Abstract
We have previously reported that particles resembling retroviral particles and possessing an RNA-directed DNA polymerase activity can be prepared from platelets. Furthermore, we and others have shown that these particles are present at higher levels in patients with essential thrombocythemia and polycythemia vera. We show here that these particles package RNA molecules that encode HERV-K–related pol genes. A subset of the RNA molecules that are packaged are likely to encode the RNA directed DNA polymerase activity and, because these RNAs possess long/full-length open reading frames for the reverse transcriptase and RNaseH (also for part of the integrase domains in genomic clones) of HERV-K, we propose that these transcripts are indeed strong candidates for encoding the enzyme activity found in these particles. Moreover, by using a modification of the polymerase chain reaction-based reverse transcriptase assay in which activated DNA is added during cDNA synthesis to suppress DNA polymerase-mediated RNA-directed DNA synthesis, we have found that the particle-associated enzyme behaves like a retroviral reverse transcriptase, further supporting the conclusion that retrovirus-like, perhaps HERV-K sequences, encode this enzyme activity.
SINCE THE DISCOVERY of the first human endogenous retroviral (HERV) sequence element in 1981 by Martin et al,1 there have been approximately 200 reports published in the field.2 The majority of these describe the identification and characterization of new sequences or sequence families (reviewed by Wilkinson at al3 ). A handful describe the detection of protein products of these elements either by immunologic, electron microscopic, or enzymatic methods (see, for example, Venables et al,4 Kurth et al,5 Dalton et al,6 Nelson et al,7 and Schlom et al8 ) or, more rarely still, the characterization of the protein product itself.9-11 None of the reports describing the molecular cloning of HERVs has ever described a functional provirus that encodes active gag, pol, and env or that has the ability, even in a model system, to retrotranspose. Indeed, all too frequently, the reports describing sequences have concentrated on such small polymerase chain reaction (PCR) products that no information regarding function could be inferred.12-14
The human genome contains large numbers of these HERV elements; indeed, the very number of these, together with the sequence structure of them, has led to the suggestion that these elements may be actively retrotransposing.15,16 More convincing than the number of these elements is the presence of perhaps 10,000 solitary LTRs derived from just one family of sequences, HERV-K, in some cases at least flanked by short direct repeats.17 Although this evidence does not prove that retrotransposition is taking place (particularly because direct repeats are also relics of transposition), it is at least strongly suggestive of a process of retrotransposition followed by homologous recombination.17 However, to date, there has been no demonstration of HERVs moving within the genome and no evidence for the existence of even one locus that possesses the enzymatic capacity to be the much sought after active HERV retrotransposon (reviewed in Wilkinson et al3 ). Two possibilities exist: either HERVs do not encode functional retrotransposons or the plethora of elements present in the genome makes finding the functional ones like looking for a needle in a haystack. The former of these possibilities, if correct, requires no further discussion. The latter, and this is of course the position taken by those who study HERV elements, presents to date, a highly refractory practical problem.
In other species, endogenous viruses were originally discovered because they conferred resistance to superinfection by related viruses.18 In fact, although we do not have evidence at this time for the existence of exogenous viruses that are related to any of the families of HERVs that have thus far been identified (with the possible exception of the spumavirinae), this does not mean that these loci do not, or have not in the recent evolutionary past, performed this function. Endogenous retroviruses of animals have also been shown to recombine to produce infectious viruses,19,20 to alter the tropism of an exogenous retrovirus by envelope pseudotyping21 or by recombination,22 and also to produce infectious virus with tropism for species other than the host.23,24 Taking all of the above into consideration, one can argue both for and against the existence of active retrotransposons encoded by HERV elements. For this reason we have attempted to characterize elements that, for a number of reasons described below, have an increased probability of encoding functional genes in general. In particular, we have tried to identify elements that might encode a functional reverse transcriptase (RT).25,26 The argument goes as follows. Retrovirus-like particles have been detected in several systems, namely in placenta,6,26,27 teratocarcinoma cells,5,28,29 the T47D mammary carcinoma cell line,25,30,31 and also in lysates of purified platelets.32-34 From this information, it can be deduced that these particles are encoded by gag-like genes present in the genome, because gag is both necessary and sufficient for retroviral particle formation.35 In each of these systems, the particles that have been detected copurified with an enzyme activity that was, to a greater or lesser extent, indistinguishable from a retroviral RT.7,25,26,32,34,36 If this enzyme is produced as part of a gagpol polyprotein, as appears to occur with all but the spumavirinae,37,38 then the same type of retroviral gene that encodes the particles likely encodes the RT. Because genome packaging specificity is a function of the interaction of the RNA packaging signal with the nucleocapsid protein,39,40 one can assume that the sequences that are packaged into these particles are genetically related to the genes that encoded the particles and the enzyme activity. Therefore, we have attempted to identify genes that encode the RNA directed DNA polymerase (RDDP) activity found in platelet lysates. To achieve this, we have used RT-PCR to amplify genomic RNAs from particles that have been purified on sucrose gradients and then cloned both the cDNA and also those proviral sequences that encode full-length sequences with the highest homology to these.25,26,32 33
MATERIALS AND METHODS
Patient samples. Patient samples were obtained by plateletpheresis from previously untreated patients diagnosed with essential thrombocythemia. Patient UPN07, a 30-year-old white woman, had a history of peptic ulcer and no history of hemorrhagic or thrombotic episodes. At the time of plateletpheresis, clinical laboratory studies showed a hemoglobin level of 13.6 g/dl, a hematocrit level of 41%, and a white blood cell (WBC) count of 8,840/μL, with a differential blood count showing 50% mature neutrophils, 2% bands, 37% lymphocytes, 7% monocytes, 3% eosinophils, and 1% basophils. Magnetic resonance imaging showed a slightly enlarged spleen. Her platelet count was 1.71 × 106/μL and her karyotype was normal. Patient UPN08, a 66-year-old white woman, had a long history of venous and arterial thrombosis. At the time of plateletpheresis, clinical laboratory studies showed a hemoglobin level of 11.5 g/dL, a hematocrit level of 41%, and WBC count of 17,260/μL, with a differential blood count showing 77% mature neutrophils, 4% bands, 10% lymphocytes, 4% monocytes, 4% eosinophils, and 1% basophils. Her spleen was not palpable. Her platelet count was 1.06 × 106/μL and her karyotype was normal. Patient UPN09, a 25-year-old white woman, had a history of transient ischemic episodes manifested periodically by neurologic symptoms. At the time of plateletpheresis, clinical laboratory studies showed a hemoglobin level of 10.9 g/dL, a hematocrit level of 34.1%, and a WBC count of 10,080/μL, with a differential blood count showing 54% mature neutrophils, 35% lymphocytes, 8% monocytes, 2% eosinophils, and 1% basophils. Her spleen was not palpable. Her platelet count was 1.24 × 106/μL and her karyotype was normal.
Sucrose gradient purification of retrovirus-like particles. Platelets for sucrose gradients were purified as described previously.34 Platelet pellets were lysed by repeated freeze-thaw cycles. Cellular debris was removed by centrifugation at 4,000g for 10 minutes at 4°C and the supernatant from this was then recentrifuged at 20,000g for 20 minutes at 4°C to remove mitochondria and other subcellular organelles. The resulting supernatant was then layered over a linear 20% to 65% (wt/vol) sucrose gradient prepared and run as described previously.34
RT-PCR, cloning, and sequencing. One-milliliter fractions from gradients prepared as described above were collected and a 20 μL solution of RNA was prepared from these as follows: to 250 μL sucrose fraction, 750 μL of RNAzol B (Molecular Research Center, Inc, Cincinnati, OH) was added, followed by 125 μL of chloroform. The rest of the preparation was performed according to the manufacturer's instructions.
Complementary DNA was synthesized from UPN07's gradient RNA prepared as described above using the 3′ PCR oligonucleotide 1506 (5′ CAT TCC TTG TGG TAA AAC TTT CCA YTG 3′) as follows: 5 μL of the RNA was incubated with 100 ng oligonucleotide for 2 minutes at 90°C in 10 mmol/L HEPES, pH 7.0, 1 mmol/L EDTA in a final volume of 10 μL and then cooled on ice. To this mixture was added 20 U of Moloney murine leukemia virus reverse transcriptase (MoMLV-RT; GIBCO-BRL, Gaithersburg, MD), and this was then incubated at 37°C for 90 minutes in a final reaction volume of 20 μL according to the manufacturer's instructions. Five microliters of this reaction product was then incubated with 2.5 U of Amplitaq DNA polymerase (Perkin-Elmer, Norwalk, CT), 800 μmol/L dNTPs, and 200 ng of each oligonucleotide 1505 (5′ TCC CCT TGG AAT ACT CCT GTT TTY GT 3′) and 1506, using the following cycles: 94°C for 30 seconds, 50°C for 45 seconds, and 72°C for 30 seconds for 30 cycles followed by a final extension at 72°C for 5 minutes.
Complementary DNA was synthesized from UPN08's gradient RNA prepared as described above and in Brodsky et al41 using the 3′ PCR oligonucleotide YIDD (5′ CTA GAA GCT TCT GCA GCA CAT AAA ATA TCA TCA ATA TA 3′) as follows: 5 μL of the RNA was incubated with 100 ng of oligonucleotide for 2 minutes at 90°C in 10 mmol/L HEPES, pH 7.0, 1 mmol/L EDTA in a final volume of 10 μL and then cooled on ice. To this mixture was added 20 U of MoMLV-RT (GIBCO-BRL) and this was then incubated at 37°C for 90 minutes in a final reaction volume of 20 μL according to the manufacturer's instructions. Five microliters of this reaction product was then incubated with 2.5 U of Amplitaq DNA polymerase (Perkin-Elmer), 800 μmol/L dNTPs, and 200 ng of each oligonucleotide LPQ (5′ TTC GGA TCC TGG AAA GTG TTA CCT CAG G 3′) and YIDD, using the following cycles: 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 30 seconds for 30 cycles followed by a final extension at 72°C for 5 minutes. RT-PCR products were cloned into a TA vector (Invitrogen, San Diego, CA) using standard methods.42 To amplify the whole of the reverse transcriptase and RNaseH domains from sucrose gradient-purified lysates, we used a nested RT-PCR approach. cDNA was synthesized from patient UPN09's gradient-purified particles as described above for UPN08, except that the oligonucleotide RT3outer (5′ CAT GAG TCA AAG CAT GAA GTT CTT G 3′) was substituted for YIDD. The first round of PCR was then performed with primers RT5outer (5′ TCA CTG TAG AGC CTC CTA AAC CC 3′) and RT3outer using the following cycles: 94°C for 30 seconds, 60°C for 30 seconds, and 68°C for 10 minutes for 30 cycles followed by a final extension at 72°C for 5 minutes using a 30:1 mixture of Amplitaq and Pfu (Stratagene, La Jolla, CA) polymerases. This product corresponds to nucleotides 3924-5678 on the HERV-K10 map (Genbank accession no. M14123). The second-round PCR reactions were performed by inoculating 0.1 μL of the product of the first round reaction into a 50 μL reaction containing the internal primers RT5Eco (5′ AGA GAT ATC CCA CTA ACT TGG AAA ACA GAA AAA CC 3′) and RT3Not (5′ GAG GCG GCC GCT TTT ATG AGT GCA GAT GAT ACC AG 3′) using the same cycling conditions. This product corresponds to nucleotides 3950-5651 on the HERV-K10 map. RT-PCR products were cloned into a TA vector (Invitrogen) using standard methods42 or after cleavage with the restriction endonucleases EcoRV and Not I and into the same sites of plasmid pBBV (created by subcloning the Black Beetle Virus [BBV] translational start signal from pBD743 into pcDNA I-Neo; Invitrogen). Plasmid inserts were sequenced using either Sequenase 2.0 (Amersham, Arlington Heights, IL) or on an ABI 377 automated sequencer (PERKIN ELMER, Norwalk, CT).
Reverse transcriptase assay. RT activity was detected by using the assay described by Silver et al44 with a few minor modifications. The template and primer used were as described. However, annealing of the primer to the template for cDNA synthesis was performed by heating both to 90°C for 2 minutes and then cooling on ice in 10 mmol/L HEPES, pH 7.0, and 1 mmol/L EDTA. Reverse transcription and PCR were performed as described. After electrophoretic separation, ethidium bromide-stained PCR products were photographed. Note that we typically obtain two bands in samples that possess detectable RNA-directed DNA polymerase activity. These are derived from the common 5′ primer producing products primed at the 3′ end with both the 3′ PCR reaction primer (139 bp) and the cDNA synthesis primer (167 bp). For experiments in which activated DNA was added to the cDNA synthesis reaction, up to 10 μg of DNaseI-treated calf thymus DNA (Sigma) was added during the cDNA synthesis step.
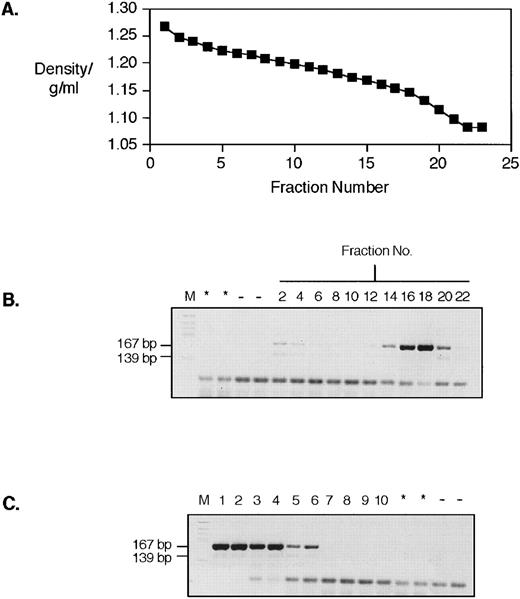
RNA with homology to HERV-K10; bands on a sucrose gradient at the typical density of retroviral particles. (A) shows the density profile of a sucrose gradient fractionation of a platelet lysate made from patient UPN07. The arrow indicates the fraction that gave the positive signal shown also arrowed in (B). (B) shows the result of RT-PCR with degenerate oligonucleotide primers. Lanes are (from left to right) M, DNA molecular weight marker; *, no cDNA synthesis step control for DNA template contamination; −, water control; 4, 8, 12, 16, 20, 24, and 28, sucrose fraction numbers; plus (+RT) or minus (−RT) reverse transcriptase.
RNA with homology to HERV-K10; bands on a sucrose gradient at the typical density of retroviral particles. (A) shows the density profile of a sucrose gradient fractionation of a platelet lysate made from patient UPN07. The arrow indicates the fraction that gave the positive signal shown also arrowed in (B). (B) shows the result of RT-PCR with degenerate oligonucleotide primers. Lanes are (from left to right) M, DNA molecular weight marker; *, no cDNA synthesis step control for DNA template contamination; −, water control; 4, 8, 12, 16, 20, 24, and 28, sucrose fraction numbers; plus (+RT) or minus (−RT) reverse transcriptase.
Amplification and cloning of HERV pol sequences from a genomic library. We screened a human genomic library (ATCC #37333; American Type Culture Collection, Rockville, MD) with an HERV-K–derived PCR product-derived riboprobe essentially as described previously.41 Screening of approximately one genome equivalent yielded 55 strongly hybridizing clones. PCR amplification from these clones with primers 516 (5′ CCA MTM ACT TGG AAA WCA GAM RAA 3′; HERV-K10 nucleotides 3948-3973) and either 518 (5′ TAM RTG KGT RAC ATC CAT TTG CCA 3′; HERV-K10 nucleotides 5825-5848) or Endk10rt3 (5′ RTA TCC WGG NSC RTT RTC WGT TTT NAT 3′; HERV-K10 nucleotides 5996-6022) yielded products of 1.9 and 2.07 kb, respectively, from 9 of the clones. Shorter products or no product were obtained from the other genomic clones.
Expression PCR and in vitro translation. PCR products from genomic clones described above were amplified using primers RGP1 (5′ CCA AGC TTC TAA TAC GAC TCA CTA TAG GGT TTT TAT TTT TAA TTT TCT TTC AAA TAC TTC CAC CAT GGT ACC AMT MAC TTG GAA AWC 3′; HERV-K10 nucleotides 3948-3966) and NewK10rt3Salmyc (5′ TCT CGT CGA CTC ACA AAT CTT CCT CTG AGA TAA GTT TCT GCT CTA MRT GYG TRA CAT CCA TTT GCC A 3′; HERV-K10 nucleotides 5825-5848), which introduced in the 5′ oligonucleotide (RGP1) a T7 promoter, an alfalfa mosaic virus ribosomal binding site, and an initiation codon fused to the amino terminal end of the RT domain of the HERV-K pol gene and introduced in the 3′ oligonucleotide (NewK10rt3Salmyc) a c-terminal 10 amino acid tag corresponding to the epitope recognized by anti–c-myc monoclonal antibody 9e10.45 For in vitro translation, 5 ng of each PCR product was incubated in an in vitro transcription reaction (Promega, Madison, WI) and subsequently in a rabbit reticulocyte-based in vitro translation reaction (Promega) essentially as recommended by the manufacturer. 35S- cysteine-labeled and methionine-labeled (NEN, Boston, MA) proteins were detected after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and fluorography with Amplify (Amersham). For HERV-K RT-PCR pol clones in pBBV, 250 ng of DNA was incubated in a 25-μL TNT (Promega)-coupled transcription-translation reaction and analyzed as described above.
RESULTS
As we have previously shown for placental tissue and the T47D mammary carcinoma cell line, we find that retroviral-like RNA sequences from sucrose gradient fractionated platelet lysates can be predominantly found in the fractions in which intact retroviral particles would be expected.25,26 Figure 1 shows the result of sucrose gradient fractionation of a platelet lysate from an individual with essential thrombocythemia (UPN07), followed by RT-PCR of RNA prepared from individual fractions using primers designed to amplify members of the HERV-K family and related sequences.12,25,26 Figure 1A shows the density profile of the gradient. Figure 1B shows the product of the PCR with and without reverse transcription. Clearly visible is an RT-dependent product from fraction 12 (indicated by an arrow) that has a density of 1.168 g/mL. This corresponds well to the expected buoyant density of intact retroviral particles of 1.14 to 1.18 g/mL. Similar products generated using primers for the RT active site41,46 were cloned and sequenced from a gradient from another patient with essential thrombocythemia (UPN08) and the results of this are shown in Fig 2. As can be seen, these sequences have significant homology (≥90% identity) to HERV-K10.47 We have previously shown that sequences with this level of homology are indistinguishable from HERV-K10 by Southern blot analysis.26 The deduced amino acid sequence derived from translation of these sequences is shown in Fig 2B. We assayed these same fractions from patient UPN08 for RT activity using the PCR-based assay described by Silver et al,44 as shown in Fig 3. This assay uses an exogenous RNA template annealed to a short DNA oligonucleotide as a substrate for cDNA synthesis. Synthesis is only performed in the presence of an RNA-directed DNA polymerase, and synthetic products are detected by PCR with primers mapping upstream of the 3′ cDNA synthesis oligonucleotide. PCR products are produced from amplification from both the 3′ cDNA synthesis oligonucleotide (167 bp) and the 3′ PCR oligonucleotide (139 bp) with a common 5′ oligonucleotide. As we have found before using a homopolymer template primer assay,32-34 the peak activity bands in the typical retroviral particle range of 1.14 to 1.18 g/mL (Fig 3B). We also noted, as we have on previous occasions, that a weak RT signal was detectable in the fractions from the bottom of the gradient.34 We suspect that this is due to particulate material that is pelleted at the bottom of the gradient, trapping the retrovirus-like particles and their associated RT. Note that, although the peak RT-PCR and RT signals from these two gradients are from fractions 12 and 16 to 18, respectively, these fractions do correspond to similar densities of 1.168 and 1.15 to 1.16 g/mL.
Comparison of sequences obtained from sucrose gradient fractionated platelet lysates. (A) Nucleotide sequences from RT-PCR from sucrose fractions 16 and 18 from patient UPN08. Six clones were sequenced and compared with HERV-K10.47 “−” indicates identity. P1 represents primer LPQ, and P2 represents primer YIDD. (B) Deduced amino acid sequences for the sequences shown in (A). “*” indicates an ochre stop codon.
Comparison of sequences obtained from sucrose gradient fractionated platelet lysates. (A) Nucleotide sequences from RT-PCR from sucrose fractions 16 and 18 from patient UPN08. Six clones were sequenced and compared with HERV-K10.47 “−” indicates identity. P1 represents primer LPQ, and P2 represents primer YIDD. (B) Deduced amino acid sequences for the sequences shown in (A). “*” indicates an ochre stop codon.
Reverse transcription of Brome Mosaic Virus (BMV) RNA by sucrose gradient fractionated platelet lysate. (A) shows the density profile of a sucrose gradient fractionation of a platelet lysate made from patient UPN08. (B) and (C) show the result of a reverse transcriptase assay performed on fractions from the gradient. (B) Lanes are (from left to right) M, marker; *, no BMV RNA control; −, no reverse transcriptase control; 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, and 22, sucrose fraction numbers. (C) Lanes are (from left to right) M, marker; 1 and 2, 2.0 U; 3 and 4, 0.2 U; 5 and 6, 0.02 U; 7 and 8, 0.002 U; 9 and 10, 0.0002 U of Moloney murine leukemia virus reverse transcriptase, respectively. “*” and “−” are as for (B).
Reverse transcription of Brome Mosaic Virus (BMV) RNA by sucrose gradient fractionated platelet lysate. (A) shows the density profile of a sucrose gradient fractionation of a platelet lysate made from patient UPN08. (B) and (C) show the result of a reverse transcriptase assay performed on fractions from the gradient. (B) Lanes are (from left to right) M, marker; *, no BMV RNA control; −, no reverse transcriptase control; 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, and 22, sucrose fraction numbers. (C) Lanes are (from left to right) M, marker; 1 and 2, 2.0 U; 3 and 4, 0.2 U; 5 and 6, 0.02 U; 7 and 8, 0.002 U; 9 and 10, 0.0002 U of Moloney murine leukemia virus reverse transcriptase, respectively. “*” and “−” are as for (B).
To determine whether these sequences were likely to encode for full-length open reading frames in the pol gene, we attempted to amplify them using primers designed to amplify the pol region of HERV-K10 (nucleotides 3948-5848 or 3948-6022 of HERV-K10; Genbank M14123). We found that these primers worked poorly on the RNA packaged into particles and therefore pursued two approaches to attempt to obtain PCR products for a larger region of the genome. The first approach was to amplify the homologous region from genomic sequences isolated from a human genomic library. We obtained 55 genomic clones that hybridized to an HERV-K10 probe at high stringency and these were then screened by PCR with the primers for HERV-K10 pol. Nine clones had a full-length product and these were therefore screened by expression coupled-PCR (E-PCR).48 Five clones (in addition to HERV-K10 itself ) could be shown to possess long- or full-length open reading frames by E-PCR, as shown in Fig 4. These were then screened for RT activity. We found that the in vitro translation reactions possessed significant endogenous RDDP activity. We attempted to remove this activity by immunoprecipitating the pol PCR products, which encoded, in addition, a 10 amino acid epitope from c-myc with an anti-myc monoclonal antibody. Unfortunately, although the precipitations did result in partial purification of the HERV-pol–derived protein (not shown), the endogenous RDDP bound nonspecifically to the beads and could not be removed. All attempts to do so were unsuccessful.
HERV-K–related genomic and platelet RT-PCR–derived clones encode an apparently full-length pol protein. (A) shows SDS-PAGE of E-PCR products detected by fluorography. Lanes are (from left to right) M, marker; −, reticulocyte lysate, no exogenous DNA added; 1 through 9, individual HERV-K–related clones; 10, MPMV control; 11, HERV-K10. The arrow indicates the full-length protein produced by HERV-K10.
HERV-K–related genomic and platelet RT-PCR–derived clones encode an apparently full-length pol protein. (A) shows SDS-PAGE of E-PCR products detected by fluorography. Lanes are (from left to right) M, marker; −, reticulocyte lysate, no exogenous DNA added; 1 through 9, individual HERV-K–related clones; 10, MPMV control; 11, HERV-K10. The arrow indicates the full-length protein produced by HERV-K10.
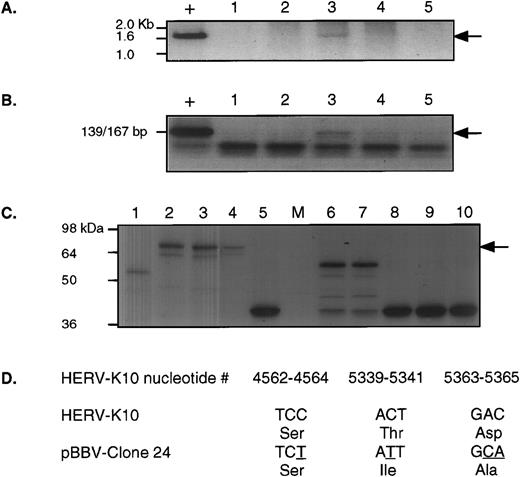
The second approach we used to obtain RT-PCR products from the gradient RNA was to use nested oligonucleotide primer pairs (RT5out with RT3out first round followed by RT5Eco with RT3Not) that would amplify the RT and RNaseH domains of the HERV-K genome (HERV-K10 nucleotides 3950-5651). Using these, we were able to amplify a 1.7-kb product by RT-PCR directly from gradient-purified platelet lysates from patient UPN09, as shown in Fig 5A. We could also demonstrate that this same fraction possessed RT activity, as shown in Fig 5B. After subcloning of the RT-PCR product into pBBV, we analyzed the in vitro translation products from these clones, as shown in Fig 5C. One clone, no. 24 (lane 2), demonstrated the capacity to produce an apparently full-length protein (by comparison with HERV-K10); we therefore sequenced it. Figure 5D shows those positions that differ from the prototypic HERV-K10 sequence and their translation. Clearly, this clone is almost identical to the prototypic HERV-K10 and, as predicted from the in vitro translation reactions, it has a full open reading frame for the whole of RT and RNaseH.
Comprehensive analysis from patient UPN09. (A) shows the 1.7-kb RT-PCR product obtained using nested PCR, first with RT5out/RT3out and then with RT5Eco/RT3Not. “+” indicates the positive control for this reaction, a clone of HERV-K10. Lanes 1, 2, 3, 4, and 5 are selected fractions from the bottom to top of a sucrose gradient preparation from this patient. The arrow indicates the position of the positive signal. Lane 3 is positive. (B) shows the same fractions analyzed for RT activity. “+” indicates the positive control for this reaction, ie, 0.002 U of MoMLV-RT. Again, lane 3 is positive and the arrow indicates the position of the positive signal. (C) shows SDS-PAGE products detected by fluorography from in vitro translation of several independent RT-PCR clones in pBBV. Lanes 1, 2, 5, and 6 through are independent clones obtained from the RT-PCR product shown in (A), lane 3 above. Lanes 3 and 4 are two different clones of HERV-K10 included for comparison. As can be seen, only lane 2 expresses a protein of apparently full length, which is indicated by the arrow. (D) shows the nucleotide sequence and the deduced amino acid sequence of this clone (pBBV-24) at only those positions that differ from the HERV-K10 prototypic sequence between nucleotides 3950 and 5651 (the RT and RNaseH domains).
Comprehensive analysis from patient UPN09. (A) shows the 1.7-kb RT-PCR product obtained using nested PCR, first with RT5out/RT3out and then with RT5Eco/RT3Not. “+” indicates the positive control for this reaction, a clone of HERV-K10. Lanes 1, 2, 3, 4, and 5 are selected fractions from the bottom to top of a sucrose gradient preparation from this patient. The arrow indicates the position of the positive signal. Lane 3 is positive. (B) shows the same fractions analyzed for RT activity. “+” indicates the positive control for this reaction, ie, 0.002 U of MoMLV-RT. Again, lane 3 is positive and the arrow indicates the position of the positive signal. (C) shows SDS-PAGE products detected by fluorography from in vitro translation of several independent RT-PCR clones in pBBV. Lanes 1, 2, 5, and 6 through are independent clones obtained from the RT-PCR product shown in (A), lane 3 above. Lanes 3 and 4 are two different clones of HERV-K10 included for comparison. As can be seen, only lane 2 expresses a protein of apparently full length, which is indicated by the arrow. (D) shows the nucleotide sequence and the deduced amino acid sequence of this clone (pBBV-24) at only those positions that differ from the HERV-K10 prototypic sequence between nucleotides 3950 and 5651 (the RT and RNaseH domains).
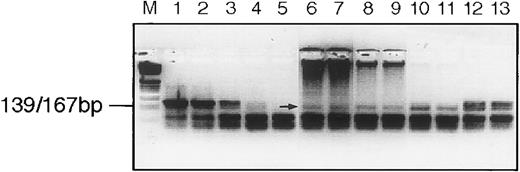
We next considered the possibility that the RDDP activity that had been detected on the gradients might not be derived from a virus-like gene, ie, that it might be due to the RDDP activity of a cellular polymerase. To test this, we examined the sensitivity of the enzyme activity to the addition of exogenous activated DNA during the cDNA synthesis step of the RT assay procedure. This method of examination of an RDDP was described by Lugert et al49 as a way of distinguishing between RDDP derived from a cellular polymerase and that from a viral RT. As shown in Fig 6, the particle-associated RDDP activity obtained from patient UPN08 fraction 17 (ρ ≈ 1.17 g/mL) is able to copy an RNA template, even in the presence of 10 μg of activated DNA, in a 20 μL cDNA synthesis reaction. This clearly suggests that the particle-associated RDDP activity behaves more like a retroviral RT than like a cellular DNA polymerase.49 Although this does not prove that the enzyme activity we detect here is derived from a virus-like RT, it does strongly suggest it.
Platelet lysate RT preferentially uses an RNA template. RT assay of sucrose fraction 17 from patient UPN08. Lanes are (from left to right) M, marker; 1 through 5, 2.0, 0.2, 0.02, 0.002, and 0.0002 U of Moloney murine leukemia virus reverse transcriptase; 6 through 13, 1.0 μL of sucrose fraction in duplicate with activated DNA added as follows: lanes 6 and 7, 10 μg; 8 and 9, 1 μg; 10 and 11, 0.1 μg; and 12 and 13, no activated DNA added. The arrow indicates the signal from the RT. Note that the activated DNA can be seen at the top of the gel at the higher concentrations.
Platelet lysate RT preferentially uses an RNA template. RT assay of sucrose fraction 17 from patient UPN08. Lanes are (from left to right) M, marker; 1 through 5, 2.0, 0.2, 0.02, 0.002, and 0.0002 U of Moloney murine leukemia virus reverse transcriptase; 6 through 13, 1.0 μL of sucrose fraction in duplicate with activated DNA added as follows: lanes 6 and 7, 10 μg; 8 and 9, 1 μg; 10 and 11, 0.1 μg; and 12 and 13, no activated DNA added. The arrow indicates the signal from the RT. Note that the activated DNA can be seen at the top of the gel at the higher concentrations.
DISCUSSION
In three different human systems we have now found evidence that suggests that HERV elements encode proteins that can assemble particles into which are packaged HERV transcripts.25 26 In each case we have also found evidence of the existence of an RDDP activity that copurified with the particles and sequences. We show here that there are HERV-K10–related sequences that are expressed in platelets and that copurify with RT that possess long- or full-length open reading frames in their pol gene that could encode for the activity that we detect. In addition, we have identified genomic clones that also possess long open reading frames that we have amplified from individual clones from a genomic library.
We have assayed these clones for the ability to encode an active RT in a variety of ways and using full-length open reading frames amplified either from the particles obtained from a variety of different tissues, viz, placenta26 and T47D mammary carcinoma cells,25 or from genomes derived from clones that are closely related to the sequences found in the particles in platelets. Although we have little difficulty identifying the long ORFs, we have been unable to show that these sequences encode a functional enzyme. Either we have not expressed the correct fragment or the RT encoded by the HERVs being studied is not active in our assays. Is this inherently so or due to some component of the system being used to study it? We do not know. Nevertheless, the evidence that there is a particle-associated RDDP polymerase is strong, and somewhere in the genome lies a gene that encodes this activity. It has been suggested that this activity is due to the presence of mitochondrial DNA polymerase, because this enzyme has the ability to copy RNA templates.50,51 We have previously assayed sucrose gradient-purified mitochondria and, using the “Silver” PCR-based RT assay we have used here, we found no detectable activity.25,44 Lugert et al49 described a very simple and elegant way to create an apparently truly RT-specific version of this assay by adding activated DNA. We have therefore used this modification to determine whether the RT-like enzyme found in platelet particles could be inhibited. As we have shown here, this enzyme activity is resistant to inhibition by the addition of activated DNA, further confirming that, by all available means of assay, it behaves as an RT.49 It is possible that an HERV other than HERV-K encodes the reverse transcriptase activity we have detected. Using primers that were described by Shih et al,46 we previously tried to amplify HERV sequences from sucrose gradients from a variety of cell types including platelets and we were either unable to do so or only saw sequences with 70% or greater homology to HERV-K.25 Therefore, it seems somewhat unlikely that these sequences are packaged at comparable levels into these particles, unless they derive from HERVs that could not be amplified by these primers.
Another alternative source for the reverse transcriptase activity in these samples is that it is encoded by a LINE element or other retrotransposon.52 Although we are unable to distinguish between a LINE and an HERV using our reverse transcriptase assay, we can argue that it is unlikely that a LINE element or other retrotransposon is the source of the activity. Previous studies performed by several groups, including our own, have shown particles possessing envelopes that morphologically resemble enveloped retroviral particles by electron microscopy in a variety of samples, including inter alia, platelets, placenta, T47D mammary carcinoma cells, and GH teratocarcinoma cells.6,27,28,32,34,53,54 These studies have shown that the particles copurified with reverse transcriptase activity at a density of 1.14 to 1.18 g/mL. We saw no other electron-dense structures copurifying with these particles. This does not mean that a LINE or similar macromolecular complex is not present, but HERV-like particles are.32 34 Thus, two possibilities need to be considered. First, is it likely that a LINE-encoded complex of ORF 1 and 2 gene products could copurify with HERV encoded particles at this density? Secondly, could a LINE encoded reverse transcriptase become packaged into particles encoded by HERV sequences?
The answer to the first question may well be no. Certainly for retroviral particles, nonenveloped cores band typically at a higher density (1.21 to 23 g/mL), although extrapolating this to LINES may not be valid. Nevertheless, when taken together with studies showing that, in GH teratocarcinoma cells in particular, the retrovirus-like particles consist of proteins whose cores are antigenically related to HERV-K29 and also package retroviral sequences related to HERV-K, it seems likely that the particles we see here (which we have on previous occasions shown to resemble enveloped retroviral particles32 34 ) are both encoded by an HERV and package HERV transcripts. This leaves the second question, could a LINE-encoded reverse transcriptase get packaged into an HERV encoded particle? This seems unlikely unless the particles are assembled from a mixture of HERV gag and LINE ORF1 proteins. Such a mixture has not been reported.
We and others have previously published data showing that the platelet-associated RT activity is higher in individuals that have a diagnosis of essential thrombocythemia or polycythemia vera.32-34 Although we have not directly addressed this issue in the present study, we do feel that the identity of the activity we previously detected is now fairly clear; it is an enzyme with many of the properties of a retroviral RT and it seems not unreasonable to propose that it may indeed be an endogenously encoded RT. In addition, the data presented here suggest to us, that the enzyme may well be encoded by a member of the HERV-K family of endogenous retroviral elements. With this in mind, it seems appropriate to now try to determine, using a more quantitative assay than that described here, how tightly linked the old observation of increased RT activity and a diagnosis of either essential thrombocythemia or polycythemia vera really are. We are aware that, if we could identify a functional locus that encodes this RT activity, we might also be able to develop an assay, based on determining the levels of expression of a specific HERV, that could enable us to examine these ideas from smaller amounts of sample. Our inability to be able to do this at this time represents one of the major hurdles obstructing a larger scale investigation of the putative RT activity/diagnosis correlation.
Ultimately, the question we have to address is whether HERVs have an impact on the human genome or on the individual. There are several levels at which this question can be tackled. First, it is incontrovertible that there are more than 1,000 partial or complete proviruses integrated into the human genome (reviewed in Wilkinson et al3 ). Moreover, there are some 10,000 solitary LTR-like sequences from the HERV-K family of sequences alone.17 How these sequences achieved their current locations and levels is unclear; whether by gene amplification, repeated cycles of infection, transposition or retrotransposition, we simply do not know. Notwithstanding this, HERVs have undoubtedly contributed to our genetic diversity and in some cases have led to altered patterns of gene expression. Specific examples of this exist, eg, comparison of parotid gland amylase expression in humans and in related primate species.55 56 The altered tissue specificity seen is the result of the presence of an HERV-E–related LTR, which is present in humans but not in other primates and which drives expression of the salivary amylase gene in human parotid gland. Fixation of such an event in the human lineage must be the result of evolutionary selection. Note however, that this does not necessitate positive selection for parotid gland amylase expression, the fixation of this altered expression pattern is likely to be due to founder effects after speciation.
At a more immediate level our question becomes whether HERVs cause human disease. To try to predict the answer to this question, we might consider any or all of the processes that have been seen to occur in animals. There is no evidence at this time to support recombination yielding either an infectious virus from two endogenous loci or from an exogenous virus recombining with an endogenous locus. There have been many attempts to isolate an infectious retrovirus from human cells and the only successful efforts have been from cells infected with an exogenous retrovirus (reviewed in Weiss et al57 ). Although incomplete, various attempts have been made to isolate an HERV-encoded infectious virus. The available evidence therefore suggests that, if HERVs either are or can become functional infectious agents, then this must indeed be a rare event because no one has observed it. If not by infection, how else might HERVs cause human disease? It is possible that HERV sequences encode for proteins that are poorly recognized as self proteins during thymic development. HERVs have been acquired only relatively recently in evolutionary terms and this may explain the observation that anti-HERV protein antibodies are present in a variety of disorders and perhaps most strikingly, in the sera of nearly all individuals with seminomas.58 It is further possible, therefore, that autoantibodies to HERV proteins may be a source of autoimmune disease (reviewed in Perl and Banki59 ). These antibodies have been described, for example, in patients with Sjögren's syndrome.60 Whether the virus like particles and proteins we detect here are immunogenic, we do not know.
Finally, given that HERVs can assemble retrovirus-like particles and that these both package RNA and possess an RT-like enzymatic activity, perhaps HERVs can retrotranspose? Since the first demonstration by Tchenio and Heidmann in 1991 of intracellular retrotransposition by a retrovirus, it has seemed possible that a noninfectious HERV element might retrotranspose.61 Furthermore, as stated above, circumstantial support for this hypothesis exists in the genome in the case of the lone HERV-K LTRs.17 Nevertheless, an HERV retrotranspositional event has yet to be described and this fact does beg questions relating to the significance of such an event, even if it should eventually be observed at some time in the future. Most likely, if such events do occur, they have an impact upon the genome that we can detect only on an evolutionary time scale. Moreover, these rare de novo events will probably only be detectable in an individual by an extraordinary piece of serendipity.
ACKNOWLEDGMENT
The authors gratefully acknowledge Dr Dale Haines for invaluable discussion and for critical review of the manuscript and Tom Nesspor for technical assistance.
Supported in part by the Institute for Cancer and Blood Diseases.
Address reprint requests to Isadore Brodsky, MD, Division of Hematology/Oncology, Department of Medicine, Mailstop 412, Allegheny University of the Health Sciences, Hahnemann Campus, Broad and Vine Sts, Philadelphia, PA 19102-1192.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal