Abstract
Homozygous p16INK4A (p16) gene deletion is frequent in primary tumor cells from acute lymphoblastic leukemia (ALL), suggesting that loss of p16 may be an important precursor to transformation in ALL. We have previously described JKB, a human ALL cell line, that contains homozygous deletion of the p16 gene. Because ectopic expression of p16 suppresses cell growth, we created a temperature sensitive p16 mutant to develop a system for inducible p16 function in human ALL. JKB cells were transfected either with a p16 gene mutated at position 119 (E119G) to confer temperature sensitivity (JKB p16MT) or with control vector. The percentage of cells in G1 phase was similar in JKB control cells or in JKB p16MT cells cultured at restrictive conditions (40°C). However, with lowering of temperature from 40°C to permissive conditions (31°C), the percentage of JKB p16MT cells in G1 phase and binding of p16 to CDK4 and CDK6 increased, with associated decreases in CDK4 and CDK6 kinase activities, and dephosphorylation of retinoblastoma protein (pRB). Culture of JKB p16MT cells at 31°C for ≥3 days irreversibly inhibited growth. Moreover, JKB p16MT cells cultured under these permissive conditions showed a less transformed morphology and more differentiated phenotype than did these cells cultured under restrictive temperatures. Finally, dexamethasone (Dex) induced apoptosis of JKB p16MT cells cultured at 40°C, but did not trigger death of these cells cultured at 31°C. These results suggest that deletion of p16 gene in JKB human ALL cells is associated with dysregulated growth of less differentiated tumor cells, which nonetheless remain susceptible to apoptosis triggered by Dex.
ABNORMALITIES OF CELL cycle regulatory proteins result in dysregulated cell growth and may facilitate cell transformation.1 The p16INK4A (p16) protein induces dephosphorylation of retinoblastoma protein (pRB) by inhibiting binding of cyclin-dependent kinase (CDK)4 and CDK6 to cyclin D, resulting in G1 growth arrest.2 The discovery that p16 gene is mutated or deleted in a striking proportion of human tumors raised the possibility that p16 might predispose to cancer development.1,3-6 For example, germline mutations of p16 gene are present in familial melanoma,7,8 and homozygous deletions of p16 gene are observed in primary tumor cells from 70% to 80% of T-acute lymphoblastic leukemia (ALL), 15% to 20% of B-ALL, non-Hodgkin's lymphomas, lymphoblastic crisis of chronic myelogenous leukemia, and disease progression of adult T-ALL from chronic to acute phase.9-16 The high frequency of spontaneous lymphomas at an early age in p16 gene-deficient mice further supports a potential role for p16 in neoplastic transformation and pathogenesis of lymphoid malignancies.17 To date, however, the role of p16 gene deletion in the pathogenesis of lymphoid malignancies in man has not been fully characterized.
We have previously described the JKB human ALL cell line, which has homozygous deletion of p16 and chromosomal translocation between the loci of p16 gene (9p21) and immunoglobulin (Ig) heavy chain gene (14q32).18 As noted above, p16 binds to CDK4 and CDK6, inhibiting activation of pRB and resulting in G1 growth arrest. Due to the lack of p16 in JKB cells, CDK4 and CDK6 form complexes with cyclin D, thereby facilitating phosphorylation of pRB and continuous proliferation of JKB cells.18 Transfection of p16 cDNA into JKB cells may therefore aid in defining the functional significance of homozygous p16 gene deletion in lymphoid leukemogenesis. However, growth inhibition in cells ectopically expressing p16, coupled with the outgrowth of clones that express low levels of this protein, complicate this approach.
To overcome this limitation previous studies, primarily in melanoma and other solid tumors, have assessed p16 function either by transient transfection,19,20 stable transfection,21 or with the use of inducible p16.22 To develop a system for inducible p16 function in human ALL, we attempted in this study to create p16 temperature-sensitive mutants. Analysis of cDNA for human erythrocyte ankyrin shows repeated sequences composed of 22 tandem 33 amino acids,23 and similar repeats are also found in yeast and human proteins involved in cell cycle control and tissue differentiation.23 In particular, ankyrin repeats within p16 gene are homologous to those in cdc10 gene, which can regulate cell cycle progression in yeast. Moreover, several point mutations within ankyrin repeats of the cdc10 gene have been shown to confer temperature-sensitive growth.24 We hypothesized that analogous mutations within the ankyrin repeats of human p16 gene may allow for p16 function at permissive temperatures and lack of p16 function under restrictive conditions. Such temperature-sensitive mutants would facilitate characterization of the role of p16 in leukemogenesis.
In this study, we mutated p16 gene at position 119 (E119G) to create a temperature-sensitive mutant, which was transfected into JKB human ALL cell line (JKB p16MT cells). Decrease from 40°C (restrictive) to 31°C (permissive) culture temperatures increased G1 growth arrest and binding of p16 to CDK4 and CDK6, associated with decreased CDK4 and CDK6 kinase activities and dephosphorylation of pRB. Importantly, growth was inhibited under permissive conditions, and cells had less transformed morphology and more differentiated cell surface phenotype. In addition, culture of JKB p16MT cells under permissive conditions inhibited the subsequent induction of apoptosis by dexamethasone (Dex). These results suggest that deletion of the p16 gene in ALL may contribute to a more transformed morphology and less differentiated phenotype, dysregulated growth, and susceptibility of tumor cells to apoptosis triggered by Dex.
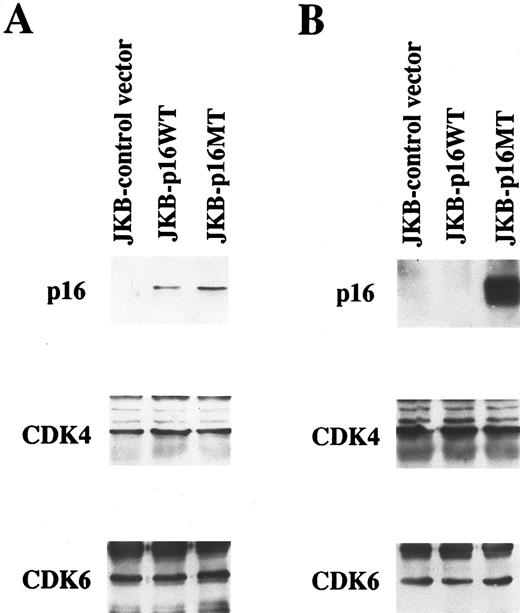
Ectopic expression of p16 protein in JKB cells. Total lysates obtained from JKB cells transfected with control vector (JKB control), wild-type p16 gene (JKB p16WT), or mutated p16 gene (E119G)(JKB p16MT) were immunoprecipitated with anti-p16 MoAb followed by Western immunoblotting with the same MoAb. IP and WB with anti-CDK4 or anti-CDK6 polyclonal Abs served as controls. Expression of p16 protein was determined after 7 days (A) and 2 months (B) of puromycin selection.
Ectopic expression of p16 protein in JKB cells. Total lysates obtained from JKB cells transfected with control vector (JKB control), wild-type p16 gene (JKB p16WT), or mutated p16 gene (E119G)(JKB p16MT) were immunoprecipitated with anti-p16 MoAb followed by Western immunoblotting with the same MoAb. IP and WB with anti-CDK4 or anti-CDK6 polyclonal Abs served as controls. Expression of p16 protein was determined after 7 days (A) and 2 months (B) of puromycin selection.
MATERIALS AND METHODS
Cell line. JKB is a pre-B ALL cell line with a chromosomal translocation between 9p21 and 14q32, on which p16 and heavy chain Ig gene, respectively, are located.18 Homozygous deletion of the p16 gene in JKB cells was confirmed by polymerase chain reaction (PCR), and the protein product was not detectable by Western Blotting (WB). Cells were cultured in RPMI-1640 medium (Sigma, St Louis, MO) containing heat inactivated 10% fetal bovine serum (FBS) (PAA Laboratories, Newport Beach, CA), L-glutamine (L-glu) (GIBCO, Grand Island, NY), 100 U/mL penicillin (pen), and 50 μg/mL streptomycin (strep) (GIBCO) in humidified air with 5% CO2 .
Mutagenesis. The construction of p16 mutants was performed with the BioRad Muta-Gene Phagemid in vitro Mutagenesis System (Biorad, Hercules, CA). Briefly, full length p16 DNA (provided by Dr Geoffrey I. Shapiro, Dana Farber Cancer Institute, Boston, MA) was introduced into CJ236 E. coli, which is deficient for dUTPase and uracil-N-glycosylase, resulting in an occasional substitution of uracil for thymidine in newly synthesized DNA. Single-stranded DNA was obtained using helper phage. Complementary DNA strand for the mutated p16 gene (E119G) was made using a synthetic oligonucleotide (5′GATGGCCCAGCTCGCCGGCCAGGTCCACGG3′ ) as primer, followed by cloning into EcoRI/Sall site of pBabe-puro retroviral vector (provided by Dr Mark Ewen, Dana Farber Cancer Institute).19 T79A and L113T p16 gene mutations were also similarly made, but not pursued because p16 function in G1 growth arrest was not altered in these mutants relative to wild-type p16.
Retroviral infection. pBabe-puro (control), pBabe-p16 wild-type-puro, and pBabe-p16 mutated type (E119G) vectors were introduced into Bing packaging cells, obtained from Dr G.I. Shapiro (Dana Farber Cancer Institute), using standard calcium phosphate transfection technique.25 26 Bing cells were cultured for 1 day posttransfection and 60% of supernatant then exchanged with fresh media for an additional 1 day. Next 60% of media was exchanged with fresh media containing JKB cells and polybrene (2.0 mg/mL) (Sigma) for 2-day cultures, followed by an additional 1-day culture in fresh media. This process was done a total of three times. Selection for transfected JKB cells was performed by culture with puromycin (2.0 mg/mL). Eighteen clones growing in 48-well plates were selected and expanded, and from these, the clone with optimal temperature-dependent effect on cell cycle was chosen for further study.
Immunoprecipitation and Western immunoblotting. Immunoprecipitation (IP) and WB were performed as previously described.27 28 For IP, cells (1 × 107 cells/sample) were washed three times with phosphate-buffered saline (PBS) and lysed for 30 minutes at 4°C in buffer: 1 mmol/L Tris-HCl (pH 7.6), 150 mmol/L NaCl, 0.5% Nonidet p-40, 5 mmol/L EDTA, 1 mmol/L phenylmethylsulfonyl fluoride (PMSF), 200 mmol/L Na3VO4, aprotinin, and 1 mmol/L NaF. Anti-p16 monoclonal antibody (MoAb) (Pharmigen, San Diego, CA); anti-CDK4, anti-CDK6, or anti-CDK2 polyclonal antibodies (Abs) (Santa Cruz Biotechnology, Santa Cruz CA); or anti-pRB MoAb (Calbiochem, San Diego, CA) were added for 16 hours at 4°C to immunoprecipitate protein complexes. Proteins were collected using protein G sepharose (PGS). Aliquots of each lysate were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred onto polyvinylidine difluoride (PVDF) membrane (NEN Dupont, Boston, MA), and nonspecific binding was blocked by incubation with 5% skim milk. The membrane was probed with Ab followed by antimouse or antirabbit Ig Abs conjugated with horseradish peroxidase (HRP) (Amersham, Arlington Heights, IL). Complexes were detected using the enhanced chemiluminescence system (Amersham).
Immune complex kinase assays. Immune complex kinase assays were performed as previously described.29 Briefly, cells were suspended at 1 × 107/mL in lysis buffer described above, and the supernatants were precipitated for 16 hours at 4°C with PGS plus rabbit anti-CDK4 or anti-CDK6 polyclonal Abs. Immunoprecipitated proteins on PGS were rinsed with lysis buffer and washing buffer (50 mmol/L HEPES, pH 7.5, containing 1 mmol/L dithiothreitol [DTT] ), and suspended in 30 μL of kinase buffer (50 mmol/L HEPES, 10 mmol/L MgCl2, 1 mmol/L DTT) containing substrate and 2.5 mmol/L EGTA, 10 mmol/L β-glycerophosphate, 0.1 mmol/L sodium orthovanadate, 1 mmol/L NaF, 20 μmol/L adenosine triphosphate (ATP), 5 μCi of γ-32P-ATP (NEN Dupont). Soluble glutathione S-transferase (GST)-RB fusion protein (Santa Cruz Biotechnology) (1 γg) was used as a substrate for assays of CDK4/CDK6-associated kinase activity. After incubation for 30 minutes at 30°C with occasional mixing, the samples were boiled in polyacrylamide gel sample buffer and separated by SDS-PAGE. Phosphorylated proteins were visualized by autoradiography of the dried slab gels.
Cell cycle analysis. The effect of ectopic p16 expression on cell cycle distribution in JKB cells was examined at different culture temperatures (40°C, 37°C, 34°C, and 31°C). Cell cycle analysis was performed using propidium iodide (PI) staining and fluorescence-activated cell sorting (FACS) analysis, as in a previous report.29 Briefly, cells were collected and suspended in 0.5 mL of 3.4 mmol/L sodium citrate, 10 mmol/L NaCl, 0.1% NP-40, and 50 mg/mL PI (Sigma) to stain nuclear DNA. Cell cycle distribution for each sample (10,000 cells) was determined using the Coulter Epics 753 cell sorter and program M Cycle software (Coulter Electronics, Hialeah, FL).
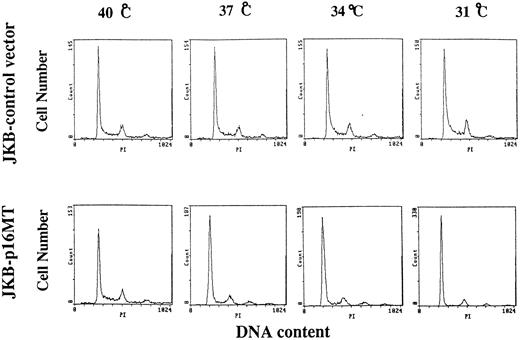
Effects of culture temperature on cell cycle distribution of JKB control cells and JKB p16MT cells. JKB control cells or JKB p16MT cells were cultured at different temperatures (40°C, 37°C, 34°C, and 31°C) for 1 day. Cell cycle distribution was determined using PI staining followed by FACS analysis.
Effects of culture temperature on cell cycle distribution of JKB control cells and JKB p16MT cells. JKB control cells or JKB p16MT cells were cultured at different temperatures (40°C, 37°C, 34°C, and 31°C) for 1 day. Cell cycle distribution was determined using PI staining followed by FACS analysis.
Morphologic and phenotypic analyses. Cell cytosmears were prepared and stained with Wright-Giemsa solution. The cell surface phenotype was determined using specific MoAbs in an indirect immunofluorescence assay, as previously described.30 31 Antibody-coated cells were enumerated by flow cytometric analysis using the Coulter Epics 753 cell sorter. MoAbs used included the following: anti-CD19 (B4), anti-CD10 (CALLA), and anti-CD20 (B1) (Coulter).
Assays for apoptosis. After incubation for 1 day at 40°C or 31°C, p16 mutated gene transfected JKB cells (1 × 106/mL) were treated with Dex (10 μmol/L) for another day. The percentage of apoptotic cells was determined by acridine orange (100 μg/mL) (Sigma) and ethidium bromide (100 μg/mL) (Sigma) staining. Apoptotic cells were enumerated by fluorescence microscopy (Microstar IV, Reichert-Jung, Buffalo, NY) at 490 nm excitation wave length.32
Assays for DNA fragmentation were performed as previously described, with minor modification.32 Briefly, pellets of 2 × 106 cells were lysed in 500 μL of lysis buffer (50 mmol/L of Tris pH 8.0, 100 mmol/L NaCl, 50 mmol/L EDTA, 1% SDS). After washing pellets with PBS and incubation with proteinase K (50 ng/mL) at 56°C for 2 hours, protein was removed from the recovered supernatants by treating with 500 μL of phenol/chloroform (pH 7.8). Nucleic acid in the upper layer was precipitated in 1 mL of ethanol and 0.3 mol/L sodium acetate (pH 5.2), centrifuged, dried, and suspended in water. After treatment with RNAse at 37°C for 10 minutes, extracted DNA was electrophoresed in 1.5% agarose, stained with ethidium bromide, and photographed under ultraviolet illumination.
Assays for cellular transformation. To determine the impact of transfection with p16MT vector on cellular transformation, JKB MT cells and JKB control cells (1 × 104/mL) were plated in 1 mL of RPMI-1640 media containing 0.8% methylcellulose, 10% FBS, and 2.0 μg/mL puromycin as a final concentration and cultured in a humidified chamber at 40°C, 37°C, 34°C, and 31°C. Macroscopically visible colony-forming units were counted after 5 days of culture. Standard deviation was calculated from three independent experiments. Student's t-test was used to determine the significant differences.
RESULTS
Ectopic expression of p16 protein in JKB cells. To characterize the effect of p16 on JKB ALL cells, this cell line was transfected with control vector, wild-type p16 gene, or mutated p16 gene, and transfectants were selected by culture with puromycin for 7 days. As can be seen in Fig 1A, p16 protein was expressed in JKB cells transfected with both wild-type p16 (JKB p16WT) or with mutated p16 (JKB p16MT) and was lacking in control transfectants (JKB control). The percentage of cells in G1 was 10% higher in JKB p16WT cells than in either JKB control or JKB p16MT cells (data not shown). After 2 months of puromycin selection, p16 protein remained highly expressed in JKB p16MT cells. In contrast, cell number decreased in >7-day cultures of JKB p16WT cells, and p16 protein was no longer detectable in the remaining viable cells (Fig 1B). The expression of both CDK4 and CDK6 were unchanged in JKB control, JKB p16WT, or JKB p16MT cells in short- and long-term cultures.
Effects of culture temperature on cell cycle distribution of JKB control and JKB p16MT cells. JKB p16MT cells and JKB control cells were cultured for 1 day at different temperatures (40°C, 37°C, 34°C, and 31°C), and cell cycle profile was determined by PI staining (Fig 2). At 40°C, the percentage of cells in G1 phase was similar (60% ± 2%) in JKB control and JKB p16MT cells. Interestingly, the percentage of JKB p16MT cells in G1 phase increased at lower culture temperatures: 66% G1 at 37°C, 75% G1 at 34°C, and 83% G1 at 31°C. In contrast, cell cycle distribution of JKB control cells did not significantly vary with culture temperature.
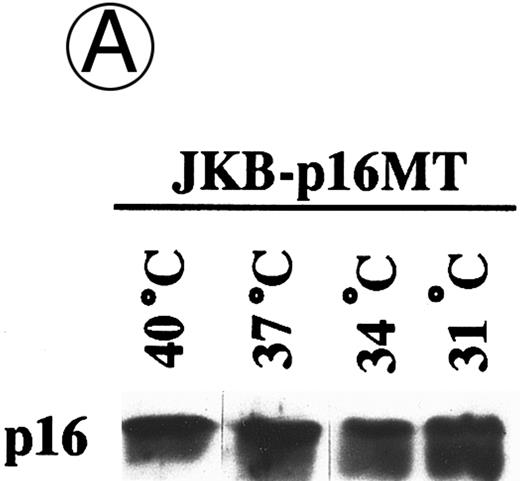
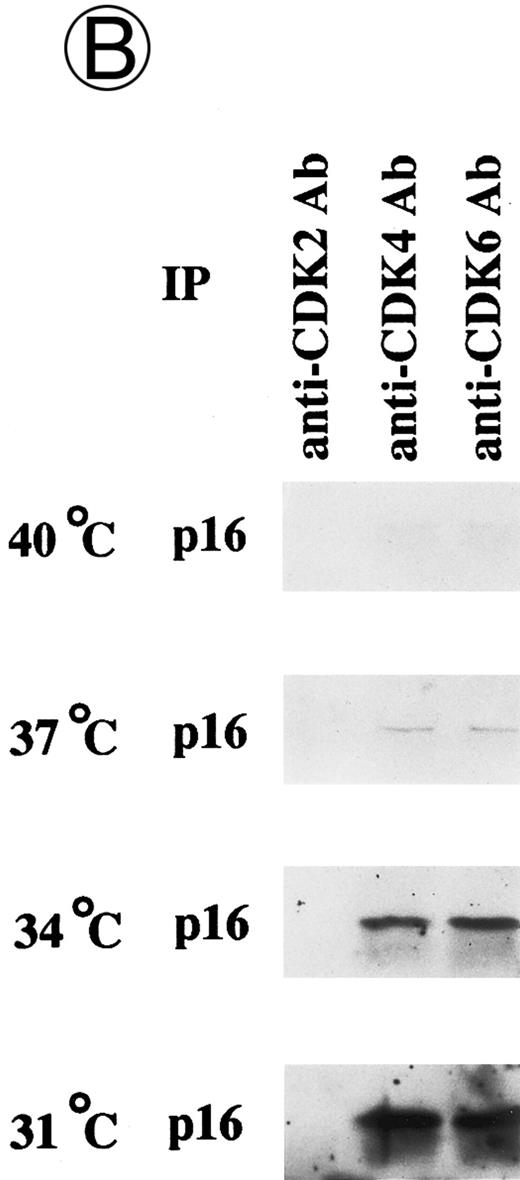
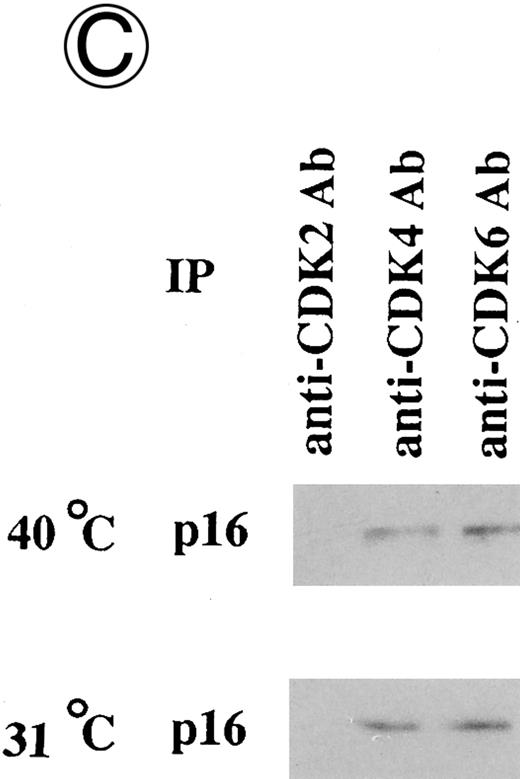
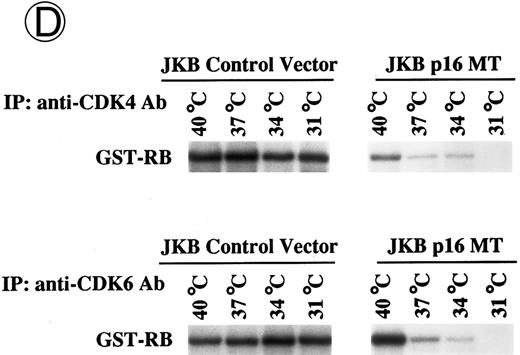
Effects of culture temperature on p16 protein expression, p16 binding to CDK4 and CDK6, CDK4 and CDK6 kinase activities, and phosphorylation of pRB in JKB p16MT cells. JKB p16MT cells, JKB p16 WT cells, and JKB p16 control cells were cultured under varying temperatures (40°C, 37°C, 34°C, and 31°C) for 1 day. (A) Total lysates of JKB p16MT cells were immunoprecipitated with anti-p16 MoAb and immunoblotted with the same MoAb. Total lysates of JKB p16MT cells (B) or JKB p16WT cells (C) were immunoprecipitated with anti-CDK2, anti-CDK4, or anti-CDK6 Abs and immunoblotted with anti-p16 MoAb. (D) Total lysates of JKB p16MT cells and JKB control cells were immunoprecipitated with anti-CDK4 or anti-CDK6 Abs. GST-RB fusion protein was used as substrate in assays of kinase activities of CDK4- or CDK6-associated immune complexes. (E) Total lysates of JKB p16MT cells and JKB control cells were immunoprecipitated with anti-pRB MoAb (Ab-1) and immunoblotted with anti-pRB MoAb (Ab-6).
Effects of culture temperature on p16 protein expression, p16 binding to CDK4 and CDK6, CDK4 and CDK6 kinase activities, and phosphorylation of pRB in JKB p16MT cells. JKB p16MT cells, JKB p16 WT cells, and JKB p16 control cells were cultured under varying temperatures (40°C, 37°C, 34°C, and 31°C) for 1 day. (A) Total lysates of JKB p16MT cells were immunoprecipitated with anti-p16 MoAb and immunoblotted with the same MoAb. Total lysates of JKB p16MT cells (B) or JKB p16WT cells (C) were immunoprecipitated with anti-CDK2, anti-CDK4, or anti-CDK6 Abs and immunoblotted with anti-p16 MoAb. (D) Total lysates of JKB p16MT cells and JKB control cells were immunoprecipitated with anti-CDK4 or anti-CDK6 Abs. GST-RB fusion protein was used as substrate in assays of kinase activities of CDK4- or CDK6-associated immune complexes. (E) Total lysates of JKB p16MT cells and JKB control cells were immunoprecipitated with anti-pRB MoAb (Ab-1) and immunoblotted with anti-pRB MoAb (Ab-6).
Effects of culture temperature on p16 protein expression, p16 binding to CDK4 and CDK6, CDK4 and CDK6 kinase activities, and phosphorylation of pRB in JKB p16MT cells. Once we had determined the permissive and restrictive temperatures for G1 growth arrest, we next examined whether p16 expression and function in JKB p16MT cells, JKB p16WT cells, or JKB control cells also varied with culture temperature. As can be seen in Fig 3A, p16 was expressed in JKB p16MT cells at all culture temperatures examined. However, binding of p16 to CDK4 and CDK6 in JKB p16MT cells increased at lower culture temperatures (Fig 3B). p16 did not bind to CDK2 at any temperature. In contrast, p16 binding to CDK4 and CDK6 in JKB p16WT cells did not vary with culture temperature (Fig 3C).
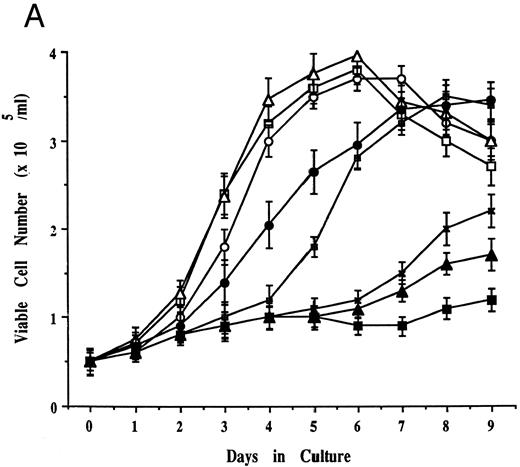
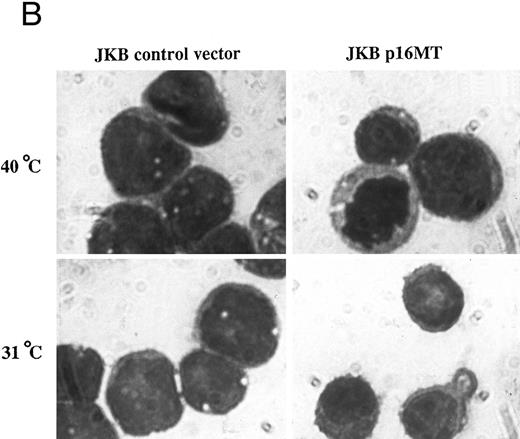
Effects of cell culture temperature on viable cell number, morphology, and cell surface phenotype of JKB p16MT cells. (A) JKB p16MT cells were cultured under the following conditions: 40°C for 7 days (□); 31°C for 7 days (▪); 31°C for 1 day, then 40°C for 6 days (○); 31°C for 2 days, then 40°C for 5 days (⊞); 31°C for 3 days, then 40°C for 4 days (×); and 31°C for 4 days, then 40°C for 3 days, ( — ▴ — ). JKB control cells were cultured at either 40°C (▵) or 31°C (•) for 7 days. Viable cell number was assessed using trypan blue staining. (B) JKB control (left) and JKB p16MT cells (right) were cultured at 40°C (upper) or 31°C (lower) for 7 days. Cell cytosmears were stained with Wright-Giemsa solution.
(C) JKB control (left) and JKB p16MT (right) cells were cultured at 40°C (upper) or 31°C (lower) for 7 days. Cell surface phenotype was determined by flow cytometric analysis.
Effects of cell culture temperature on viable cell number, morphology, and cell surface phenotype of JKB p16MT cells. (A) JKB p16MT cells were cultured under the following conditions: 40°C for 7 days (□); 31°C for 7 days (▪); 31°C for 1 day, then 40°C for 6 days (○); 31°C for 2 days, then 40°C for 5 days (⊞); 31°C for 3 days, then 40°C for 4 days (×); and 31°C for 4 days, then 40°C for 3 days, ( — ▴ — ). JKB control cells were cultured at either 40°C (▵) or 31°C (•) for 7 days. Viable cell number was assessed using trypan blue staining. (B) JKB control (left) and JKB p16MT cells (right) were cultured at 40°C (upper) or 31°C (lower) for 7 days. Cell cytosmears were stained with Wright-Giemsa solution.
(C) JKB control (left) and JKB p16MT (right) cells were cultured at 40°C (upper) or 31°C (lower) for 7 days. Cell surface phenotype was determined by flow cytometric analysis.
We next examined the kinase activities of CDK4 and CDK6 associated immune complexes, assayed by phosphorylation of GST-RB fusion protein, at different temperatures (Fig 3D). Activities of these kinases in lysates of JKB p16MT cells decreased at lower culture temperatures. In contrast, activities of these kinases in JKB control cells did not change significantly under these culture conditions.
Phosphorylation state of pRB was next analyzed in JKB p16MT cells and JKB control cells cultured at different temperatures (Fig 3E). pRB was constitutively phosphorylated at 40°C. In JKB p16MT cells, dephosphorylated pRB was induced at 37°C, and both phosphorylated and dephosphorylated pRB were expressed at 37°C and 34°C. Complete disappearance of phosphorylated pRB was noted in JKB p16MT cells cultured at 31°C. In contrast, no changes in pRB phosphorylation in JKB control cells were observed at varying temperatures.
Effects of cell culture temperature on viable cell number, morphology, and cell surface phenotype of JKB p16MT cells. We next attempted to correlate the above changes in p16 expression and function in JKB p16MT cells under permissive and restrictive culture conditions with changes in viable cell number, cell morphology, and cell surface phenotype. JKB p16MT cells (5 × 105/mL) cultured at 40°C demonstrated an exponential increase in cell number to day 7 (Fig 4A). In contrast, no significant changes in cell number were seen in JKB p16MT cells cultured at 31°C. Because G1 growth arrest occurred after culture of JKB p16MT cells at 31°C for 1 day, we next tested the growth of JKB p16MT cells cultured initially at 31°C and then transferred to culture at 40°C. As seen in Fig 4A, JKB p16MT cells transferred to 40°C after initial culture at 31°C for 1 day or 2 days reacquired exponential growth. However, after culture of JKB p16MT cells at 31°C for 3 or 4 days, recovery of growth was delayed and incomplete.
We next examined the effect of functional p16 expression on cellular morphology using Wright-Giemsa staining (Fig 4B). JKB p16MT cells cultured at 40°C showed basophilic cytoplasm with vacuoles, as well as immature nuclei with condensed chromatin and prominent nucleoli. This finding was similar to the appearance of JKB control cells cultured at either 40°C or 31°C. In contrast, JKB p16MT cells cultured at 31°C for 7 days were smaller, without vacuoles in cytoplasm, with nuclei containing less condensed chromatin and fewer nucleoli.
Flow cytometric analysis was used to correlate changes in cell surface phenotype with functional p16 expression (Fig 4C). JKB p16MT cells were cultured under either restrictive (40°C) or permissive (31°C) conditions for 7 days and examined for expression of cell surface CD19, CD10, and CD20 antigens. JKB p16MT cells cultured at 40°C strongly expressed CD19 (100%, ++) and CD10 (60%, ++), but only weakly expressed CD20 (18%,+). JKB p16MT cells cultured at 31°C also more strongly expressed CD19 (100%, +++). However, CD10 was more weakly expressed (11%, +) and CD20 was expressed at greater intensity (98%, ++) on cells cultured at 31°C than on cells cultured at 40°C. The phenotype of JKB control cells was not altered at varying temperatures.
To assay for the effect of transfection with p16MT on cellular transformation, JKB cells transfected with p16MT or control vectors were cultured for 5 days in methylcellulose and colonies enumerated at 5 days. As can be seen in Table 1, colonies of JKB control cells did not change significantly with culture temperatures, whereas JKB p16MT colonies were significantly reduced at permissive temperatures. Importantly, although the number of colonies did not differ between JKB control cells and JKB p16MT cells cultured at 40°C, the number of colonies of JKB p16MT cells was significantly reduced compared with colonies of JKB control cells (P < .001, n = 3) when cultures were done at 31°C.
Effects of p16MT on JKB Colony Formation
| Culture Temperature . | No. of Colonies . | |
|---|---|---|
| . | Control Vector . | p16 MT . |
| 40°C | 228 ± 24 | 190 ± 22 |
| 37°C | 247 ± 27 | 143 ± 20 |
| 34°C | 217 ± 32 | 75 ± 11 |
| 31°C | 206 ± 17 | 1 ± 1 |
| Culture Temperature . | No. of Colonies . | |
|---|---|---|
| . | Control Vector . | p16 MT . |
| 40°C | 228 ± 24 | 190 ± 22 |
| 37°C | 247 ± 27 | 143 ± 20 |
| 34°C | 217 ± 32 | 75 ± 11 |
| 31°C | 206 ± 17 | 1 ± 1 |
JKB MT cells (1.0 × 104/mL) were plated in 1 mL of RPMI-1640 containing 0.8% methylcellulose, 10% FBS, and puromycin 2.0 μg/mL at a final concentration, and cultured in a humidified chamber at 40°, 37°, 34°, and 31°C. Macroscopically visible colony-forming units were counted after 5 days of culture. Standard deviation was calculated from three independent experiments. Student's t-test was used to determine the significant difference.
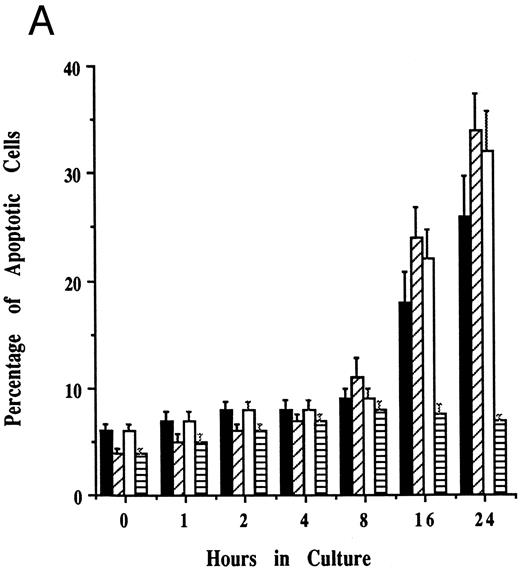
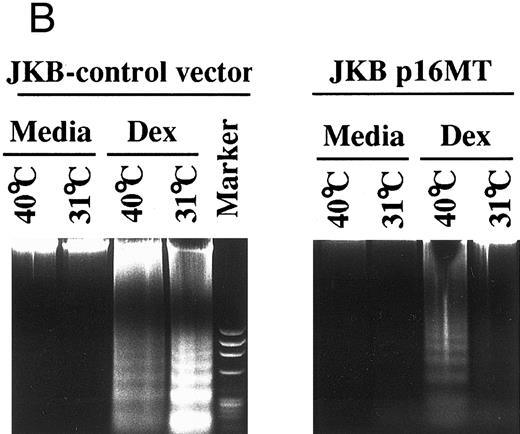
Effects of cell culture temperature on susceptibility of JKB p16MT cells to apoptosis triggered by dexamethasone. To assess whether functional p16 protein expression affects susceptibility to apoptosis, JKB control and JKB p16MT cells were cultured at either restrictive (40°C) or permissive (31°C) conditions for 1 day followed by culture with Dex (10 μmol/L) for 1 day to induce apoptosis. The percentage of apoptotic cells was determined by acridine orange/ethidium bromide staining (Fig 5A) and by DNA fragmentation (Fig 5B). Dex-induced apoptosis of JKB control and JKB p16MT cells cultured at 40°C, resulting in >30% apoptotic cells at 1 day. In contrast, Dex did not trigger apoptosis of JKB p16MT cells cultured at 31°C. Dex did trigger apoptosis of JKB control cells cultured at either 31°C or 40°C.
Effects of cell culture temperature on susceptibility of JKB p16MT cells to apoptosis triggered by dexamethasone. JKB control (▪) and JKB p16MT (□) cells were cultured for 1 day at 40°C; JKB control (▨) or JKB p16MT (▤) were also cultured for 1 day at 31°C. Cells were then cultured for 1 day with Dex (10 μmol/L) at either temperature to induce apoptosis. The percentage of apoptotic cells was determined by acridine orange/ethidium bromide staining (A) and DNA fragmentation (B).
Effects of cell culture temperature on susceptibility of JKB p16MT cells to apoptosis triggered by dexamethasone. JKB control (▪) and JKB p16MT (□) cells were cultured for 1 day at 40°C; JKB control (▨) or JKB p16MT (▤) were also cultured for 1 day at 31°C. Cells were then cultured for 1 day with Dex (10 μmol/L) at either temperature to induce apoptosis. The percentage of apoptotic cells was determined by acridine orange/ethidium bromide staining (A) and DNA fragmentation (B).
DISCUSSION
In the present report, we were able to study the role of p16 in ALL for two principal reasons. The first was the recent derivation of the JKB ALL cell line, with translocation between 9p21 and 14q32, the locations of the p16 and heavy chain Ig genes, respectively.18 Homozygous deletions of p16 gene in JKB cells was confirmed by PCR, and p16 protein product was not detectable by WB. Therefore, JKB is the first example of an Ig heavy chain translocation associated with deletion of p16. The second component essential for this study was the development of a system for inducible p16 function in JKB cells. This was necessary because stable transfection of p16 into cells suppresses their growth and confounds further mechanistic study. Prior studies have used an isopropyl-1-thiol-β-D-galacto-pyranoside-inducible p16 construct to stably transfect a human melanoma cell line and establish inducible and reversible G1 growth arrest.22 We chose in this study to attempt to develop temperature-sensitive p16 constructs by making point mutations in ankyrin repeats within p16 gene, based on prior reports that mutations in the ankyrin repeats of the yeast cdc10 gene confer temperature sensitivity (Fig 6). Of the mutations attempted in our study, a mutation at position 119 (E119G) was found to be restrictive at higher temperature and permissive at lower temperature for binding to CDK4 and CDK6, inhibiting CDK4 and CDK6 complex kinase activities, decreasing phosphorylation of pRB, and inhibiting growth. Moreover, the expression of p16 in JKB p16MT cells at permissive temperatures was greater than in JKB p16WT cells, allowing us to examine the phenotype of overexpressed p16 in this study.
E119G mutation in fourth ankyrin repeat of p16 gene. The amino acid sequence of the fourth ankyrin repeat (110-141) is compared with the homologous sequence of cdc10. Point mutation (E119G) in p16 gene was made corresponding to the mutation (D354G), which confers temperature sensitivity of cdc10.
E119G mutation in fourth ankyrin repeat of p16 gene. The amino acid sequence of the fourth ankyrin repeat (110-141) is compared with the homologous sequence of cdc10. Point mutation (E119G) in p16 gene was made corresponding to the mutation (D354G), which confers temperature sensitivity of cdc10.
The mechanism whereby mutations within ankyrin repeats result in temperature sensitivity remains undefined. However, structural-prediction algorithms indicate a helix-turn-helix motif with amphipathic helices in these repeats, which may enable interaction with other proteins by protein folding.23,33,34 It is, therefore, possible that the mutations identified in this region interfere with p16 function at the restrictive temperature by modulating binding sites to target proteins CDK4 and CDK6. Studies to date have suggested a functional significance to distinct sites within p16. For example, Parry et al35 have demonstrated that the sequences between E-120 and T-137 are essential for the functional integrity of p16. In addition, Fahreaus et al36 have shown that the sequence between D-84 and R-103 binds to and inhibits the activities of CDK4/CDK6. Yang et al37,38 demonstrated that mutations of p16 were observed throughout the entire gene with no apparent mutational patterns or hot spots; however, deletion of the fourth ankyrin repeat abolished p16 activity completely. Naturally-occurring G101W and V126D mutations within the ankyrin repeats of familial melanoma confer temperature-sensitive growth,35 supporting a functional significance for these sites. Our study shows a similar biologic outcome related to a mutation at E119G. Ongoing studies will further define those regions of p16 required for its function, as well as the mechanisms underlying the temperature sensitivity conferred by mutations within ankyrin repeats of p16 gene.
In this study, we used the JKB ALL cell line and the p16 temperature-sensitive mutant to investigate the role of homozygous deletion of p16 gene in lymphoid transformation. Specifically, induction of functional p16 protein in JKB p16MT cells cultured at permissive temperatures attenuated their growth rate and transforming activity. Interestingly, in JKB p16MT cells cultured under permissive conditions for ≤48 hours, the growth arrest was reversible, as evidenced by reacquisition of exponential growth when changing to restricted culture temperature. However, JKB p16MT cells cultured for ≥72 hours at permissive temperatures showed growth suppression, with only delayed and incomplete recovery. This suggests that prolonged p16 expression at permissive temperatures and related growth arrest may be associated with loss or degradation of key cell cycle regulatory proteins, and that cells thereafter cannot enter cycle even when p16 function is inhibited under restrictive conditions. Alternatively, cell differentiation induced by p16 might be associated with irreversible growth suppression.
In the present study, culture of JKB p16MT cells at permissive temperatures also induced a less transformed morphology, fewer colonies in methylcellulose, and more differentiated cell surface phenotype. In previous studies, Serrano et al39 have shown that expression of p16 not only suppressed proliferation, but also inhibited cellular transformation of primary rat embryo fibroblasts triggered by Ras oncogene. Ectopic expression of p16 in other tumor cell lines similarly induced both growth arrest and morphologic changes in lung and brain tumors.19 20 These studies all support the view that loss of p16 gene may predispose to transformation of lymphocytes.
Recent reports suggest that CDK inhibitors including p16 not only suppress cell cycle progression from G1 to S phase, but may also protect from apoptosis. For example, Wang et al40 showed resistance to apoptosis conferred by CDK inhibitors during myocyte differentiation. Similarly, p21 prevents apoptosis during differentiation of neuroblastoma cells.41 Melanoma cells expressing p16 are significantly more resistant to apoptosis due to methotrexate, vinblastine, and cisplatin compared with melanoma cells lacking p16.22 In our study, JKB p16MT cells cultured at permissive temperatures expressed functional p16 and were resistant to apoptosis triggered by Dex. The mechanism of this protective effect is undefined. However, the observation that ectopic expression of p16 or p21 in myocytes lacking RB does not protect against apoptosis during mitogen deprivation-induced differentiation42 suggests that RB, and related growth arrest, is required for the anti-apoptotic effect of p16 or p21. Furthermore, the demonstration that ectopic expression of wild-type RB in the human SAOS-2 osteosarcoma cell line, which lacks RB, results in increased resistance to radiation-induced apoptosis43 also supports this view. In our study of JKB p16MT cells, functional p16 at permissive conditions induced dephosphorylated pRB, which was associated with resistance to Dex-induced apoptosis. Finally, homozygous RB mutant embryos die at days 12 to 15 of gestation with a number of developmental defects, including prominent apoptosis in nervous and hematologic systems,44 further suggesting the importance of pRB in the anti-apoptotic effect of p16 or p21. These studies are consistent with the notion that RB inactivated by CDK inhibitors may suppress apoptosis via induction of G1 growth arrest.
What are the therapeutic implications of the current and prior studies? Our study strongly suggests that p16 impacts not only cell growth, but also cell differentiation and survival. In our study, proliferating JKB p16MT cells were susceptible to apoptosis triggered by Dex, but growth-arrested JKB p16MT cells acquired resistance to Dex. This observation is consistent with the fact that Dex induces apoptosis in immature or activated lymphocytes and is an essential component in the therapy of ALL.45 46 Moreover, our results also suggest that those ALL cells which express p16 may have relative resistance to apoptosis. These data are consistent with the clinical observation that ALL cells with a high proliferative rate are often chemosensitive, whereas cells with a lower growth fraction may be relatively resistant to therapy. Finally, our data also support the view that treatment approaches based on inducing p16 and related G1 growth arrest in tumor cells that lack p16 are unlikely to offer therapeutic benefit, as such strategies may also confer resistance to chemotherapy and radiotherapy treatments and thereby enhance tumor cell survival.
Supported by National Institutes of Health Grant No. CA 50947 and Uehara Memorial Foundation, Japan.
Address reprint requests to Kenneth C. Anderson, MD, Dana-Farber Cancer Institute, 44 Binney St, Boston, MA 02215.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal