity, cotransactivation experiments were performed in Drosophila SL cells. These cells are known to lack Sp1, and we determined using Western blotting that they also lack PU.1. These cells were therefore used to determine whether PU.1 and Sp1 might function synergistically in transactivating the MR promoter.
The wild-type 300-bp reporter gene construct alone was inactive in these Drosophila cells (Fig 7). Transactivation with PU.1 alone resulted in only a twofold to threefold increase in activity. Similarly, the addition of Sp1 increased activity fivefold. However, cotransfection of the wild-type plasmid with PUpECE and pPacSP1 resulted in a 90-fold increase in activity (Fig 7).
Constructs in which PU.1 sites at −177, −22, and −32 were mutated could readily be coactivated to the 90-fold levels of the wild-type by the addition of PU.1 and Sp1 (Fig 7). In contrast, the −164 PU.1 mutant could be transactivated only 10-fold by these expression plasmids. These results confirm the functional significance of the unmutated PU.1 site at −164. Similarly, it can be seen that the mutation of the Sp1 site rendered such constructs less responsive (only 5-fold) to transactivation by these expression vectors. These experiments therefore provide further evidence of the importance of the PU.1 site at −164 and the Sp1 site at −104 in the regulation of the murine MR promoter.
DISCUSSION
We report here the cloning of the 5′ DNA flanking sequence of the murine macrophage MR gene and describe some of the key cis-acting elements and transcription factors involved in its regulation. Inspection of the sequence within this region showed a number of noteworthy features. Among the potentially functional transcription factor binding sites, we have examined in particular the role of PU.1 and Sp1 and we demonstrate that they are important regulators of the promoter. Previously, in the CD64 promoter, we showed that PU.1 plays a critical role in the maintenance of lineage specificity as well as in the interferon-γ inducibility of the gene.21 PU.1 has not previously been reported to have a functional role in the regulation of the MR gene.
Our results indicate a prominent role for two of four potential PU.1 sites (located at −164 and −177) and for the single Sp1 site (at −104). In gel mobility shift assays, we have shown that the −164 PU.1 site has the greatest binding activity. The −177 PU.1 site bound to a lesser extent. However, in transient transfection assays in U937 cells, mutation of the −164 PU.1 site greatly reduced promoter activity, whereas mutation of the −177 PU.1 site abrogated promoter activity (Fig 5). In the transactivation experiments, a clear role for the −164 PU.1 site was observed (Fig 7). Our results indicate that both the −177 PU.1 site and the −164 PU.1 play a role in the transcriptional activity of the mannose receptor promoter, but their binding activity may be dependent on a second unidentified factor necessary to stabilize the interaction of PU.1 with the DNA. Such secondary factors have been shown to facilitate the binding of PU.1 to the Ig λ 2-4 enhancer18 and to the Ig κ gene 3′ enhancer.46 Alternatively, the −177 bp site may not be a true PU.1 binding site and might bind another ETS like factor.
Ray-Gallet et al47 used the PCR-mediated binding site selection procedure to define the following best consensus sequence for Spi-1/PU.1: AAAA A/T/(G) G/C A/C/G GGAA G/C T A/G G/C. Of the four potential PU.1 binding sites, P164 conforms most closely to this suggested consensus.
We have shown by gel-shift assay that the Sp1 site located at −104 binds Sp1 protein (Fig 6) and that mutation of this site results in loss of reporter gene activity in transfections of the constructs into U937 cells (Fig 5). Furthermore, such a mutation of the Sp1 consensus resulted in a marked reduction in transactivation by Sp1 expression vectors in Drosophila SL cells, thereby confirming the significance of this site (Fig 7).
Our transactivation assays also provide evidence of synergism between PU.1 and Sp1 in this promoter. Whereas, singly, the Sp1 and PU.1 expression vectors could transactivate the promoter only a few fold, in combination there was a 90- to 100-fold increase in activity (Fig 7). More particularly, Fig 7 indicates that it is the PU.1 site at −164 and the −104 Sp1 site that contain most of the activity. The synergistic effect, reflected in the 90-fold increase in activity, may therefore function through these two sites. However, it is not clear whether either factor recruits the other to the promoter. In the CD11b promoter, it has been suggested that PU.1 and Sp1 function synergistically to regulate myeloid cell differentiation.31 PU.1 binds first before recruiting Sp1 to the promoter. However, in some erythroid-specific genes, Sp1 binds first and then recruits the factor GATA-1.29
Our transfection data show that the inclusion of promoter sequence upstream of −300 silences reporter gene activity in U937 cells (Fig 2). Preliminary experiments to determine whether the human CCAAT displacement protein (CDP), which acts as a repressor in the myeloid gp91 phox promoter,48 49 might bind to this region of the mannose receptor promoter were inconclusive. We found that serial 100-bp deletion constructs (Fig 2) behaved differently in U937 and RAW cells. We suspect that the assembly of combinations of transcription factors that differ in these cell lines may account for the difference.
In conclusion, the murine MR promoter contains PU.1 and Sp1 transcription factor binding sites that appear to play a critical role in the regulation of this gene. Further studies examining the interplay between these factors and other factors binding this promoter should provide a clearer understanding of the transcriptional regulation of this myeloid-specific receptor and may contribute to the evolving paradigm of the regulation of myeloid-specific genes.
ACKNOWLEDGMENT
The authors are grateful to Dr R. Maki for the gifts of the PU.1 expression vector, anti-PU.1 antibody, and cDNA. We thank Dr David Hume, Dr Dan Tennen, and members of our laboratory for valuable discussions.
Supported by National Institutes of Health Grants No. HL43510 and RO1HL48929. R.A.E. was an Established Investigator of the American Heart Association. D.H. is a Fulbright Cancer Research Fellow. J.E. is funded by the Pediatric Scientist Development Program.
Address reprint requests to R. Alan Ezekowitz, MD, PhD, Chief, Pediatric Service, Department of Pediatrics, Massachusetts General Hospital, 15 Parkman St, Boston, MA 02114.



![Fig. 6. Identification of Sp1 binding to the MR promoter by electrophoretic mobility shift analysis. A double-stranded MR promoter oligonucleotide (bp −90 to −125) that included the single Sp1 site was labeled with [γ-32P]-dATP and incubated with either nuclear extract (U) or with purified Sp1 protein (Promega) in the presence of 2 μg of double-stranded poly (dI:dC) under conditions described in the Materials and Methods. Unlabeled double-stranded competitor oligonucleotides of each promoter segment were added at a 50-molar excess over the probe oligonucleotide (lanes 2 and 5). The antibody supershift assay was performed by preincubating the binding protein for 15 minutes with anti-Sp1 antibody (Santa Cruz Biotechnology) before the addition of the probe for an additional 20 minutes. The arrows on the left of the diagram indicate the positions of the Sp1 binding complex (lanes 1 and 4) and the antibody supershifted complex (lane 3).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/10/10.1182_blood.v90.10.4135/3/m_bl_0033f6.jpeg?Expires=1763590161&Signature=PqaR6HlXZhAhLjlJu-D9okDrfQ-d0q52K4esqekrrcIhOoQZII-eko6gjLQVebm80ctJWSpvgVFJxnxcFDuZVwY9vv6aRmzajON7X~1NAla0WGauz2I0Bucz6XUhhXYz-0e61WrG0~b9upAqDBOLMiaf9m~~B47SS3XcG6h84QLV7CUViCv7U8Dn2P1MgsNoHcDKmV5p4uSAYiWpiKSV1l7tC~JOCQJJeb8ITN1T1Gqsb4jsX56yzpEN797Ejth2N3qYnTr~G5~7SnI2y1OB-AsTkzVcgVbBw8MHHht~4rp0JR6shYEtIlQxZZKdbO1zWeUcCcFiIJN90aUOOcRm5A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal